Abstract
Illumina BeadArrays are among the most popular and reliable platforms for gene expression profiling. However, little external scrutiny has been given to the design, selection and annotation of BeadArray probes, which is a fundamental issue in data quality and interpretation. Here we present a pipeline for the complete genomic and transcriptomic re-annotation of Illumina probe sequences, also applicable to other platforms, with its output available through a Web interface and incorporated into Bioconductor packages. We have identified several problems with the design of individual probes and we show the benefits of probe re-annotation on the analysis of BeadArray gene expression data sets. We discuss the importance of aspects such as probe coverage of individual transcripts, alternative messenger RNA splicing, single-nucleotide polymorphisms, repeat sequences, RNA degradation biases and probes targeting genomic regions with no known transcription. We conclude that many of the Illumina probes have unreliable original annotation and that our re-annotation allows analyses to focus on the good quality probes, which form the majority, and also to expand the scope of biological information that can be extracted.
INTRODUCTION
Illumina BeadArrays are microarrays consisting of randomly positioned beads. A specific 50-mer oligonucleotide sequence is assigned to each bead type, which is replicated a random number of times on each array (∼40 times on average for the most common arrays). The BeadArray technology can be used in a wide range of applications, including gene expression studies, single-nucleotide polymorphism (SNP) genotyping, methylation profiling, and copy number variation analysis. Illumina arrays have played an important role in large international projects such as HapMap/GENEVAR (1), the Cancer Genome Atlas (2) and large-scale transcriptional profiling for the discovery of expression quantitative trait loci (3).
We, and others, have devoted attention to the improvement of methods of preprocessing and statistical analysis of Illumina microarray data (4–9), but probe annotation is also a fundamental issue in data reliability. No biologically meaningful interpretation can be made without detailed knowledge of what transcriptomic or genomic sequences the microarray probes map to, and problems associated with probe identity can cause misleading interpretation of data. Early expression microarray platforms adopted a gene-centric approach for probe design. This led to probes that fail to target some biologically relevant isoforms, and cannot distinguish between those that are targeted. Even the current platforms, with a few exceptions, provide limited information in this regard. Furthermore, the relevance of exon and exon junction levels of expression analysis has been acknowledged in several studies (10–13).
Recent studies revealed that the high experimental reproducibility between Affymetrix GeneChips and Illumina BeadArrays can be improved when the analysis is restricted to probes on the two platforms that target the same set of transcripts of a given gene (4,14). Several efforts in re-annotating Affymetrix 25-mer probe sequences have been shown to improve the reliability of differential expression analysis studies (15–19), and the importance of probe-level analysis of GeneChip data has also been reported (20,21). The annotation of Illumina probes poses a different problem from Affymetrix, as the replicated observations for the same bead type all have the same probe sequence attached. For an Affymetrix probeset, if one probe is defective then there are still multiple probes that can be used to interrogate the gene. However, if an Illumina probe is defective, then all measurements for that bead type are compromised, which is a concern because many genes are represented by only one bead type. Despite the incorporation of annotation information in Bioconductor (22), published re-annotation efforts for BeadArrays have not gone beyond the redefinition of non-redundant and universal oligonucleotide identifiers (23), the assignment of genes and transcripts to probes on BioMart (24), the genomic remapping of probes for visualization on the Ensembl and UCSC genome browsers (25,26) or the alignment of probes with RefSeq (27) and Ensembl transcripts for platform comparison (28). In a previous study, we have already discussed some of the implications of probe annotation in the statistical analysis and summarization of BeadArray data (5). We revealed, for the Mouse WG-6 version 1 platform, the existence of a large number of probes mapping to intronic or intergenic regions, mainly among those based on UniGene, which are likely to give no meaningful signal. This was consistent with another study reporting higher detectability of RefSeq transcripts, when compared with non-RefSeq transcripts (3).
Here, we present a pipeline for the complete genomic and transcriptomic re-annotation of Illumina probe sequences, which can also be applied to other platforms. Its output is publicly and interactively available online, and incorporated in the current Bioconductor annotation packages. We also compare the re-annotation with the original from Illumina and evaluate its impact on the interpretation of data from different experiments.
METHODS
Annotation pipeline
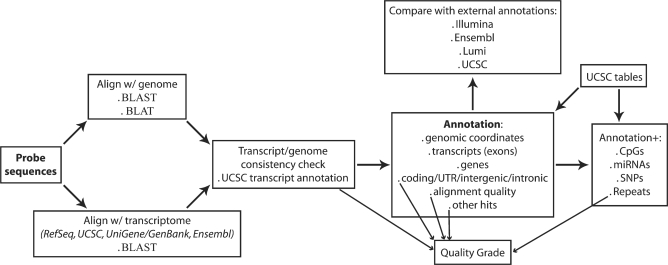
The pipeline for re-annotation of microarray probes relies on a Perl script and its key steps are illustrated in Figure 1. Probe sequences provided by Illumina (http://www.switchtoi.com/annotationfiles.ilmn) are BLASTed (29) (blastn, e-value = 10−6, DUST filter off—see Supplementary Data for details) and BLATed (30) (Web-based parameters) against the corresponding genome (Human hg18—NCBI 36.1, Mouse mm9—NCBI 37, Rat rn4—RGSC 3.4) and BLASTed against all the transcripts in RefSeq (27), UCSC Known Genes (31), UniGene/GenBank (32) and Ensembl (25). The transcriptomic alignment reports are then parsed and the selected transcripts are mapped to the genome and annotated at the gene level, based primarily not only on UCSC annotation tables (33) but also on UniGene (34) and Ensembl (25). The genomic coordinates thereby obtained are compared with the output of the genomic BLAST and BLAT reports, to check for consistency and to detect misalignments between the annotated transcripts and the genome. A probe is considered to be specific if all its transcriptomic matches align to one single genomic location, irrespective of the number of gene isoforms targeted and discrepancies between different sources of gene models. The described procedure also allows for the identification of probes mapping to intergenic or intronic sequences.
Figure 1.
Annotation pipeline. Schematics of the computational pipeline flow—see Methods section for details.
Annotation of cytobands, sequence repeats, CpGs, SNPs and overlapping micro RNAs (miRNAs) relies on UCSC annotation tables (33). A quality grade is assigned to each probe: ‘Perfect’ if the probe perfectly and uniquely matches the target transcript; ‘Good’ if the probe, although imperfectly matching the target transcript, is still likely to provide considerably sensitive signal [up to two mismatches are allowed, based on empirical evidence that the signal intensity for 50-mer probes with <95% identity to the respective targets is <50% of the signal associated with perfect matches (35)]; ‘Bad’ if the probe matches repeat sequences, intergenic or intronic regions, or is unlikely to provide sensitive and specific signal for any transcript (e.g. if the probe has three or more mismatches with targets, or if it targets multiple transcripts encoded from different loci) and ‘No match’ if the probe does not (according to the criteria defined above) significantly match any genomic region or transcript. We generally describe ‘Perfect’ and ‘Good’ probes as being reliable, and probes in all other categories as unreliable.
We have re-annotated probes from 8 BeadArray platforms: HumanWG-6 versions 1, 2 and 3; MouseWG-6 versions 1, 1.1 and 2; Rat Ref-12 version 1 and Human DASL. For Human and Mouse, Ref-8 probes are a subset of the corresponding WG-6 probes. The repertoire of probes for Human HT-12 is identical to those for HumanWG-6 version 3.
ReMOAT, re-annotation tables and Bioconductor
ReMOAT (Re-annotation and Mapping for Oligonucleotide Array Technologies) is a Web-based interface to the re-annotation data generated by the described pipeline. Data generated were processed using Perl and stored in a relational database. ReMOAT uses an Apache Web server, in a Linux environment, and a collection of PHP and Perl CGI scripts provide the user interface coupled to a MySQL RDBMS (http://www.mysql.com). The Web interface provides access to a tool capable of converting Illumina probe IDs, Entrez gene IDs or Ensembl gene IDs to several other formats such as Illumina probe IDs, Ensembl gene IDs, Entrez gene IDs, HGNC gene symbols, Unigene IDs or Lumi IDs. The resulting Web pages provide access to further information via links to Wikigenes (36), iHOP (37), HGNC nomenclature (38), Entrez Gene (39) and Ensembl (25) databases for each gene. A second search page enables extraction of the probe sequence, probe type, repeat masking results, GC contents, associated CpG islands, miRNAs and SNPs and quality assessment, generated by the pipeline for submitted Illumina IDs. Further search pages provide information on probe location, transcript and non-specific genomic hits (Supplementary Figure S1), and these are documented online.
Full re-annotation tables and their detailed description, as well as a link to ReMOAT, can be found online on http://www.compbio.group.cam.ac.uk/Resources/Annotation/.
A Bioconductor annotation package for each re-annotated platform has been built using the AnnotationDbi package. We chose reliable probes (i.e. with ‘Good’ or ‘Perfect’ annotation) and retrieved the RefSeq IDs as determined by our re-annotation. These IDs were then used by AnnotationDbi to create a database schema with predefined mappings. The packages can be installed by the same means as any other Bioconductor package and are named according to the platform annotated. For instance, the Human WG-6 V3 chip is available as the illuminaHumanv3.db (for data summarized using Illumina’s BeadStudio software) or illuminaHumanv3BeadID.db (for those starting from raw Illumina data) packages.
Annotation tables and ReMOAT are updated at least every 6 months, concomitantly with Bioconductor, and consistent version labelling is used for tracking old annotations.
Data sets
In order to evaluate the impact of probe annotation on the analysis and interpretation of Illumina expression data, we have looked at 27 Human WG-6 version 1 data sets (926 arrays) derived from a wide variety of conditions and tissue types (including breast, blood, artery, stem cell, sperm). These were obtained from GEO (40) on 18 November 2008, searching for platform GPL2507 and using the GEOquery Bioconductor package (41) to read the data into R (42).
The MAQC project (43,44) selected two human RNA samples for the comparison of differential expression on microarray platforms. These were the Universal Human Reference RNA (UHRR) from Stratagene (http://www.stratagene.com/manuals/740000.pdf) (a pool of 10 cancer cell lines from multiple tissues) and Human Brain Total RNA from Ambion (http://www.ambion.com/catalog/CatNum.php?AM6050). The original MAQC publication analyzed a dilution series of the two samples hybridized at three different locations. The Illumina portion of the data was generated using the Human WG-6 V1 platform and available in GEO (GSE5350) and ArrayExpress (E-TABM-132). For this study, we only consider the two extreme (pure) dilution levels. The same samples have since been run on Human WG-6 V2 BeadArrays by Asuragen Inc. and made publicly available on Illumina’s Website (http://www.switchtoi.com/datasets.ilmn). Finally, we have run those same samples in-house on Human WG-6 V3 BeadArrays.
Another illustrative data set consisted of Human WG-6 V2 arrays from the study in (45), which compared gene expression in hepatocytes between trisomic mice carrying human chromosome 21 and their wild-type litter-mates.
To illustrate possible effects of a SNP on the performance of a probe, we have used the Japanese HapMap (46) population. These 45 individuals have been examined using Illumina expression BeadArrays, and additionally genotyped, making them ideal for this purpose. Expression information comes from the Human WG-6 V1 platform and was obtained from the GENEVAR (1) project, and the corresponding genotypes were obtained from BioMart (24).
Two Illumina training samples were run on the DASL platform (47) using the standard cancer panel of 1506 probes. These consisted of RNA from the Illeum, and corresponding DNA (run in duplicate and triplicate, respectively).
Finally, we have looked at the preprocessed data from Miranda and colleagues, GEO series GSE13733. Samples consisted of MCF7 cells and were run on 10 Human WG-6 V3 arrays. There were two different drug treatments (DZNep and 5-Aza-CdR), with four technical replicates each, and two replicates of an untreated control sample.
Throughout the rest of the manuscript, these data sets are referred to as GEO, MAQC (V1, V2 and V3), Trisomy, HapMap, DASL and Miranda, respectively.
Microarray data analysis
All the analysis of Illumina microarray data was performed in R (42). The analysis of the GEO data set is described in more detail in (48). As the arrays in this data set were generated from a diverse set of tissues and processed using different normalization methods, direct comparisons between all arrays were not possible. Therefore, the intensities on each array were ranked to investigate the relative expression levels of the probes. The average rank for each bead type across the entire data set was then calculated and used to assess differences between the different annotation categories.
Two separate analyses were performed on the MAQC data. First, a comparison of the MAQC samples run on different Illumina platforms was performed. Normalized data generated from V1 and V2 platforms were taken directly from the published summarized values and the V3 data were BASHed (49) and summarized using beadarray (50), and then median normalized. We were then able to look at how particular probes of interest evolved over different versions of the annotation.
The MAQC data generated using Human WG-6 V1 BeadArrays were analyzed to look at the interaction between filtering, differential expression and the annotation categories. Non-normalized MAQC V1 data were read into R and a series of different filtering approaches (Supplementary Data) were applied to all probes. A differential expression analysis was also performed on non-filtered, quantile-normalized, data. The limma (51) package was used to find differentially expressed genes, and the log-odds scores given by empirical Bayes moderation of variances (52) were used to rank probes.
For the Trisomy study, the data were summarized, quantile-normalized and log2-transformed using beadarray (50). Differential expression was again quantified by the log-odds after empirical Bayes moderation of variances.
For DASL, data were analyzed and summarized using default BeadStudio settings to provide a 5 × 1506 matrix of observations.
RESULTS
Summary of annotation results
Table 1 summarizes the results of our re-annotation. Illumina’s reported efforts on improving the design and annotation of human probes (http://www.illumina.com/) have proven to be successful, as the percentage of unreliable probes has substantially decreased: 44% for WG-6 V1, 34% for V2 and 28% for V3. This improvement, consistent with a previous report (28), is essentially due to the increase in the proportion of RefSeq-derived probes and in the reliability of the UniGene-derived ones (Supplementary Figure S2). Illumina has indeed substantially redefined the RefSeq probes: only 16% of such probes are conserved between versions 1 and 3, 3647 probes are completely novel in version 3 when compared to version 2 and 5720 probes were redesigned (i.e. same target, different location) in the updating. Of the probes chosen for the human DASL platform, 95.6% are reliable (a much higher percentage than for the other platforms). This is not surprising, as there are fewer probes for DASL and they target known and well-curated cancer-related genes.
Table 1.
Results of re-annotation of Illumina probe sequences
| Platform | Source | Total number of probes | Perfect | Good | Bad | No match | Intronic | Intergenic | Splice junctions | Repeat sequences |
|---|---|---|---|---|---|---|---|---|---|---|
| Human WG-6 v3 | RefSeq | 36 079 73.5% | 28 154 78.0% | 909 2.5% | 6737 18.7% | 279 0.8% | 749 2.1% | 1306 3.6% | 2791 7.7% | 4254 11.8% |
| UniGene | 12 997 26.5% | 6389 49.2% | 88 0.7% | 5801 44.6% | 719 5.5% | 37 0.3% | 160 1.2% | 58 0.4% | 3987 30.7% | |
| Total | 49 076 | 34 543 70.4% | 997 2.0% | 12 538 25.5% | 998 2.0% | 786 1.6% | 1466 3.0% | 2849 5.8% | 8241 16.8% | |
| Human WG-6 v2 | RefSeq | 30 808 63.3% | 23 755 77.1% | 747 2.4% | 6028 19.6% | 278 0.9% | 747 2.4% | 1320 4.3% | 2246 7.3% | 3580 11.6% |
| UniGene | 17 894 36.7% | 7698 43.0% | 117 0.7% | 8435 47.1% | 1644 9.2% | 284 1.6% | 1211 6.8% | 57 0.3% | 5598 31.3% | |
| Total | 48 702 | 31 453 64.6% | 864 1.8% | 14 463 29.7% | 1922 3.9% | 1031 2.1% | 2531 5.2% | 2303 4.7% | 9178 18.8% | |
| Human WG-6 v1 | RefSeq | 26 098 55.2% | 20 769 79.6% | 783 3.0% | 4421 16.9% | 125 0.5% | 238 0.9% | 871 3.3% | 1681 6.4% | 2921 11.2% |
| UniGene | 21 198 44.8% | 4949 23.3% | 130 0.6% | 15 847 74.8% | 272 1.3% | 4113 19.4% | 9405 44.4% | 106 0.5% | 3463 16.3% | |
| Total | 47 296 | 25 718 54.4% | 913 1.9% | 20 268 42.9% | 397 0.8% | 4351 9.2% | 10 276 21.7% | 1787 3.8% | 6384 13.5% | |
| Human DASL | Total | 1506 | 1402 93.1% | 37 2.5% | 66 4.4% | 1 0.1% | 0 0.0% | 2 0.1% | 17 1.1% | 40 2.7% |
| Mouse WG-6 v2 | RefSeq | 23 031 50.9% | 19 115 83.0% | 556 2.4% | 3150 13.7% | 210 0.9% | 383 1.7% | 926 4.0% | 1080 4.7% | 1567 6.8% |
| MEEBO | 16 591 36.6% | 15 267 92.0% | 318 1.9% | 863 5.2% | 143 0.9% | 47 0.3% | 6 0.04% | 384 2.3% | 692 4.2% | |
| Riken | 5659 12.5% | 3574 63.2% | 292 5.2% | 1749 30.9% | 44 0.8% | 0 0.0% | 0 0.0% | 292 5.2% | 1636 28.9% | |
| Total | 45 281 | 37 956 83.8% | 1166 2.6% | 5762 12.7% | 397 0.9% | 430 0.9% | 932 2.1% | 1756 3.9% | 3895 8.6% | |
| Mouse WG-6 v1.1 | RefSeq | 32 342 69.3% | 28 560 88.3% | 394 1.2% | 3140 9.7% | 248 0.8% | 431 1.3% | 961 3.0% | 1026 3.2% | 1556 4.8% |
| MEEBO | 9512 20.4% | 7306 76.8% | 290 3.0% | 1322 13.9% | 594 6.2% | 191 2.0% | 19 0.2% | 113 1.2% | 953 10.0% | |
| Riken | 4784 10.3% | 2832 59.2% | 261 5.5% | 1644 34.4% | 47 1.0% | 0 0.0% | 0 0.0% | 171 3.6% | 1542 32.2% | |
| Total | 46 638 | 38 698 83.0% | 945 2.0% | 6106 13.1% | 889 1.9% | 622 1.3% | 980 2.1% | 1310 2.8% | 4051 8.7% | |
| Mouse WG-6 v1 | RefSeq + MEEBO | 34 159 85.5% | 29 196 85.4% | 530 1.6% | 4080 11.9% | 353 1.0% | 582 1.7% | 967 2.8% | 866 2.5% | 2211 6.5% |
| Riken | 5809 14.5% | 3665 63.1% | 294 5.1% | 1800 31.0% | 50 0.9% | 0 0.0% | 0 0.0% | 302 5.2% | 1694 29.1% | |
| Total | 39 968 | 32 861 82.2% | 824 2.1% | 5880 14.7% | 403 1.0% | 582 1.5% | 967 2.4% | 1168 2.9% | 3905 9.8% | |
| Rat RefSeq v1 | RefSeq | 22 523 | 15 534 69.0% | 322 1.4% | 6189 27.5% | 478 2.1% | 295 1.3% | 2533 11.2% | 933 4.1% | 1187 5.3% |
The number of probes in each annotation category is indicated; percentages (italics) indicate the corresponding proportion for the associated source.
The outstanding wealth of mouse transcriptomic sequences and respective annotation in the databases reflects on the quality of the probes for mouse, superior to human’s. Interestingly, the increase in reliability from version to version has been very subtle (84% reliable probes for V1, 85% for V1.1, 86% for V2).
The percentage of reliable RefSeq-derived probes for rat is 70%, lower than that observed for RefSeq human (81%) and mouse (85%) probes. This can be explained by the relative scarcity of transcriptomic information available for rat, when compared with the other two species.
Our transcriptomic annotation of probes is very similar to that of Du et al. (23) for all platforms and to Ensembl’s (25) for Human versions 1 and 2. However, they fail to take into account probes matching the reverse strand of the target transcript, which is the cause of most of the ∼2% discrepancy. Our pipeline rejects such probes, as probes are supposed to be strand specific. The strand specificity is of particular importance given the large number of human antisense transcripts (53,54).
Filtering and differential expression analysis
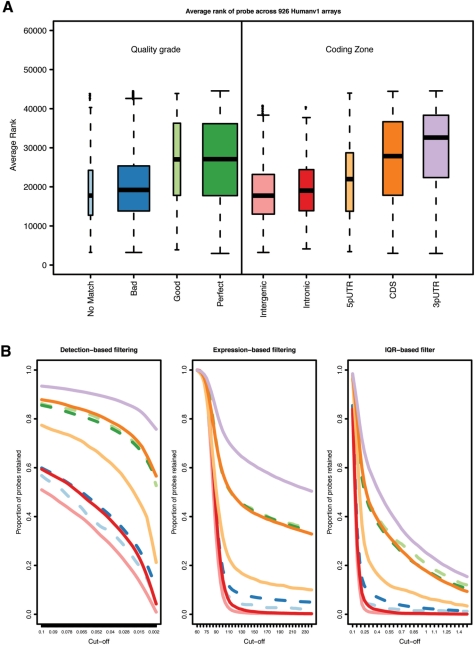
Figure 2A shows how, for the GEO data set, the average expression ranks of genes vary between different annotation categories of probes. As could be expected, probes annotated as reliable are observed to have a high mean rank (∼27 000 for ‘Perfect’ and ‘Good’), whereas other probes (e.g. matching to an intronic or intergenic region, or with no matches) have a much lower mean rank of below 20 000. The figure also shows a decreased mean rank of probes mapping closer to the 5′ end of transcripts, consistent with the instability of RNA molecules and their degradation typically starting from the 5′ end. This effect has been addressed in the probe-level models for the analysis of Affymetrix (20) and motivates most of the manufacturers, including Illumina, to design their gene expression platforms with probes targeting the 3′ end of transcripts.
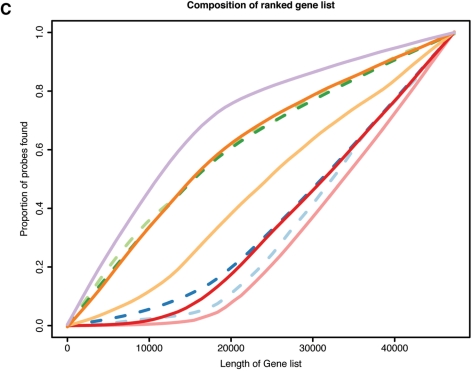
Figure 2.
Impact of annotation on expression analysis. (A) Box plots of the average expression ranks of probes, calculated across the GEO arrays, for each annotation category (box widths proportional to the number of probes in the respective category); (B) proportions of probes retained by each filtering method applied to the MAQC V1 data set (y-axis) as a function of the chosen cut-off (x-axis); (C) proportions of probes of each category (y-axis) found in a gene list of certain length arising from a differential expression analysis of the MAQC V1 data (x-axis). For panels (B) and (C), the colour code is the same as used in (A); dashed lines are associated with quality grade categories and solid lines with coding zones.
Among the 100 bead types with the highest average rank (>46 891), 39 are found to target ribosomal proteins that function in protein biosynthesis and some of which have been previously found to be housekeeping or reference genes (55,56). Thus, these genes are often considered useful for normalization. Indeed, 4 genes from this list are also found in the table of top 15 candidate housekeeping genes presented in (55), which includes 13 ribosomal proteins. Other interesting features of these 100 probes are that 53 have multiple matches to the genome, 24 map to repeat regions and 41 map to SNPs (Supplementary Table S1).
For the MAQC V1 data set, Figure 2B shows that, for filtering based both on expression level (‘Detection’ and ‘Expression’) and variability (‘IQR’), probes annotated as ‘Perfect’ or ‘Good’ are more readily retained than those classified as ‘Bad’ or not matching.
Figure 2C shows, in terms of our annotation categories, the composition of the ranked gene lists from the differential expression analysis of the MAQC V1 data. As expected, bead types annotated as reliable occur towards the top of the list. On the other hand, genes mapping to intronic and intergenic regions, or having no match at all, are ranked lower in the list.
SNPs and mismatches
Having presented the general good performance of Illumina probe design, we now discuss some specific cases where the performance requires careful interpretation.
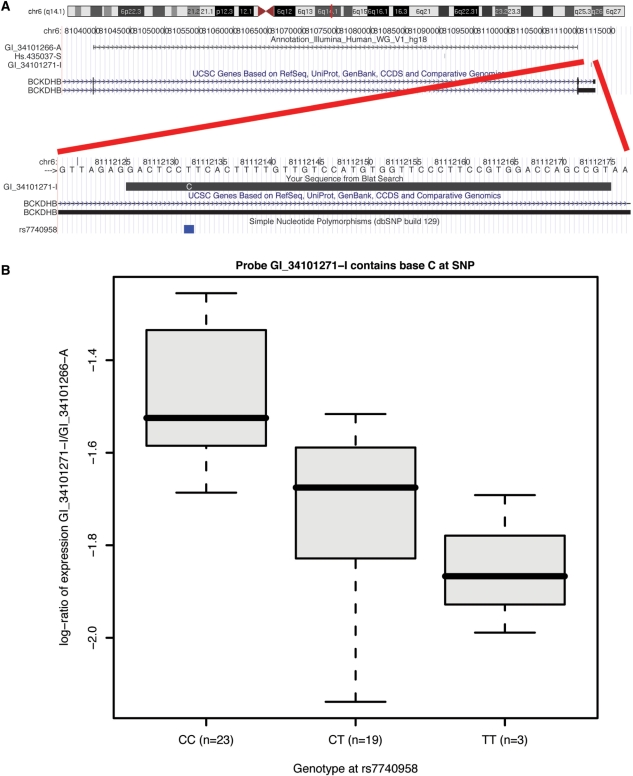
The retention, by the filtering methods, of probes with mismatches (named ‘Good’) and their generally high expression ranks suggest that hybridization is still possible when probe and target sequences do not match perfectly. Furthermore, for Human V2 over 100 of the detected mismatches can be explained by SNPs and 5791 probes map to annotated SNPs. Therefore, allelic ambiguity needs to be taken into account when interpreting gene expression data. Figure 3 and Supplementary Figure S3 illustrate an association between genotype and measured expression in the Japanese HapMap population that is consistent with a mismatch inducing a significant decrease in registered intensity. Similar effects have been previously reported for Affymetrix (57).
Figure 3.
Association between genotype and measured expression. (A) The sequence we examine is in the BCKDHB gene on chromosome 6, which contains a SNP (rs7740958—T/C) at position 7. The Illumina probe targeting this sequence (GI_34101271-I) contains a C at this location. There is also a probe (GI_34101266-A) targeting a constitutive splice junction of BCKDHB and matching no known SNPs. [Figure built based on UCSC Genome Browser graphics (26).] (B) Box plots of the log2 expression ratios in the Japanese HapMap population according to the rs7740958 genotype.
As another example, human probes ILMN_1692545 and ILMN_1670800 (WG versions 2 and 3) differ in only one nucleotide (C/A at position 19) that corresponds to SNP rs13082444 (G/T for the genomic Watson strand). A striking difference in expression between the two is found for all the samples in the Miranda data set (median of 48 fold). According to the current human genome annotation, these probes primarily target an intergenic region and, therefore, no known transcript. However, these probes were designed to target transcripts XM_933970.1 and XM_944901.1 that perfectly map to the two genomic variants and have since been removed from the databases (Supplementary Figure S4).
We have listed, for the three Human WG-6 platforms, all the pairs of probes differing in 1, 2 and 3 nucleotides (Supplementary Table S2). There is a substantial increase in the number of probe pairs differing in one nucleotide from V1 (9) to V2 (177), suggesting that attention to SNPs and allele specific expression has been given in Illumina’s redesigning of probes.
Repeat sequences
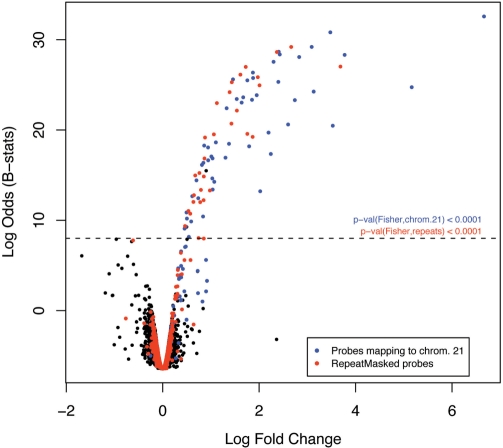
It is not expected that intergenic and intronic regions of the genome will feature in gene expression studies. However, there are a few examples of probes that fall into these categories yet are highly ranked on all GEO arrays (outliers in categories ‘Bad’ and ‘Intergenic’ in Figure 2A). Most of such probes include repeat sequences and are partially or totally ‘masked’ by RepeatMasker (http://www.repeatmasker.org). They are likely to cross-hybridize and provide non-specific signal. We have, therefore, classified all ‘masked’ probes as ‘Bad’. A good example of how these probes can impact on the analysis of gene expression is provided by the study in (45), where our annotation has been used. As illustrated in Figure 4, all human transcripts but one overexpressed in Tc1 mouse livers (whose cells contain one human chromosome 21, apart from the normal mouse karyotype), when compared to their wild-type litter-mates, are either from chromosome 21 (as expected) or targeted by probes containing repeat sequences. Moreover, the number of overexpressed ‘masked’ transcripts is statistically significant. These results suggest that repetitive probes, although not originally annotated as mapping to chromosome 21, are non-specifically binding to repetitive transcripts from that chromosome. Assuming transcripts from human chromosome 21 are those truly differentially expressed between Tc1 and normal litter-mates, ignoring the information about repeat sequences would have led us to a false discovery rate of 37%, instead of the current 2%.
Figure 4.
Effect of repetitive sequences on gene expression measurements. Volcano plot (y-axis: empirical Bayes log-odds of differential expression; x-axis: log2 fold change in expression) comparing the expression of human transcripts in livers between Tc1 mice carrying a human chromosome 21 and their wild-type litter-mates (Tc0). Blue dots depict probes targeting genes on human chromosome 21 and red dots probes comprising human RepeatMasker sequences. The dashed line is an arbitrary cut-off for differential expression based on the assumption that there are no human sequences transcribed in the wild-type mouse and, therefore, no human transcript should be overexpressed in Tc0. The P-values (Fisher’s exact test) show that the numbers of differentially expressed transcripts from human chromosome 21 (blue) and comprising human repeats (red) are both highly significant.
Another interesting example is provided by a set of six genomically clustered human probes, two of which were designed to target the PSMC1 gene while the other four match spurious transcripts that have since been removed from the databases. The six probes exhibit a good match not only with the PSMC1 locus but also with several other regions in different chromosomes (including an intronic sequence of the PLCZ1 locus) and the relative position of the matches is generally conserved (Supplementary Figure S5).
An even more striking example of potential non-specificity of hybridization is given by a set of 15 probes, each targeting transcripts from multiple genes from the GAGE cluster on chromosome X (Supplementary Figure S6).
Alternative splicing
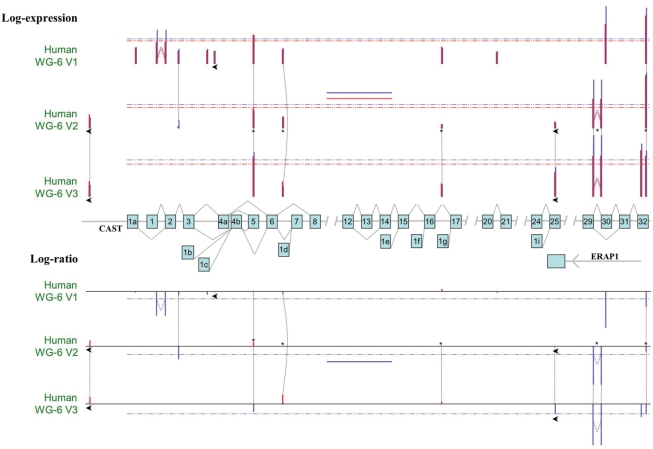
Although for Illumina the majority of genes are targeted by one unique reliable probe type, there are still a few thousand genes covered by more than one probe type (Supplementary Figure S7). Almost all multi-exon genes undergo alternative splicing (58,59) and, therefore, comprise multiple isoforms. Probes targeting different transcripts for the same gene can generate ambiguity in the definition of gene expression but they also provide an opportunity for the detection of alternative splicing. This is well illustrated in Figure 5 and Supplementary Figure S8 for the human CAST (calpastatin) gene, covered by at least six different probe types for any of the platforms. The probes target the constitutive last exon, several alternative first exons and alternative splice junctions. The differences between probes, not only in expression levels but also in differential expression ratios, show the extra insight brought by a transcript/exon-centric analysis and how misinformative a gene-centric summary of data can be. This example is also an illustration of the importance of strand specificity of probes, as ILMN_1752145 targets the 3′-most exon of an antisense ERAP1 (endoplasmic reticulum aminopeptidase 1) transcript and genomically overlaps with a constitutive exon of CAST.
Figure 5.
Importance of alternative splicing on the interpretation of gene expression data. Schematics of the structure of the human CAST gene, with exons depicted by numbered boxes in light blue and all annotated alternative splicing events represented by the associated thin black dashed lines. The three upper tracks represent the logged expression levels for all the probes targeting the CAST locus for each of the three human Illumina WG platforms. Bars are positioned according to the relative position of the respective probes in the locus. Bar height is proportional to the average log gene expression in the MAQC data: red for brain samples and blue for reference samples. The coloured dashed lines indicate the respective average expression levels of probes targeting CAST for each sample type. The short coloured full lines in the middle of the V2 track indicate the expression level of CAST estimated by the original Illumina gene summarization procedure for each sample type; black stars identify V2 probes targeting CAST according to the original Illumina annotation. Black arrows identify probes mapping to the reverse strand. The three lower tracks represent the corresponding log-ratios. This figure shows that a gene-centric analysis indicating underexpression of the CAST gene in brain (dashed blue line) might be biased by a brain-specific skipping of exon 30 and/or an alternative first exon.
Probes matching splice junctions can be particularly effective in the detection of alternative splicing events. We clearly show, for the DASL platform, the increase in relative signal intensity for such probes from hybridization against genomic DNA to hybridization against transcriptomic RNA (Supplementary Figure S9).
The interpretation of expression data can also be affected by ambiguity in the definition of a gene or, in other words, by distinct genes sharing the same locus. For example, two different human genes, CPNE1 (copine I) and RBM12 (RNA-binding motif protein 12), share their most 5′ exons (as well as the promoter region) (60). Three Illumina probes (ILMN_1701229, ILMN_2276000, ILMN_2276002) target both an alternative non-coding first exon of CPNE1 and the last and only coding exon of RBM12 (Supplementary Figure S10). An even more striking example is the PCDHG locus on human chromosome 5, which seems to cluster several distinct genes and different isoforms of the same gene (generally characterized by alternative first exons) (Supplementary Figure S11). Illumina designed probes to target virtually all the different transcripts and annotates each of them as a distinct gene. In contrast, UniGene (34) considers them to be all part of the PCDHGC3 gene cluster. This illustrates well the difficulty in choosing a suitable universal gene identifier and that even a system like UniGene, specifically designed for organizing transcripts into non-redundant gene-oriented clusters and, particularly, popular in meta-analyses (61), cannot deal with all the idiosyncrasies associated with defining what a gene is.
Another example of the particular importance of analyzing expression at the transcript level for genes containing alternative promoters and first exons is given by the CDKN2A (cyclin-dependent kinase inhibitor 2A) locus for both human and mouse. The gene has two alternative first exons, separated by over 10 Kb, that give rise to two structurally and functionally distinct isoforms: p16INK4a (inhibitor of CDK4 kinase) and p14ARF (alternate open reading frame, involved in the stabilization of p53 by sequestering of MDM2). All the Human and Mouse WG-6 platforms have three probes targeting CDKN2A: one for each of the alternative first exons and one for the common last exon (Supplementary Figure S12). Annotating the trio of probes as having the same gene target would, for most experiments, lead to the observation of inconsistency between the associated three gene expression measurements.
Any signal given by probes matching an intron or exon junction and, therefore, partially intronic, like human ILMN_1690644 (Supplementary Figure S13), is very difficult to interpret. In particular, this holds when there is no evidence for splice variants involving an alternative splice site or a retained intron.
Generally, probes targeting constitutively transcribed sequences are expected to exhibit stronger signal than probes matching only alternative transcripts. The former are, therefore, more suitable for a gene-centric analysis of data. We support this assumption by showing that there is a positive correlation between the proportion of isoforms in a gene targeted by a probe and its intensity (Supplementary Figure S14).
Other interpretation issues
There are examples of other aspects of the design of Illumina probes that might affect the interpretation of expression data.
Human WG-6 V3 probes ILMN_2311089 and ILMN_1738027 both perfectly target a constitutive exon of BRCA1 (breast cancer 1) and overlap in 49 out of their 50 nucleotides (Supplementary Figure S15A). However, for all samples in the Miranda data set (GEO series GSE13733), ILMN_1738027 exhibits higher intensities (Supplementary Figure S15B). Hybridization dynamics are particularly sensitive to poly-C/poly-G tracts at the end of the probes, and it is plausible that the one-nucleotide increase in length of the poly-C tract, in this case, would account for the observed discrepancy.
Probes ILMN_1726308 and ILMN_1756139 were designed to have distinct gene targets yet differ in only one nucleotide. The first perfectly and uniquely targets the 3′UTR of the mono-exonic FAM10A4 transcript on chromosome 13. The second perfectly targets the predicted transcript XR_038903.1 on chromosome X. Some signal generated by unwanted cross-hybridization, due to the probes’ insufficient specificity, is inevitable.
Finally, we have found transcripts targeted by many (more than 10) probes, namely the major isoforms of HYDIN (Supplementary Figure S16) and FAM90A1 (Supplementary Figure S17). Illumina probes were designed so that the majority of genes would be targeted by only one probe each (Supplementary Figure S6). Therefore, contrary to Affymetrix, no probe-level models have been developed to summarize ‘probeset’ information. In particular, the lack of a model taking into account the relative matching position along the transcript might reduce the precision of the gene expression measurements.
DISCUSSION
Designing the Illumina 50-mer probes might be considered harder than the Affymetrix 25-mers. Although designing shorter probes is conditioned by a higher chance of random matches to non-target sequences, it is known that specificity of long probes decreases with growing probe length, due to a higher probability of a probe fragment matching an unwanted target (or even to fold, self-hybridize and form a ‘hairpin’), thereby neutralizing the gain in sensitivity associated with higher binding energies (62). This is supported by our observation of generally high expression ranks of probes with mismatches (‘Good’). Avoiding SNPs, which is more difficult with longer probes, is, therefore, important. Moreover, as the GeneChip technology has been well established for over a decade, there is more literature on thermal and hybridization dynamics for Affymetrix probes (63). It has been shown that the base composition of 25-mers can affect the observed intensity and summarization methods have been developed to deal with such effects (64). To our knowledge, only one study has investigated such effects for BeadArrays (5).
Nonetheless, BeadArrays have become increasingly popular, not only because of their reliable measures of what is targeted by the highly sensitive beads but also due to their low bias and high precision. Our work suggests that future studies should not overlook the importance of annotating probe sequences and checking if they target the desired features. Two previous re-annotation efforts (23,25) have mapped Illumina probes to transcripts with reasonable success (despite failing to recognize the strand specificity) but they are not as extensive in investigating other factors that we have shown to be important.
Our re-annotation depends on the accuracy of not only the probe sequences provided by Illumina but also the transcriptomic information in the public databases. The impact of database content on microarray performance and reproducibility has previously been shown (28). For example, UCSC annotates the genomic locus of GenBank mRNA AK024373 as spanning more than 2 Mb of human chromosome 19 and many genes. Any intergenic probe in that region (e.g. ILMN_1719185) will be spuriously classified as intronic as a result of this.
Some ‘poorly designed’ probes can, on occasion, be useful. Some intergenic probes may actually target novel non-coding RNAs and intronic probes that can be used in the detection of alternative splicing events involving the retention of introns. Others can be seen as negative controls. Their utility demands accurate annotation, as the probes are not targeting the features they were supposed to. Most filtering approaches would likely exclude these probes from the gene expression analysis, otherwise it would be questionable whether they could still be considered reliable or informative. There are more suitable technologies for such applications anyway (e.g. Affymetrix Exon ST Arrays for the analysis of alternative splicing).
Preliminary filtering to remove uninformative probes is regarded as an essential part of the analysis and has been shown to decrease the false discovery rate in differential expression assessment (65). However, such filtering is typically performed using measures based on expression level, variability and ad hoc cut-offs that can vary between experiments. Such approaches also bias the downstream analysis against transcripts that are relatively lowly expressed and may still be biologically relevant. On the other hand, probes with high signal caused by spurious cross-hybridization would still be included. A clear message from our investigations is that uninformative probes may be identified by their re-annotation, and that filtering based on the quality of design and annotation is an unbiased way of improving the specificity of an analysis. It is worth bearing in mind that a probe perfectly and uniquely matching its target may still be unreliable due to its particular composition and hybridization dynamics (5).
With the exception of Affymetrix (57,66), very little is reported about the impact of SNPs on the analysis of expression data. The increase in the number of probe pairs differing in one nucleotide from older to recent versions of BeadArray platforms suggests that the aspect is now being given attention by Illumina. Even when we can match up SNP/expression pairs, there may be no expression registered (either due to probe design issues or naturally), or no variation seen at the SNP within this population. Furthermore, the location of the SNP within the probe sequence will undoubtedly influence its impact, as it has been reported for several SNP association and expression quantitative trait loci studies (57, 67–69).
Many recent high impact studies have used BeadArrays for gene expression analysis. The majority of Illumina probes are of good quality and filtering has enriched analyses for these good quality probes. Therefore, most of the biological inferences from those analyses should be reliable. However, our work shows the possibility for misinterpretation of results, as a consequence of analyzing probes that should have been removed due to poor design. For example, one of the key findings in (70) is that the pluripotent cells share a protein–protein network (PluriNet) that has a 299-gene signature. Our re-annotation reveals that ∼8% of the associated 370 probes (Supplementary Table S3) can be considered unreliable (‘Bad’) for targeting repeats, intronic regions or annotated transcripts that cannot be aligned to the genome. Furthermore, 80 of the reliable probes target known SNPs. This has clear implications on the interpretation of this signature.
Finally, efforts like the present study should help the microarray manufacturers to improve the probe design of their new platforms. Nonetheless, it is important to keep up-to-date annotation of probes for the existing ones, as it provides the opportunity for improved analysis of old data sets and can be an important tool in large studies involving meta-analyses of data sets generated from different platforms. For instance, 6976 human and 31 307 mouse probes have identical sequences across all WG-6 version numbers (Supplementary Figure S18), and, thus, have the potential to be combined reliably in a meta-analysis. Using nuID (23) or even the sequence itself as an universal probe identifier can aid in the creation of a single library of probes for each species. Transcriptomic annotation would, therefore, be particularly important in the meta-analysis of probes from different versions with the same target.
The Web interface and the periodic update of Bioconductor packages will allow us to provide updated re-annotation of the covered platforms, as the genomic and transcriptomic databases also get updated. Moreover, the described computational pipeline can be used to re-annotate probes for virtually any microarray application.
In summary, using the accurate, comprehensive and up-to-date re-annotation of probes described herein improves the accuracy of the analysis of Illumina microarray data (including the re-analysis of previous studies) and widens the scope of biological information that can be extracted from BeadArray experiments.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Cancer Research UK (to N.L.B.M., M.J.D., S.A.S., A.G.L. and S.T.); Medical Research Council (to M.J.D.); European Molecular Biology Organization (to J.F.J.D); Isaac Newton Trust (to M.E.R.). Funding for open access charge: Cancer Research UK.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge the support of the University of Cambridge, Cancer Research UK, and Hutchison Whampoa Limited. We thank Illumina, Bin Liu, Carlos Caldas, Natalie Thorne, Leonard Goldstein, Inma Spiteri, David Allard, Michelle Osborne, James Hadfield, Mahesh Iddawela, Ana Teresa Maia, Ashraf Ibrahim, Roslin Russell, Christina Curtis, Mike Wilson, Mike Smith, Dan Tomlinson, Gos Micklem, Gordon Smyth and Lynn Amon for their support and feedback.
REFERENCES
- 1.Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, Thorne N, Redon R, Bird CP, de Grassi A, Lee C, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goring HH, Curran JE, Johnson MP, Dyer TD, Charlesworth J, Cole SA, Jowett JB, Abraham LJ, Rainwater DL, Comuzzie AG, et al. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat. Genet. 2007;39:1208–1216. doi: 10.1038/ng2119. [DOI] [PubMed] [Google Scholar]
- 4.Barnes M, Freudenberg J, Thompson S, Aronow B, Pavlidis P. Experimental comparison and cross-validation of the Affymetrix and Illumina gene expression analysis platforms. Nucleic Acids Res. 2005;33:5914–5923. doi: 10.1093/nar/gki890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunning MJ, Barbosa-Morais NL, Lynch AG, Tavaré S, Ritchie ME. Statistical issues in the analysis of Illumina data. BMC Bioinformatics. 2008;9:85. doi: 10.1186/1471-2105-9-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunning MJ, Ritchie ME, Barbosa-Morais NL, Tavaré S, Lynch AG. Spike-in validation of an Illumina-specific variance-stabilizing transformation. BMC Res. Notes. 2008;1:18. doi: 10.1186/1756-0500-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunning MJ, Thorne NP, Camilier I, Smith ML, Tavaré S. Quality control and low-level statistical analysis of Illumina BeadArrays. Rev. Stat. 2006;4:1–30. [Google Scholar]
- 8.Lin SM, Du P, Huber W, Kibbe WA. Model-based variance-stabilizing transformation for Illumina microarray data. Nucleic Acids Res. 2008;36:e11. doi: 10.1093/nar/gkm1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xie Y, Wang X, Story M. Statistical methods of background correction for Illumina BeadArray data. Bioinformatics. 2009;25:751–757. doi: 10.1093/bioinformatics/btp040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bitton DA, Okoniewski MJ, Connolly Y, Miller CJ. Exon level integration of proteomics and microarray data. BMC Bioinformatics. 2008;9:118. doi: 10.1186/1471-2105-9-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, Shoemaker DD. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 12.Okoniewski MJ, Hey Y, Pepper SD, Miller CJ. High correspondence between Affymetrix exon and standard expression arrays. Biotechniques. 2007;42:181–185. doi: 10.2144/000112315. [DOI] [PubMed] [Google Scholar]
- 13.Robinson MD, Speed TP. A comparison of Affymetrix gene expression arrays. BMC Bioinformatics. 2007;8:449. doi: 10.1186/1471-2105-8-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maouche S, Poirier O, Godefroy T, Olaso R, Gut I, Collet JP, Montalescot G, Cambien F. Performance comparison of two microarray platforms to assess differential gene expression in human monocyte and macrophage cells. BMC Genomics. 2008;9:302. doi: 10.1186/1471-2164-9-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, Bunney WE, Myers RM, Speed TP, Akil H, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33:e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gautier L, Moller M, Friis-Hansen L, Knudsen S. Alternative mapping of probes to genes for Affymetrix chips. BMC Bioinformatics. 2004;5:111. doi: 10.1186/1471-2105-5-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harbig J, Sprinkle R, Enkemann SA. A sequence-based identification of the genes detected by probesets on the Affymetrix U133 plus 2.0 array. Nucleic Acids Res. 2005;33:e31. doi: 10.1093/nar/gni027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sandberg R, Larsson O. Improved precision and accuracy for microarrays using updated probe set definitions. BMC Bioinformatics. 2007;8:48. doi: 10.1186/1471-2105-8-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu H, Wang F, Tu K, Xie L, Li YY, Li YX. Transcript-level annotation of Affymetrix probesets improves the interpretation of gene expression data. BMC Bioinformatics. 2007;8:194. doi: 10.1186/1471-2105-8-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 21.Okoniewski MJ, Miller CJ. Hybridization interactions between probesets in short oligo microarrays lead to spurious correlations. BMC Bioinformatics. 2006;7:276. doi: 10.1186/1471-2105-7-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome. Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Du P, Kibbe WA, Lin SM. nuID: a universal naming scheme of oligonucleotides for Illumina, Affymetrix, and other microarrays. Biol. Direct. 2007;2:16. doi: 10.1186/1745-6150-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smedley D, Haider S, Ballester B, Holland R, London D, Thorisson G, Kasprzyk A. BioMart—biological queries made easy. BMC Genomics. 2009;10:22. doi: 10.1186/1471-2164-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hubbard TJ, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Clarke L, et al. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eggle D, Debey-Pascher S, Beyer M, Schultze JL. The development of a comparison approach for Illumina bead chips unravels unexpected challenges applying newest generation microarrays. BMC Bioinformatics. 2009;10:186. doi: 10.1186/1471-2105-10-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 30.Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hsu F, Kent WJ, Clawson H, Kuhn RM, Diekhans M, Haussler D. The UCSC known genes. Bioinformatics. 2006;22:1036–1046. doi: 10.1093/bioinformatics/btl048. [DOI] [PubMed] [Google Scholar]
- 32.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2008;36:D25–D30. doi: 10.1093/nar/gkm929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karolchik D, Kuhn RM, Baertsch R, Barber GP, Clawson H, Diekhans M, Giardine B, Harte RA, Hinrichs AS, Hsu F, et al. The UCSC Genome Browser Database: 2008 update. Nucleic Acids Res. 2008;36:D773–D779. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pontius JU, Wagner L, Schuler GD. The NCBI Handbook. Bethesda, MD: National Center for Biotechnology Information; 2003. [Google Scholar]
- 35.He Z, Wu L, Li X, Fields MW, Zhou J. Empirical establishment of oligonucleotide probe design criteria. Appl. Environ. Microbiol. 2005;71:3753–3760. doi: 10.1128/AEM.71.7.3753-3760.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoffmann R. A wiki for the life sciences where authorship matters. Nat. Genet. 2008;40:1047–1051. doi: 10.1038/ng.f.217. [DOI] [PubMed] [Google Scholar]
- 37.Hoffmann R. Using the iHOP information resource to mine the biomedical literature on genes, proteins, and chemical compounds. Curr. Protoc. Bioinformatics. 2007 doi: 10.1002/0471250953.bi0116s20. Chapter 1, Unit1 16. [DOI] [PubMed] [Google Scholar]
- 38.Bruford EA, Lush MJ, Wright MW, Sneddon TP, Povey S, Birney E. The HGNC Database in 2008: a resource for the human genome. Nucleic Acids Res. 2008;36:D445–D448. doi: 10.1093/nar/gkm881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sayers EW, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2009;37:D5–D15. doi: 10.1093/nar/gkn741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Edgar R. NCBI GEO: mining tens of millions of expression profiles—database and tools update. Nucleic Acids Res. 2007;35:D760–D765. doi: 10.1093/nar/gkl887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sean D, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 2007;23:1846–1847. doi: 10.1093/bioinformatics/btm254. [DOI] [PubMed] [Google Scholar]
- 42.R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2009. [Google Scholar]
- 43.Shi L, Reid LH, Jones WD, Shippy R, Warrington JA, Baker SC, Collins PJ, de Longueville F, Kawasaki ES, Lee KY, et al. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat. Biotechnol. 2006;24:1151–1161. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shippy R, Fulmer-Smentek S, Jensen RV, Jones WD, Wolber PK, Johnson CD, Pine PS, Boysen C, Guo X, Chudin E, et al. Using RNA sample titrations to assess microarray platform performance and normalization techniques. Nat. Biotechnol. 2006;24:1123–1131. doi: 10.1038/nbt1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilson MD, Barbosa-Morais NL, Schmidt D, Conboy CM, Vanes L, Tybulewicz VL, Fisher EM, Tavaré S, Odom DT. Species-specific transcription in mice carrying human chromosome 21. Science. 2008;322:434–438. doi: 10.1126/science.1160930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bibikova M, Talantov D, Chudin E, Yeakley JM, Chen J, Doucet D, Wickham E, Atkins D, Barker D, Chee M, et al. Quantitative gene expression profiling in formalin-fixed, paraffin-embedded tissues using universal bead arrays. Am. J. Pathol. 2004;165:1799–1807. doi: 10.1016/S0002-9440(10)63435-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dunning MJ. University of Cambridge; 2008. Genome-wide analyses using bead-based microarrays. Ph.D. Thesis. [Google Scholar]
- 49.Cairns JM, Dunning MJ, Ritchie ME, Russell R, Lynch AG. BASH: a tool for managing BeadArray spatial artefacts. Bioinformatics. 2008;24:2921–2922. doi: 10.1093/bioinformatics/btn557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dunning MJ, Smith ML, Ritchie ME, Tavaré S. beadarray: R classes and methods for Illumina bead-based data. Bioinformatics. 2007;23:2183–2184. doi: 10.1093/bioinformatics/btm311. [DOI] [PubMed] [Google Scholar]
- 51.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 52.Lonnstedt I, Speed TP. Replicated microarray data. Statistica Sinica. 2002;12:31–46. [Google Scholar]
- 53.Lehner B, Williams G, Campbell RD, Sanderson CM. Antisense transcripts in the human genome. Trends. Genet. 2002;18:63–65. doi: 10.1016/s0168-9525(02)02598-2. [DOI] [PubMed] [Google Scholar]
- 54.Yelin R, Dahary D, Sorek R, Levanon EY, Goldstein O, Shoshan A, Diber A, Biton S, Tamir Y, Khosravi R, et al. Widespread occurrence of antisense transcription in the human genome. Nat. Biotechnol. 2003;21:379–386. doi: 10.1038/nbt808. [DOI] [PubMed] [Google Scholar]
- 55.de Jonge HJ, Fehrmann RS, de Bont ES, Hofstra RM, Gerbens F, Kamps WA, de Vries EG, van der Zee AG, te Meerman GJ, ter Elst A. Evidence-based selection of housekeeping genes. PLoS ONE. 2007;2:e898. doi: 10.1371/journal.pone.0000898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thorrez L, Van Deun K, Tranchevent LC, Van Lommel L, Engelen K, Marchal K, Moreau Y, Van Mechelen I, Schuit F. Using ribosomal protein genes as reference: a tale of caution. PLoS ONE. 2008;3:e1854. doi: 10.1371/journal.pone.0001854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Benovoy D, Kwan T, Majewski J. Effect of polymorphisms within probe-target sequences on olignonucleotide microarray experiments. Nucleic Acids Res. 2008;36:4417–4423. doi: 10.1093/nar/gkn409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 59.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang W, Ng P, Zhao M, Wong TK, Yiu SM, Lau YL. Promoter-sharing by different genes in human genome—CPNE1 and RBM12 gene pair as an example. BMC Genomics. 2008;9:456. doi: 10.1186/1471-2164-9-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ramasamy A, Mondry A, Holmes CC, Altman DG. Key issues in conducting a meta-analysis of gene expression microarray datasets. PLoS Med. 2008;5:e184. doi: 10.1371/journal.pmed.0050184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chou CC, Chen CH, Lee TT, Peck K. Optimization of probe length and the number of probes per gene for optimal microarray analysis of gene expression. Nucleic Acids Res. 2004;32:e99. doi: 10.1093/nar/gnh099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Skvortsov D, Abdueva D, Curtis C, Schaub B, Tavaré S. Explaining differences in saturation levels for Affymetrix GeneChip arrays. Nucleic Acids Res. 2007;35:4154–4163. doi: 10.1093/nar/gkm348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu Z, Irizarry RA. Stochastic models inspired by hybridization theory for short oligonucleotide arrays. J. Comput. Biol. 2005;12:882–893. doi: 10.1089/cmb.2005.12.882. [DOI] [PubMed] [Google Scholar]
- 65.Scholtens D, Heydebreck AV. In: Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Gentleman R, Carey VJ, Huber W, Irizarry RA, Dudoit S, editors. New York: Springer; 2005. pp. 229–248. [Google Scholar]
- 66.Kumari S, Verma LK, Weller JW. AffyMAPSDetector: a software tool to characterize Affymetrix GeneChip expression arrays with respect to SNPs. BMC Bioinformatics. 2007;8:276. doi: 10.1186/1471-2105-8-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Doss S, Schadt EE, Drake TA, Lusis AJ. Cis-acting expression quantitative trait loci in mice. Genome Res. 2005;15:681–691. doi: 10.1101/gr.3216905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huang GJ, Shifman S, Valdar W, Johannesson M, Yalcin B, Taylor MS, Taylor JM, Mott R, Flint J. High resolution mapping of expression QTLs in heterogeneous stock mice in multiple tissues. Genome Res. 2009;19:1133–1140. doi: 10.1101/gr.088120.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stranger BE, Forrest MS, Clark AG, Minichiello MJ, Deutsch S, Lyle R, Hunt S, Kahl B, Antonarakis SE, Tavaré S, et al. Genome-wide associations of gene expression variation in humans. PLoS Genet. 2005;1:e78. doi: 10.1371/journal.pgen.0010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Muller FJ, Laurent LC, Kostka D, Ulitsky I, Williams R, Lu C, Park IH, Rao MS, Shamir R, Schwartz PH, et al. Regulatory networks define phenotypic classes of human stem cell lines. Nature. 2008;455:401–405. doi: 10.1038/nature07213. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.