Abstract
B-DNA flexibility, crucial for DNA–protein recognition, is sequence dependent. Free DNA in solution would in principle be the best reference state to uncover the relation between base sequences and their intrinsic flexibility; however, this has long been hampered by a lack of suitable experimental data. We investigated this relationship by compiling and analyzing a large dataset of NMR 31P chemical shifts in solution. These measurements reflect the BI ↔ BII equilibrium in DNA, intimately correlated to helicoidal descriptors of the curvature, winding and groove dimensions. Comparing the ten complementary DNA dinucleotide steps indicates that some steps are much more flexible than others. This malleability is primarily controlled at the dinucleotide level, modulated by the tetranucleotide environment. Our analyses provide an experimental scale called TRX that quantifies the intrinsic flexibility of the ten dinucleotide steps in terms of Twist, Roll, and X-disp (base pair displacement). Applying the TRX scale to DNA sequences optimized for nucleosome formation reveals a 10 base-pair periodic alternation of stiff and flexible regions. Thus, DNA flexibility captured by the TRX scale is relevant to nucleosome formation, suggesting that this scale may be of general interest to better understand protein-DNA recognition.
INTRODUCTION
The DNA genomic sequence of numerous organisms is now available. Nevertheless, this considerable amount of data does not by itself illuminate the genetic messages contained in these genomes. It is largely because these messages must be deciphered by numerous DNA-binding proteins such as transcription factors. The DNA also influences chromatin folding via its recruitment of histone octamers to form the nucleosomes. So, the cellular DNA is continuously ‘read’ by proteins. Specific and non-specific DNA–protein binding are known to depend on the conformational preferences of the nucleotidic sequences (1–10). Thus, understanding DNA–protein interactions requires knowledge of the intrinsic flexibility of DNA sequences.
Quantifying the sequence-dependence of DNA flexibility remains a major goal of molecular biology. The flexibility of DNA can be studied at various levels of details, from base-pair step properties to the long range behavior of the double-helix considered as a flexible rod. Most DNA–protein contacts engage 15–20 base pair (bp) DNA fragments (11), apart from cases such as the nucleosome where the interface covers 146–147 bp. Thus, dinucleotide or tetranucleotide sequences provide an appropriate (and ultimately required) resolution to address intrinsic DNA flexibility as a determinant of DNA/protein interaction. One approach to quantify the dinucleotide/tetranucleotide flexibility is to observe their conformational spread on populations large enough for meaningful statistics. Assembling such a dataset is not trivial.
A range of experimental methods has been used to capture DNA deformability (12). Analyses of the variability in X-ray structures of B-DNA devoid of proteins contributed to characterize the conformational properties of the ten dinucleotides in such context. This approach has well-known limitations, notoriously the crystal packing forces which impact the details of DNA structures (13–16) and the effective temperature of the X-ray statistical ensemble (12). A large set of crystallized DNA–protein complexes was also analyzed (5), assuming that deformations in bound DNA are likely to reflect its intrinsic structural properties, and that protein-bound DNA may be less subjected to crystal packing than free DNA. The dataset was deemed large enough to average the DNA distortions imparted by the proteins. Extracting the fluctuations and correlations of bound DNA, structural parameters yielded a complete set of sequence-dependent empirical energy functions, providing a quantification of the dinucleotide step malleabilities, aimed at predicting the energetic cost of DNA deformations upon protein binding. The ranking of step malleability depended on the helicoidal parameter considered, but YpR appeared the most easily deformed steps. This approach was applied to the question of DNA folding in chromatin, accounting quite well for nucleosome locations in six nucleosome-positioning sequences (17,18).
However, a comparison of free and bound X-ray DNA (19) showed that, in contrast with free DNA, all DNA/protein oligomers exhibited numerous north sugars and a large range of α/β/γ backbone conformations. In addition, the range of helical parameter values are larger in bound than in free DNA (5). Yet, the couplings between various helical parameters (5) or between helical parameters and BI/BII backbone states (19) were qualitatively retrieved in free and bound DNA ensembles. Thus, free and bound X-ray DNA structures obey the same overall intrinsic mechanics. This reinforces the idea that proteins exploit the free DNA dynamical properties upon binding. At the same time, proteins are able to extend the DNA energy landscape, probably altering some conformational energetic barriers, in particular in the DNA backbone (20). It is thus unclear to which extent the sequence-dependent flexibility of free B-DNA in solution can be extrapolated in practice from the distortions observed in X-ray bound DNA structures.
Two recent reports analyzed datasets mixing X-ray and NMR DNA structures (21,22), but the accuracy of DNA NMR structures is still an issue. NMR provides experimental restraints which are combined with molecular simulations. With unlabeled DNA, the low density of protons, together with relatively frequent overlaps in the NOE peaks, limit the number of measured distances. Hence, the derived structures may depend on the refinement protocols and the force-fields (23). Developments requiring labeled DNA (24–27) increase the number of observables, and improve the quality of the refined structures, although the interpretation of these additional data is not straightforward and may introduce specific biases (28). Approaches using numerous restraints combined with potentials of mean-force (29) or extensive MDs in explicit solvent (23) seem to provide another avenue to produce reliable structures of DNA free in solution, with a representation of their dynamics. Unfortunately, such accurate structures are too few to provide a general guide to DNA intrinsic flexibility.
Computer modeling methods have frequently been used to study DNA deformability, based on simple potential energy calculations or molecular dynamics (MD) in explicit solvent (1,30), reviewed in (12). In particular, the elastic properties of the ten complementary dinucleotides were evaluated from MD trajectories (30–35). Studies in the same vein were performed with a variety of MD protocols: (i) with different force-fields [AMBER parm94: (31,32,35,36); parm99: (1,33,34); parmbsc0 and CHARMM27: (30)], (ii) unconstrained (1,30–33,35,36) or under stress (34), and (iii) considering the six helicoidal parameters [twist, roll, tilt, rise, shift and slide (31–33,36)] or focusing on specific parameters [twist: (34); roll: (35)]. These studies reached convergent conclusions in identifying three distinct groups of steps in terms of purine (R)/pyrimidine (Y) classification: the flexible YpR, intermediate RpR•YpY, and stiff RpY steps. However, force-field limitations still exist (23,37–40). The comparison of NMR observables to their MD counterparts has unveiled shortcomings in the representation of salient details of B-DNA structure and dynamics by the commonly used DNA force-fields (23). It showed that the best current force-fields do not reproduce the modulation of DNA dynamics by its sequence, although they implement much of the intrinsic mechanical couplings within B-DNA. Therefore, the characterization of DNA sequence-dependent intrinsic flexibility still requires experimental input.
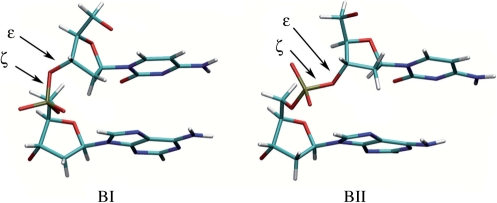
Here, we approach the intrinsic flexibility of DNA by probing its backbone properties in solution, in absence of proteins. The phosphate groups in B-DNA can adopt two conformations, the BI and BII states (Figure 1). They differ in the torsion angles ε and ζ which are trans/g- in BI (ε−ζ centered at −90°) and g-/trans in BII (ε − ζ centered at +90°). These two states, initially identified from crystallographic studies (41), were then detected in NMR by measurements of 3JH3′-P spin-spin coupling constants and/or 31P chemical shifts (δP) (42,43). Backbone and deoxyribose conformations are correlated (19,44,45). Most importantly, BI and BII conformations are also mechanically coupled to the DNA helicoidal parameters of roll, twist and X-disp, related to curvature, winding and groove dimensions, respectively (19,23,46–52). Furthermore, the propensity to undergo the BI/BII transition is sequence dependent (19,48,53). A method was recently devised to quantify for the first time the BI and BII populations in solution, from the very sensitive and accurate measurements of δP (48). Indeed, NMR studies highlighted strong linear correlations between δP and the three measured internucleotide distances H2′i−H6/8i + 1, H2″i−H6/8i + 1 and H6/8i−H6/8i+1 (48,54). These correlations, also present in very high resolution X-ray structures where δP can be substituted by (ε−ζ) values, allowed to establish a quantitative relation between δP and BI/BII ratios. They also confirm that the BI/BII ratio is associated to inter base pair parameters. The couplings between the BI↔BII equilibrium, deoxyribose conformational exchange (44) and helicoidal parameters (48) establish that the BI↔BII motions reflect the overall B-DNA structure and dynamics in solution.
Figure 1.
Illustration of the BI (left) and BII (right) phosphate linkage conformations with a CpA dinucleotide. BI and BII backbone conformations differ in the torsion angles ε and ζ which are respectively trans/g- in BI (ε − ζ = −90°) and g-/trans in BII (ε − ζ = +90°).
In the present work, 328 δP values previously reported for various B-DNA sequences in solution were collected. The influence of the dinucleotide and tetranucleotide sequence context on δP was analyzed. It provides evidence that DNA flexibility of a given step is primarily dominated by the corresponding dinucleotidic sequence, although secondary sequence effects beyond this immediate environment are observed on several steps. This forms the basis of a new and general experimental scale that characterizes the intrinsic properties of all 10 DNA complementary dinucleotides in terms of Twist, Roll and X-disp parameters, and is thus called the TRX scale.
Applying this new concept to sequences identified to preferentially bind to histone octamers and form nucleosomes (55–57) illustrates how the TRX scale provides new insights into the structural organization of DNA and its biological importance.
MATERIALS AND METHODS
BI/BII propensities in solution, from NMR
We previously (48) explained how 31P NMR chemical shifts could be converted to BII percentages by the equation BII(%) = 143 δP + 85 for δP referenced to phosphoric acid, or BII(%) = 143 δP + 621 for δP referenced to trimethylphosphate. This conversion is based on the linear cross-correlations observed between the three H2′i − H6/8i + 1, H2″i − H6/8i + 1, and H6/8i − H6/8i + 1 sequential distances and δP in NMR or ε − ζ values in X-ray structures. Both δP and ε − ζ reflect the interconversion between the backbone BI (ε − ζ < 0°) and BII (ε − ζ > 0°) states. The same method was used to compile the BII percentages per dinucleotide presented here. These statistics were gathered from a total of 328 31P chemical shifts reported in the literature for B-DNA without mismatches, chemical modifications or ligands (see Supplementary Table S1). The BII propensities per complementary dinucleotides (NipNi + 1•NjpNj + 1) are the half-sum of the BII percentages for the two corresponding phosphates. BI propensities are equal to 100-(BII propensities).
Crystallographic datasets
The X-ray structures of free B-DNA include the 37 DNA oligomers (2 octamers, 23 decamers, 11 dodecamers, 1 oligomer with 15 bp) of 2.0 Å resolution or better, the same dataset used in a previous analysis (19). These structures are all double-stranded DNAs, and do not include mismatches or modified bases, sugars or backbones.
Numerous nucleosome core particle (NCP) X-ray structures are available, but, to benefit from reliable structural details, we examined only the structures with resolution ≤2.5 Å, crystallized without histone mutants or groove ligands [PDB codes: 1EQZ (58), 1KX5 and 1KX3 (59), 1M19 (60), 2CV5 (61)]. 1EQZ and 1KX3 structures are quasi identical (RMSD ∼0.8 Å); the significance of the 2CV5 DNA fine structure appeared problematic, due to tight DNA–DNA contacts within the crystal that notably altered the DNA conformation (61). Therefore, the NCP DNA structures analyzed here are those from entries 1KX3, 1M19 and 1KX5.
Nucleosomal sequences
The sequences used in the ‘Implications for DNA recognition by proteins’ section came from Dr. Segal’s web site http://genie.weizmann.ac.il/pubs/nucleosomes06/index.html. The sequences are given in Supplementary Table S4.
Structure analysis
Analyses of DNA structures were carried out using CURVES (62), which calculates the optimal helical axis and a complete set of conformational and helicoidal parameters. Analyses were made in terms of both local and global parameters; however, since both analyses give quasi-identical values, only the global values are reported here. X-disp values are given for base pairs, i.e. for strand 1 with strand 2.
The phosphate conformations were analyzed in terms of BI and BII states, defined by the values of the two torsion angles ε (C4′–C3′–O3′–P) and ζ (C3′–O3′–P–O5′), with BI corresponding to ε/ζ = t/g- (ε − ζ < 0° and centered around −90°), and BII corresponding to ε/ζ = g-/t (ε − ζ > 0° and centered on +90°).
Fourier analysis
Values from the TRX scale for all the dinucleotidic steps along the DNA sequences were taken as input signal. Subtracting the average value <trx> = (trx(1) + … + trx(N))/N gives a centered signal b(n) = trx(n) – <trx>. The Fourier series is the sum, over the N steps of the segment, of plane waves
The function f is periodic of period 1; as such, it is completely specified by its values in the unit interval [0,1], best seen as a unit circle. All our Fourier analyses (Figure 6 and Table 2) represent the Fourier intensity or power spectrum |f(k)|2.
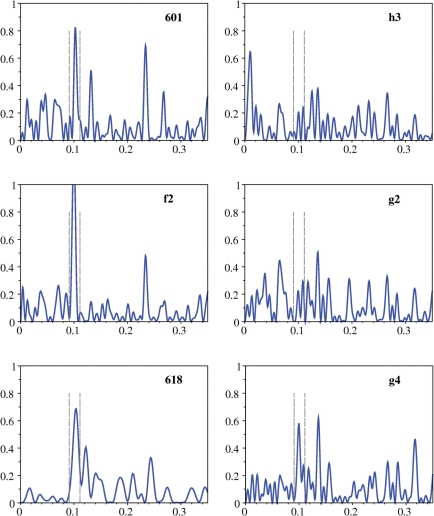
Figure 6.
Fourier analysis of the TRX signal along DNA nucleosomal sequences. Fourier transform (intensity in arbitrary units) as a function of wave vector k (bp−1) of the TRX signal along sequences representative of high (601 and f2), moderate (618) and low (h3, g2 and g4) propensity for forming nucleosome (Table 2). The Fourier analysis was performed considering the TRX scores (Table 1) as a signal along the innermost 140 dinucleotides of the nucleosomal sequences, apart from sequence 618, analyzed on the 72 central dinucleotides. The dotted vertical lines indicate the window corresponding to the period 10 ± 1 bp.
Table 2.
Nucleosomal sequences characterized according to the TRX scale
| sequences | %s | 140 dinucleotide steps |
72 dinucleotide steps |
ΔΔG | RA | RA | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| POW10 | ξs | ξf | POW10 | ξs | ξf | |||||
| 601 | 46 | 0.0065 | 2.2 | 2.6 | 0.0064 | 2.4 | 2.8 | −2.9 | 1 | |
| 618 | 51 | 0.0054 | 1.9 | 2.0 | 0.0086 | 1.7 | 1.9 | −2.3 | ||
| f2 | 58 | 0.0094 | 2.9 | 2.1 | 0.0102 | 2.5 | 2.3 | 2.47 | ||
| f3 | 55 | 0.0065 | 2.4 | 2.3 | 0.0102 | 2.5 | 2.3 | 1.60 | ||
| h2 | 47 | 0.0035 | 2.0 | 2.3 | 0.0029 | 2.4 | 2.1 | 0.11 | ||
| h3 | 58 | 0.0021 | 2.7 | 2.0 | 0.0001 | 4.1 | 1.5 | 0.15 | ||
| g1 | 45 | 0.0076 | 1.8 | 2.5 | 0.0082 | 2.0 | 2.6 | 1 | ||
| g2 | 39 | 0.0025 | 1.8 | 3.2 | 0.0007 | 2.1 | 4.4 | 0.16 | ||
| g3 | 40 | 0.0031 | 1.6 | 2.7 | 0.0027 | 1.5 | 2.8 | 0.15 | ||
| g4 | 36 | 0.0054 | 1.5 | 2.7 | 0.0047 | 1.4 | 2.8 | 0.14 | ||
| g5 | 37 | 0.0020 | 1.4 | 3.1 | 0.0005 | 1.1 | 3.7 | 0.12 | ||
The 147 bp sequences (Supplementary Table S4), their nomenclature and the relative affinities (RA) are from Dr Segal’s web site http://genie.weizmann.ac.il/pubs/nucleosomes06/index.html. The relative affinities are referenced to either 601 (values in columns 9 and 10) or g1 (values in last column) sequences. The central regions of f2 and f3 are identical. The ΔΔG (kcal/mol) values are referenced to that measured for the 5S sequence (55). The percentage of stiff complementary dinucleotides (%s) is given for each sequence. FT analyses were performed considering the TRX scores as a signal along either the 140 innermost or the 72 central dinucleotide steps of the DNA sequences. POW10 is the integrated power of the FT peak at 0.1 ± 0.01 frequency (periodicity of 10 ± 1 bp). The characteristic lengths were calculated for the stiff (ξs) and flexible (ξf) blocks defined by their TRX scores (Table 1).
Correlation length
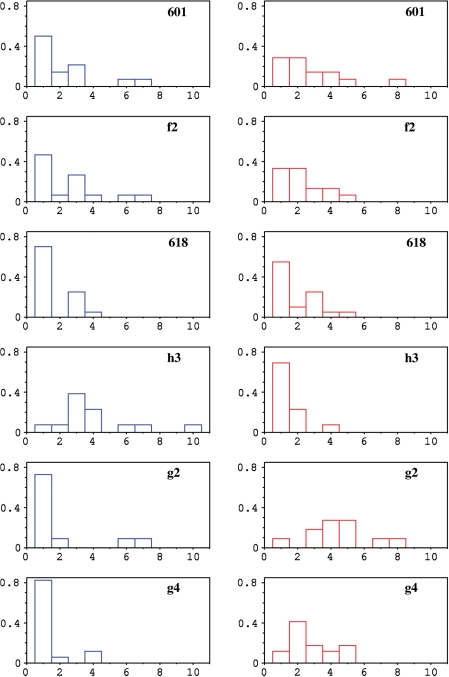
The Fourier intensity at period ∼10 is not enough to match the relative affinities for nucleosome formation. Some coherent flexibility/stiffness is also required. The dinucleotides can be classified into two categories delimited by the average TRX score of 21 (Table 1): those, flexible, with a significant propensity to adopt BII conformation (TRX score > 21, coded as +1) and those, stiffer, predominantly confined to BI (TRX score < 21, code −1). Then the DNA sequence is naturally subdivided into blocks of consecutive equal values, the flexible blocks of consecutive +1 and the stiff blocks of consecutive −1. The mean length of these blocks, readily computed from histograms of the type depicted in Figure 7, defines a characteristic length, one for each category: the flex ξf and the stiff ξs characteristic lengths. The overall average is the correlation length ξ = (ξf + ξs)/2.
Table 1.
Influence of DNA base sequence on the BII percentages in free DNA in solution
| N | %BII | TRX score | |
|---|---|---|---|
| CpG•CpG | 25 | 43•43 | 43 |
| CpA•TpG | 28 | 52•31 | 42 |
| GpG•CpC | 11 | 47•37 | 42 |
| GpC•GpC | 22 | 25•25 | 25 |
| GpA•TpC | 25 | 33•11 | 22 |
| TpA•TpA | 12 | 14•14 | 14 |
| ApG•CpT | 19 | 18•0 | 9 |
| ApA•TpT | 17 | 11•0 | 5 |
| ApC•GpT | 23 | 8•0 | 4 |
| ApT•ApT | 22 | 0•0 | 0 |
The DNA sequence is expressed in terms of the 10 complementary dinucleotides, of frequency N in the NMR data collected from the literature. The BII percentages (%BII) are given for each partner in a complementary dinucleotide. The average standard deviation of %BII is ±8. The TRX scores are the half-sums of the %BII observed for two facing phosphates in a complementary dinucleotide. The %BII and TRX scores higher (in bold) and lower than average (which is 21) correspond to enhanced and restricted flexibilities, respectively. A maximal flexibility on this scale corresponds to a TRX score of 50.
Figure 7.
Histograms of the length (expressed in number of dinucleotides) of stiff (left panels, in blue) and flexible (right panels, in red) blocks in the central 72 dinucleotides of the nucleosomal DNA sequences. The sequences presented here are the same as in Figure 6, with nucleosomal affinity ranging from very high (f2) to low (h3, g2, g4).
RESULTS
BI/BII dinucleotide sequence effect in B-DNA
The phosphate linkages in B-DNA adopt both the BI (ε − ζ < 0) and BII (ε − ζ > 0) states (Figure 1). 31P chemical shifts (δP), very accurately measured in solution by NMR, reflect the BI↔BII equilibrium (48,63). We collected from the literature a large set of 328 δP values previously reported for various B-DNA sequences in solution, excluding the phosphate linkages submitted to end effects at the sequence termini (Supplementary Table S1). Although monovalent counterions can modulate the BI/BII equilibrium (54), these subtle effects are expected to be averaged in our data-set, the δP being measured in variations of the physiological conditions.
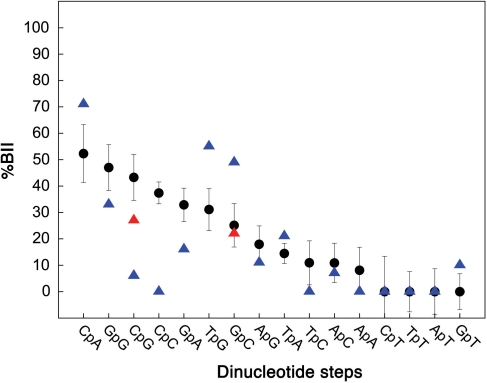
Each of the 16 B-DNA dinucleotides is characterized by a specific δP average value (Supplementary Figure S1), with small standard deviations (0.06 ppm on average). This influence of dinucleotide sequences on the BI↔BII equilibrium is presented in Figure 2, translating the δP from our data set to BII percentages with an established method (48) (see ‘Materials and Methods’ section). This analysis shows that (i) the averaged BII percentage in solution is 21% and (ii) 7 out of the 16 dinucleotides exhibit BII populations higher than average. These steps can thus be regarded as BII-rich steps. The maximal flexibility is observed for the phosphates of CpG, CpA and GpG, with a BI/BII ratio close to 1 (50% BI, 50% BII).
Figure 2.
Influence of the dinucleotide sequences on BII percentages in B-DNA. BII percentages (%BII) of free B-DNA inferred from δP in solution (black circles, with vertical bars for standard deviations) compiled from data published in the literature (see Supplementary Table S1). The conversion of δP in terms of BII percentage, using a published procedure (48), is detailed in ‘Materials and Methods’ section. BII percentages in solution are compared with those extracted from X-ray structures (triangles; blue: all structures, red: decamers only).
Considering the BII propensities of the 10 possible complementary dinucleotide sequences NipNi + 1•NjpNj + 1 (Table 1; N: any base) highlights that the facing phosphates tend to exhibit similar behaviors overall. In a first group containing ApN•NpT and TpA•TpA, the phosphates are very rarely in BII. This group, mainly confined in BI•BI, is thus characterized by a restricted backbone flexibility. In another group of dinucleotide steps (GpG•CpC, CpG•CpG, GpC•GpC and CpA•TpG) the facing phosphates can adopt BI and BII conformations, with a higher-than-average BII percentage. This family exhibits an enhanced backbone flexibility, potentially able to explore the three possible combinations, BI•BI, BI•BII (and its counterpart form BII•BI) or BII•BII. GpA•TpC is the only case where the two facing phosphates exhibit behaviors in marked contrast, with GpA able to significantly populate BII, but not TpC.
The BII percentages in solution can be compared with those extracted from crystal structures of free B-DNA (Figure 2). Considering the 16 dinucleotides, the backbone of ApA, (A/T)pC and NpT is overwhelmingly in BI in both environments. In contrast, the solution and crystal states often differ for steps with a significant propensity to populate BII. These propensities are either lower (CpG, CpC, GpG, GpA) or higher (CpA, TpG, GpC) in crystals than in solution. In X-ray structures, the behavior of the phosphates in GpG•CpC remains ambiguous since these steps are underrepresented (6 cases). Dinucleotides GpC and CpG most likely illustrate crystalline packing biases. Indeed, a large majority of GpC and CpG (34/44 and 51/69, respectively) are located at dodecamer ends, where adjacent molecules are in contact via their grooves (64). There are no such contacts in the decamers that are stacked end-to-end, where the GpC and CpG BII percentages are very close to those inferred from NMR data (Figure 2). The remaining apparent discrepancy concerns CpA•TpG, due to a tetranucleotide sequence bias in the crystal structures, explained in the next section.
In addition, the NMR BII propensities do not fit with those from the protein-bound DNA X-ray structures. Actually, a previous study (19) comparing high resolution X-ray structures of free and protein-bound DNA showed that, although the global DNA intrinsic mechanics is preserved (for instance the coupling between several helical parameters and the backbone states), the BII populations are globally reduced in protein-bound DNA.
For free B-DNA, taking the solid state biases into account allows to sufficiently reconciliate sequence effects in solution with their crystal counterpart. The fact that the BII propensities follow the same trends overall in both environments is a very strong indication that these trends represent intrinsic properties of the DNA molecule.
BI/BII tetranucleotide sequence effect in B-DNA
Experimental (65–68) and theoretical (21,33,53,69) results indicated that the behavior of some dinucleotides in B-DNA is influenced by their two bracketing nearest neighbors (flanking bases). The size of our NMR data set does not allow the analysis of all the 136 tetrameric combinations. A reasonable simplification is to consider instead the 3′- and 5′-contexts of the dinucleotides in terms of purine R or pyrimidine Y (Y–Y, R–R, Y–R, R–Y). In our NMR data set, these four categories are significantly populated for most dinucleotides (Supplementary Table S2).
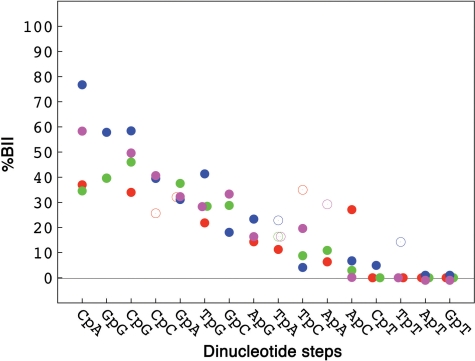
The flanking bases do influence the BI ↔ BII equilibrium of the central phosphate in most BII rich dinucleotides (Figure 3). Indeed, a Y–R context reinforces the BII populations in CpA, GpG, CpG, TpG. The BI-rich steps, together with GpA, appear mainly indifferent to their flanking bases (Figure 3). Yet, in this category, it seems possible to obtain substantial BII populations in YApAR, RTpCY and RApCY (Figure 3), but YApAY and RTpCY are represented by only two cases, and the four δP collected for RApCY are especially scattered (from −0.39 to −0.19 ppm).
Figure 3.
Influence of the tetranucleotide sequence on the BII percentages of the central phosphate group. BII percentages (%BII), compiled and translated from δP values published in the literature (see ‘Materials and Methods’ section and Supplementary Table S1), are given for the dinucleotide steps in four different tetranucleotide contexts: R–R in green, R–Y in red, Y–R in blue and Y–Y in magenta. Categories that contain more or less than three cases are represented by filled and open circles, respectively.
Overall, the four possible sequence contexts are so unequally populated in crystals that it prevents a systematic analysis of the tetranucleotide effect in this environment. However, the tetrameric sequence effect can account for the differences observed in Figure 2 between the NMR and X-ray BII percentages of CpA•TpG and CpG. Indeed, 14 out of 21 CpA•TpG steps in the X-ray data belong to the Y(CpA/TpG)R pattern that enhances BII and thus leads to dinucleotide average BII percentages higher than those found in solution. Conversely, 13 out of 18 CpG•CpG in crystals are surrounded by R–Y•R–Y or R–R•Y–Y, reducing their apparent global BII propensity.
More work is needed to understand the tetranucleotide effects, probably under-estimated, although potentially crucial for DNA–protein recognition (23,68). For the present purpose, this secondary influence does not disrupt the flexibility rankings detected at the dinucleotide level. In sum, the BI/BII propensities of phosphate groups are primarily controlled at the dinucleotide level, finely modulated at the tetrad level.
Relationship between helical parameters and backbone states
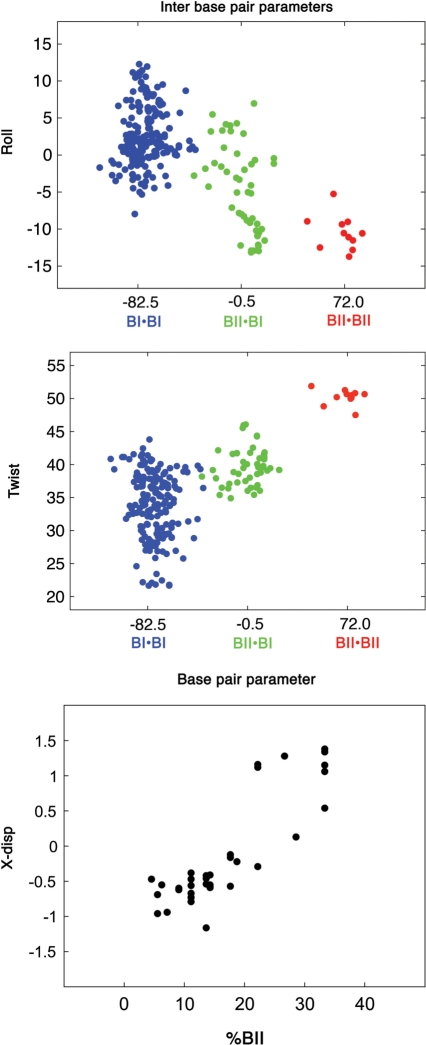
X-ray B-DNA structures are useful to demonstrate the general intrinsic mechanics of DNA. The principles underlying this mechanics do not depend on the sequence, and sound statistical analyses can be undertaken on a large set of high resolution oligomers. Such approaches have established that roll and twist values are correlated in both free and protein-bound DNA (5,46). Crucially, these inter-base descriptors depend on the conformations of the two phosphates facing one another across the strands (19,23,46) (Figure 4). BI•BI is associated with positive rolls (2.7 ± 4.1°), whereas BI•BII (−4.2 ± 5.1°) and especially BII•BII (−10.8.7 ± 2.1°) correspond to negative rolls. The twist values associated to BI•BI (33.9 ± 5.0°) are lower than those with BI•BII (39.4 ± 2.8°) or BII•BII (48.1 ± 4.1°). The only exception is for the semi-flexible GpA•TpC steps, their twist and roll appearing rather uncorrelated, as previously noticed (5). Phosphate conformational combinations are also correlated to the slide (Supplementary Figure S2) that becomes positive in BII•BII steps. This parameter is very strongly correlated to the roll (5,18). For instance, roll and slide variations work in concert to generate sharp bending in DNA/protein complexes (18). Due to this equivalence, we do not elaborate further on the slide in the following.
Figure 4.
Influence of BII phosphates on helical descriptors. Inter base pair parameters: Twist (°) and roll (°) plotted versus (ε − ζ)av, averaged on the two facing phosphates. BI•BI (blue, (ε − ζ)av = −82.5°), BI•BII (green, (ε − ζ)av = −0.5°) and BII•BII (red, (ε − ζ)av = +72°) are the three possible conformational combinations of facing phosphates. Base pair parameter: X-disp (Å), averaged along whole oligomer sequences plotted versus the BII percentage (%BII) per oligomer (number of BII phosphates/total number of phosphates). The data were extracted from high resolution X-ray structures of free DNA (19).
BII phosphates also act on groove dimensions, as previously shown by solid state NMR studies on fibrous DNA (51) and by analyzing X-ray data (19,46). Indeed, they displace base pairs towards the major groove, influencing the X-disp parameter. X-disp is defined for one base pair, positive and negative values reflecting displacement towards the major and minor grooves, respectively. Analysis of X-ray structures detects no more than two BII among the four phosphate linkages pIp•pJp surrounding one base pair I•J. This is consistent with previous X-ray analysis (19,46) and with a NMR study of S2 order parameters that highlighted that a BII rich phosphate precludes high BII populations in its 5′ and 3′ neighbors (44). Examining the X-disp values in the present X-ray data set yields X-disp = −0.6 ± 0.4, −0.2 ± 0.5 and +0.85 ± 0.7 Å when zero, one and two BII linkages surround the central base pair I•J. Several proximal (but not successive) BII phosphates enhance this effect, preserving an optimal base stacking (47,53). So, the average X-disp is ≥1.0 Å for deca- and dodecamers in which more than 22% of phosphates are simultaneously in BII (for instance, 22% correspond to 4 BII on the 18 phosphates encountered in a decamer) (19,23) (Figure 4). Such cumulative shifts in X-disp values mediate the influence of BII rich regions on the major groove overall shape, making it convex (depth of −1.0 ± 0.5 Å) instead of concave (usual depth being +5 ± 2 Å).
The helical parameters are not directly accessible by NMR, but their mechanical couplings are revealed through linear correlations between the δP values and the three sequential internucleotide distances H2′i−H6/8i + 1, H2″i−H6/8i + 1 and H6/8i−H6/8i + 1 (48,54), related to the twist and roll parameters (49). Also, the relationship between the BI/BII conformations and the base pair displacements was confirmed by NMR on fibrous DNA (51). In addition, these couplings were retrieved by modeling of DNA in explicit solvent (46,52), and in simulations under NMR constraints (23).
So, different facing backbone BI/BII conformations define sub-states in B-DNA, influencing both the local (e.g. roll and twist) and neighboring structure (e.g. X-disp) of the double helix. In particular, strong negative roll and very high twist appear distinctive of synchronous facing BII phosphates.
The TRX scale
The strong correlations between BI ↔ BII phosphate populations and helical parameters mean that the BII propensities characterizing each of the ten complementary dinucleotides reflect the intrinsic flexibility of the corresponding twist, roll and X-disp. Consequently, the scale of BII propensities at the dinucleotide level (Table 1) is a realistic and general representation of the sequence dependency of DNA intrinsic plasticity. Considering the above-mentioned similar average behavior of facing phosphates, this flexibility can be quantified by the half-sum of the BII propensities of the two facing phosphates in a complementary dinucleotide (right-most column in Table 1). This constitutes an experimental scale of local intrinsic flexibility, called ‘TRX’ in reference to the Twist, Roll and X-disp parameters that it represents.
The steps with a low TRX score (low BII propensity) have a restricted flexibility: low or moderate twists associated with null or positive rolls, respectively; those bases tend to be minimally displaced away from the axis of the double helix. The steps associated with higher than average TRX score (>21) are characterized by plurimodal conformational populations. They can explore a larger conformational space, from low twists/positive rolls (‘BI state’) to high twists/negative rolls (‘BII state’); the movement of these bases towards the major groove is facilitated. High plasticities are observed for steps CpA•TpG, CpG•CpG, GpG•CpC, with TRX scores of 42–43. These values are close to 50, the maximal flexibility in the context of the TRX scale (50% of the time in BI, 50% of the time in BII). Thus, the value of the TRX score has a physical meaning, directly related to the BII populations in free B-DNA in aqueous solution. These populations are a signature of the intrinsic mechanical flexibility of the associated base steps.
The examination of the variability of inter-base helical parameters in the X-ray datasets of free DNA [our dataset and another reported on http://rutchem.rutgers.edu/∼olson/pdna.html (5)] globally agree with the TRX scale. The only serious discrepancy is the rank of step TpA. According to the X-ray data, this step is more flexible than GpG•CpC, CpG•CpG and GpG•CpC. Conversely, the δP based observations classify TpA in the ‘stiff’ category while GpG•CpC, CpG•CpG and GpG•CpC are ranked flexible (Table 1). But in the X-ray dataset GpG•CpC are poorly represented (less than 2% of the dinucleotide steps) and GpC•GpC and CpG•CpG are subjected to intermolecular contacts, as explained above. Another source of discrepancy could come from the sugar north↔south exchange, coupled to the BI↔BII equilibrium (44), but possibly underestimated in solid phase, in particular on cytidine residues (27). So, the TpA flexibility rank deduced from X-ray structures could be biased by an incorrect estimation of the properties of GpG•CpC, CpG•CpG and GpG•CpC steps.
The TRX scale promises to be helpful for deciphering the intrinsic flexibility of free DNA underpining DNA–protein interactions. Indeed, intrinsic structural preferences of free DNA, and its possible pre-organization, are expected to limit the energetic and entropic penalties upon protein binding.
Implications for DNA recognition by proteins
To demonstrate the concrete use of the TRX scale, we illustrate how it can be applied to DNA sequences bound preferentially by the histone core. The nucleosome core particle (NCP), consisting of 146–147 bp of DNA wound twice around an octameric core of four histone proteins, is the fundamental building block of packaged DNA in eukaryotic cells. Earlier studies on chicken nucleosomal DNA established that some DNA sequences favored the NCP formation (70,71). Sequences having an exceptionally high affinity for histone octamers were then identified using the SELEX technique (55–57), leading to identify a consensus for the central 73 bp (56). Using large pools of natural sequences, the nucleosome intrinsic sequence preferences were recently characterized primarily by ∼10 bp periodicities of specific dinucleotides along the nucleosome length, thought to facilitate the sharp bending of DNA around the nucleosome (56,57,71–78). However, in absence of reliable descriptors of dinucleotide properties, the physical basis of this organization remains obscure.
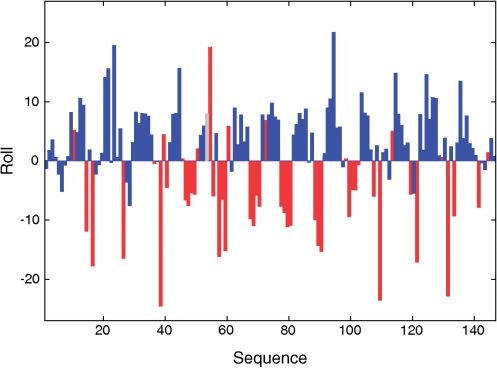
The NCP X-ray structures revealed a periodic alternation of positive and negative roll tracts along the double helix resulting in the DNA superhelical path (79). Our analysis of several high resolution NCP X-ray structures [PDB codes 1KX3 (59), 1M19 (60) and 1KX5 (59)] shows that positive and negative rolls are mainly associated to BI and BII phosphates, respectively (Figure 5 for 1KX5). Some steps escape this rule, as illustrated by steps 53 and 54 in 1KX5 (Figure 5). Step 53 is associated with an atypical (ε − ζ) value of ∼170°, and in step 54 BI•BII is associated to a strong positive roll. Such peculiar behaviors, together with unusual α/β/γ angles, help to remember that the backbone conformational landscape of bound DNA is not exactly as in free DNA (19). Nevertheless, rolls and (ε − ζ) averaged on facing phosphates are as well correlated in 1KX5, 1KX3 and 1M19 (correlation coefficients from −0.72 to −0.65) as in free X-ray DNA (correlation coefficient of −0.67). Thus, the relationship between BI/BII and roll appears robust, as well as those involving twist and X-disp (not shown). So, BI and BII rich regions are the centers of 10 bp fragments curved towards the major and the minor grooves, respectively. Furthermore, the NCP BII steps are in majority encountered on CpA•TpG, GpG•CpC and GpC•GpC (Supplementary Table S3), consistent with the TRX scale. That both BII structural and sequence effects in free DNA are transferable to the NCP structures makes this system particularly relevant to analyze how DNA intrinsic flexibility, quantified with the TRX scale, influences its assembly with proteins.
Figure 5.
Relationship between roll and backbone conformations in NCP X-ray structures. Roll (°) profile along the DNA sequence in nucleosome X-ray structure 1KX5. The bars giving the roll values are colored according to the state of the facing phosphates in the complementary dinucleotide steps: blue for BI•BI and red for BI•BII or BII•BII. The grey bar represents an unclassifiable step (BI on strand 1 and (ε − ζ) ∼ 170°, i.e. neither BI nor BII, on strand 2). Structures 1KX3 and 1M19 give similar patterns.
We analyze here the structural properties of synthetic sequences related to the SELEX approach for which affinity measurements are available (Table 2 and Supplementary Table S4). Artificial and natural sequences exhibit the same lexicographic periodicities: ApA•TpT, TpA and ApT oscillate out of phase with GpG•CpC, CpG•CpG and GpG•CpC, with a period of ∼10 bp (56). The related SELEX pool contains very high affinity sequences that do not have their equivalent in genomic positioning DNA. We wondered if the characteristics of these very high affinity sequences would emerge particularly clearly from a TRX analysis and contrast with those of lower affinity sequences. So, we first considered the SELEX sequence best at forming NCP (sequence 601), and one of the worst (sequence 618) (57). These sequences 601 and 618 present a weak similarity, with ∼30% of identical bases. A set of sequences deriving from sequence 601 were also analyzed (http://genie.weizmann.ac.il/pubs/nucleosomes06/index.html). f2 and f3 correspond to sequence 601 enriched in 10 bp periodic TpA steps either along the whole sequence (f2) or only in its central part (f3). Thus the central regions of f2 and f3 are identical; their affinities for the histone core are higher than that of sequence 601. h2–3 and g1–5 differ from sequence 601 by ∼50 mutated bases between positions 31 and 122, i.e. in the central region of the 147 bp segment. Apart from g1, these sequences display low propensities to form nucleosomes. Overall, a representative range of affinities is covered by these 11 sequences.
Considering the TRX scores as signals along the DNA sequence, Fourier transform (FT) analyses were performed on i) the 140 dinucleotides (141 bp) corresponding to the DNA length in an NCP less its outermost base pairs and ii) the central 72 dinucleotides (73 bp), given that most of the considered sequences differ from the 601 reference by mutations in this region. Regarding the NCP, the most interesting spectrum section covers the frequencies from 0.09 to 0.11, corresponding to periodicities of ∼11 and 9 bp, respectively. Indeed, the NCP sequence periodicities inferred from various biochemical experiments vary within this range [(80) and references herein]. The direct estimates of periodicity offered by high-resolution crystal structures of NCP reconstituted on a unique DNA sequence give periodicity values from 10.15 to 10.3 bp (59,79).
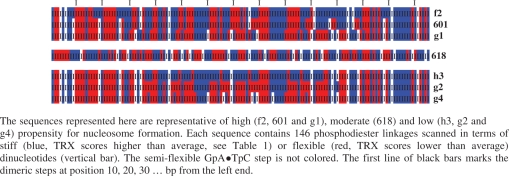
Sequences 601, f2, f3 and g1 are characterized by a strong FT signal corresponding to a ∼10 bp periodicity (see Figure 6 for 601 and f2). The corresponding powers, i.e. integrated the FT signal between frequencies 0.09 and 0.11, show maximal scores (Table 2) that are further strengthened in the central regions for f2, f3 and g1 (Table 2). This means that these preferential DNA sites for assembling into nucleosome are characterized by ∼10 bp alternations of blocks of enhanced and restricted flexibility. Considering that enhanced and restricted flexibilities correspond to TRX scores higher and lower than the average, respectively, we find that the average lengths of each type of block are globally equivalent (characteristic lengths in Table 2), especially in the central region (Table 2 and Figure 7). This organization, illustrated in Schema 1 for the sequences f2, 601 and g1, correlates the X-ray NCP structures in which the BI or BII tracts include at least three consecutive dinucleotides (Figure 5).
Scheme 1.
Nucleosomal sequences coded by the TRX flexibility scale.
Incidentally, considering TpA as flexible as CpA•TpG, as indicated by the X-ray data, results in a net decrease of all the ∼10 bp FT signals. For instance, the power of the 10 bp FT peak of f2 drops from 0.0094 (TpA stiff) to 0.003 (TpA flexible). When assuming a flexible TpA, the FT analysis detects a strong 15 bp periodicity on these high affinity sequences, not compatible with the NCP. This reinforces the notion developed above that the flexibility of TpA estimated from X-ray datasets could be inappropriate to DNA in solution.
Sequence 618, less attractive than 601 for the histone core, gives a less intense TRX-FT signal at frequency of 0.1 ± 0.01 when the entire sequence is taken into account, but the 10 bp periodicity is noticeably enhanced in its central part (Figure 6 and Table 2). Nevertheless, the block lengths are globally shorter than those of very high affinity sequences (Table 2, Figure 7 and Scheme 1), implying that the number of steps pushed at the inverse of their intrinsic structural trends in NCP is greater with 618 than with 601.
The ∼10 bp periodicity is even more subdued for the low affinity sequences h2, h3 and g2–5 (Figure 6 for h3, g2 and g4). FT peaks at 0.1 ± 0.01 are still observable but their intensities are very low (Table 2). In addition, the average lengths of stretches of restricted and enhanced flexibility are not balanced (Table 2). Either stiff (h3) or flexible (g2–5) dinucleotides are dominant in the central region of these low affinity sequences (Table 2, Figure 7 and Scheme 1 for h3, g2 and g4). Such configuration is likely to increase the energetic penalty in NCP formation, with a large deformation cost (excess of stiff dinucleotides) or a loss of entropy (excess of flexible dinucleotides). This idea is supported by the very poor affinity of sequence g4 that presents a significant 10 bp periodicity (Figure 6, Table 2) but with almost all its central blocks of restricted flexibility corresponding to only one dinucleotide (Figure 7, Table 2, Schema 1).
In sum, the TRX scale reveals that the preferential DNA sites for assembling into nucleosome are primarily characterized by a 10 bp periodicity that corresponds to an alternation of blocks characterized by restricted and enhanced flexibility. Our analysis also suggests that blocks of minimal length are required to define a high affinity sequence. The central 72 dinucleotides may be more crucial for the positioning than their flanking external parts since both the periodicity and the block length balance found for the best assembling sequences are especially marked in the central parts of the 147 bp fragments analyzed here.
DISCUSSION AND CONCLUSION
In B-DNA, any phosphate group is potentially able to oscillate between the BI and BII states, the BI/BII ratio reflecting the helicoidal twist, roll and X-disp values of the corresponding step (19,23,46–52). The BI/BII ratio is accessible by the solution NMR measurements of 31P chemical shifts (δP) (48).
The present work establishes firmly the dinucleotide sequence effect on the BI↔BII equilibrium and its populations, based on a new and extensive compilation of the available δP measured on free B-DNA in solution. The influence reflecting the category (purine or pyrimidine) of the nearest neighbors bracketing the dinucleotide is detected but this secondary effect does not disrupt the flexibility rankings detected at the dinucleotide level. Due to the predominant impact of the dinucleotide sequence, each complementary dinucleotide step can be characterized by a number corresponding to the experimental average BII population of its facing phosphates (Table 1) and thus the phosphate group flexibility. Crucially, this number also represents the malleability in terms of Twist, Roll and X-disp (base pair displacement), due to the tight coupling between helical parameters and backbone states. Thus, the numbers associated to the ten complementary dinucleotide steps result in a scale of intrinsic flexibility that we call ‘TRX’. The most inherently flexible and rigid steps correspond to TRX scores of 50 (50% BI/50% BII) and 0 (100% BI), respectively.
Broadly speaking, two dinucleotide categories emerge from the TRX scale. The first has restricted flexibility, i.e. low or moderate twists associated with positive and null rolls. Their corresponding bases are preferentially located close to the axis of the double helix. The second group exhibits an enhanced flexibility where the dinucleotides can adopt either low twists/positive rolls or high twists/negative rolls. Such bases are more easily displaced towards the major groove, away from the helical axis. According to the TRX scale, ApA•TpT, ApT•ApT, TpA•TpA have restricted flexibility. In contrast, GpG•CpC, GpC•GpC, CpG•CpG have enhanced flexibility. The steps which are mixed in terms of base composition have flexibilities either restricted (ApC•GpT and ApG•CpT) or enhanced (CpA•TpG), GpA•TpC being intermediate. Thus, the DNA dynamical properties are not easily assigned solely by reading the lexicographic nucleotide chaining.
Our approach of flexibility may go beyond the intrinsic plasticity of DNA in its B form, since the contrasted behavior of GpG•CpC, GpC•GpC, CpG•CpG on one hand, and ApA•TpT, ApT•ApT, TpA•TpA on the other hand, according to δP echoes the influence of sequences on double helix polymorphism. Indeed, contrary to A•T rich sequences, G•C and CpA•TpG rich DNAs can adopt the Z form (81–83) and facilitate the B to A transition (84–86). The A and Z forms correspond to large molecular reorganizations occurring generally in non-physiological environments, but their formation may take advantage of the dinucleotide properties highlighted here.
Most importantly, the TRX scale emerges as useful to understand how the DNA/protein interactions depend on DNA sequence-dependent intrinsic malleability. Here, the TRX scale was used to investigate the properties of nucleosomal positioning sequences. Indeed, it is established that the nucleosome reconstitutes better on certain DNA sequences than others (56,77,87). Our analysis of the high resolution NCP crystal structures reveals that the tight coupling between BI and BII backbone states and helical parameters, together with the marked sequence dependency of the BI/BII propensities, echoes our findings on free DNA. This supports the idea that intrinsic DNA properties reduce the cost of marked distortions required by wrapping in the NCP complex (56,77,87).
Our TRX analysis was performed on artificial sequences that offered a range from very high to low affinity for nucleosome reconstitution. We show that the high affinity sequences are characterized by a ∼10 bp periodicity, corresponding to a regular alternation of stiff (limited to positive and null rolls) and flexible stretches (able to adopt negative rolls). Thus, high affinity sequences consist of a succession of blocks phased to distort as required in the NCP structure. This periodicity is maximal when considering the central 73 bp. It appears also that, in these 73 bp central parts, a minimal length of both stiff and flexible blocks is required to obtain optimal nucleosomal sequences.
These findings are consistent with several observations. First, our analysis explains why sequences considerably enriched in A•T, unable to adopt negative rolls without conformational strain, function as nucleosome exclusion signals (70,71,88,89), likely due to a large deformation energy penalty. Also, it suggests that too much DNA flexibility (e.g. with a flexible dinucleotide at every step) may lower the DNA/protein affinity because of a loss of entropy when going from the flexible free form to the nucleosomal structure (90,91). Second, the importance of the sequence central part is consistent with (i) the consensus found in the central part of artificial nucleosomal sequences (56); (ii) the nucleosome reconstitution in vivo and in vitro that begins with the central (H3–H4)2 tetrasomal part, followed by incorporation of the two terminal (H2A–H2B) dimers (92,93) and (iii) a recent study showing that 73 bp fragments are efficient to analyze the nucleosome occupancy along the genomes of Saccharomyces cerevisiae and Drosophila melanogaster (94). Third, the TRX scale allows to rationalize the dinucleotide position frequencies found on both artificial (56,57,87) or natural nucleosomal DNA sequences in vitro (72,75–78) or in vivo (95). These studies converge to characterize the nucleosome sequence preferences primarily by a ∼10 bp periodicity of ApT, TpA and ApA•TpT that oscillate out of phase with GpC, CpG, GpG•CpC and CpA•TpG along the nucleosomal DNA constituent. Both groups are each composed by a mixture of YpR, RpR•YpY and RpY. Neither the modeling nor the crystallographic approaches do properly account for DNA plasticity since they all conclude to flexible YpR, intermediate RpR•YpY, and stiff RpY steps. From the TRX point of view, ApT, TpA and ApA•TpT are characterized by a restricted plasticity, contrary to the flexible GpC, CpG, GpG•CpC and CpA•TpG. Thus, the TRX approach clarifies the lexicographic periodicities encountered in nucleosomal sequences, artificial and natural.
In sum, examination of free DNA backbone motions in solution, monitored by δP, correlated to several helical parameters, allows to uncover the intrinsic B-DNA flexibility. This flexibility appears dominated by the dinucleotide sequence, although finely modulated at the tetranucleotide level. The quantification of the DNA intrinsic malleability of the ten complementary dinucleotides yields an experimental scale, called TRX (Table 1). We started to decipher the affinity of artificial DNA sequences for nucleosome formation in terms of their TRX signals. This shows that the TRX scale offers an understanding of the preferred recognition of DNA sequences by proteins, based on flexibility criteria.
A key strength of the TRX scale is its experimental foundation, directly inferred from very precise δP NMR measurements in solution, free from dependence on any structural models or limitations (e.g. force-field influences or crystal packing). To our knowledge, the TRX scale is currently the only experimental quantitative scale characterizing the ten B-DNA complementary dinucleotide steps in terms of average structure and flexibility, thus offering a conceptual and practical framework to make progress regarding the role of DNA dynamics in DNA–protein recognition mechanisms.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Institut National de la Transfusion Sanguine.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Arauzo-Bravo MJ, Fujii S, Kono H, Ahmad S, Sarai A. Sequence-dependent conformational energy of DNA derived from molecular dynamics simulations: toward understanding the indirect readout mechanism in protein-DNA recognition. J. Am. Chem. Soc. 2005;127:16074–16089. doi: 10.1021/ja053241l. [DOI] [PubMed] [Google Scholar]
- 2.Hegde RS. The papillomavirus E2 proteins: Structure, function, and biology. Annu. Rev. Biophys. Biomol. Struct. 2002;31:343–360. doi: 10.1146/annurev.biophys.31.100901.142129. [DOI] [PubMed] [Google Scholar]
- 3.Jayaram B, McConnell K, Dixit SB, Das A, Beveridge DL. Free-energy component analysis of 40 protein-DNA complexes: a consensus view on the thermodynamics of binding at the molecular level. J. Comput. Chem. 2002;23:1–14. doi: 10.1002/jcc.10009. [DOI] [PubMed] [Google Scholar]
- 4.Lavery R. Recognizing DNA. Quart. Rev. Biophys. 2005;38:339–344. doi: 10.1017/S0033583505004105. [DOI] [PubMed] [Google Scholar]
- 5.Olson WK, Gorin AA, Lu XJ, Hock LM, Zhurkin VB. DNA sequence-dependent deformability deduced from protein-DNA crystal complexes. Proc. Natl Acad. Sci. USA. 1998;95:11163–11168. doi: 10.1073/pnas.95.19.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rhodes D, Schwabe JW, Chapman L, Fairall L. Towards an understanding of protein-DNA recognition. Philos. Trans. Roy. Soc. London. 1996;351:501–509. doi: 10.1098/rstb.1996.0048. [DOI] [PubMed] [Google Scholar]
- 7.Sarai A, Kono H. PROTEIN-DNA recognition patterns and predictions. Annu. Rev. Biophys. Biomol. Struct. 2005;34:379–398. doi: 10.1146/annurev.biophys.34.040204.144537. [DOI] [PubMed] [Google Scholar]
- 8.Suck D. DNA recognition by structure-selective nucleases. Biopolymers. 1997;44:405–421. doi: 10.1002/(SICI)1097-0282(1997)44:4<405::AID-BIP5>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 9.Widom J. Role of DNA sequence in nucleosome stability and dynamics. Q. Rev. Biophys. 2001;34:269–324. doi: 10.1017/s0033583501003699. [DOI] [PubMed] [Google Scholar]
- 10.Zakrzewska K. DNA deformation energetics and protein binding. Biopolymers. 2003;70:414–423. doi: 10.1002/bip.10476. [DOI] [PubMed] [Google Scholar]
- 11.Luscombe NM, Austin SE, Berman HM, Thornton JM. An overview of the structures of protein-DNA complexes. Genome Biol. 2000;1:REVIEWS001. doi: 10.1186/gb-2000-1-1-reviews001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lankas F. DNA sequence-dependent deformability – insights from computer simulations. Biopolymers. 2004;73:327–339. doi: 10.1002/bip.10542. [DOI] [PubMed] [Google Scholar]
- 13.Hahn M, Heinemann U. DNA helix structure and refinement algorithm: comparison of models for d(CCAGGCm5CTGG) derived from NUCLSQ, TNT and X-PLOR. Acta Crystallogr. 1993;49:468–477. doi: 10.1107/S0907444993004858. [DOI] [PubMed] [Google Scholar]
- 14.Hartmann B, Lavery R. DNA structural forms. Q. Rev. Biophys. 1996;29:309–368. doi: 10.1017/s0033583500005874. [DOI] [PubMed] [Google Scholar]
- 15.Heinemann U, Alings C, Hahn M. Crystallographic studies of DNA helix structure. Biophys. Chem. 1994;50:157–167. doi: 10.1016/0301-4622(94)85028-3. [DOI] [PubMed] [Google Scholar]
- 16.Valls N, Wright G, Steiner RA, Murshudov GN, Subirana JA. DNA variability in five crystal structures of d(CGCAATTGCG) Acta Crystallogr. Sect. D Biol. Crystallogr. 2004;60:680–685. doi: 10.1107/S0907444904002896. [DOI] [PubMed] [Google Scholar]
- 17.Balasubramanian S, Xu F, Olson WK. DNA sequence-directed organization of chromatin: structure-based computational analysis of nucleosome-binding sequences. Biophys. J. 2009;96:2245–2260. doi: 10.1016/j.bpj.2008.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tolstorukov MY, Colasanti AV, McCandlish DM, Olson WK, Zhurkin VB. A novel roll-and-slide mechanism of DNA folding in chromatin: implications for nucleosome positioning. J. Mol. Biol. 2007;371:725–738. doi: 10.1016/j.jmb.2007.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Djuranovic D, Hartmann B. Conformational characteristics and correlations in crystal structures of nucleic acid oligonucleotides: evidence for sub-states. J. Biomol. Struct. Dyn. 2003;20:771–788. doi: 10.1080/07391102.2003.10506894. [DOI] [PubMed] [Google Scholar]
- 20.Varnai P, Djuranovic D, Lavery R, Hartmann B. Alpha/gamma transitions in the B-DNA backbone. Nucleic Acids Res. 2002;30:5398–5406. doi: 10.1093/nar/gkf680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bharanidharan D, Gautham N. Principal component analysis of DNA oligonucleotide structural data. Biochem. Biophys. Res. Commun. 2006;340:1229–1237. doi: 10.1016/j.bbrc.2005.12.127. [DOI] [PubMed] [Google Scholar]
- 22.Marilley M, Sanchez-Sevilla A, Rocca-Serra J. Fine mapping of inherent flexibility variation along DNA molecules: validation by atomic force microscopy (AFM) in buffer. Mol. Genet. Genomics. 2005;274:658–670. doi: 10.1007/s00438-005-0058-8. [DOI] [PubMed] [Google Scholar]
- 23.Heddi B, Foloppe N, Oguey C, Hartmann B. Importance of accurate DNA structures in solution: the Jun-Fos model. J. Mol. Biol. 2008;382:956–970. doi: 10.1016/j.jmb.2008.07.047. [DOI] [PubMed] [Google Scholar]
- 24.Getz M, Sun X, Casiano-Negroni A, Zhang Q, Al-Hashimi HM. NMR studies of RNA dynamics and structural plasticity using NMR residual dipolar couplings. Biopolymers. 2007;86:384–402. doi: 10.1002/bip.20765. [DOI] [PubMed] [Google Scholar]
- 25.Lipsitz RS, Tjandra N. Residual dipolar couplings in NMR structure analysis. Annu. Rev. Biophys. Biomol. Struct. 2004;33:387–413. doi: 10.1146/annurev.biophys.33.110502.140306. [DOI] [PubMed] [Google Scholar]
- 26.MacDonald D, Lu P. Residual dipolar couplings in nucleic acid structure determination. Curr. Opin. Struct. Biol. 2002;12:337–343. doi: 10.1016/s0959-440x(02)00328-7. [DOI] [PubMed] [Google Scholar]
- 27.Wu Z, Delaglio F, Tjandra N, Zhurkin VB, Bax A. Overall structure and sugar dynamics of a DNA dodecamer from homo- and heteronuclear dipolar couplings and 31P chemical shift anisotropy. J. Biomol. NMR. 2003;26:297–315. doi: 10.1023/a:1024047103398. [DOI] [PubMed] [Google Scholar]
- 28.Kuszewski J, Schwieters C, Clore GM. Improving the accuracy of NMR structures of DNA by means of a database potential of mean force describing base-base positional interactions. J. Am. Chem. Soc. 2001;123:3903–3918. doi: 10.1021/ja010033u. [DOI] [PubMed] [Google Scholar]
- 29.Schwieters CD, Clore GM. A physical picture of atomic motions within the Dickerson DNA dodecamer in solution derived from joint ensemble refinement against NMR and large-angle X-ray scattering data. Biochemistry. 2007;46:1152–1166. doi: 10.1021/bi061943x. [DOI] [PubMed] [Google Scholar]
- 30.Perez A, Lankas F, Luque FJ, Orozco M. Towards a molecular dynamics consensus view of B-DNA flexibility. Nucleic Acids Res. 2008;36:2379–2394. doi: 10.1093/nar/gkn082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beveridge DL, Barreiro G, Byun KS, Case DA, Cheatham TE, 3rd, Dixit SB, Giudice E, Lankas F, Lavery R, Maddocks JH, et al. Molecular dynamics simulations of the 136 unique tetranucleotide sequences of DNA oligonucleotides. I. Research design and results on d(CpG) steps. Biophys. J. 2004;87:3799–3813. doi: 10.1529/biophysj.104.045252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dixit SB, Beveridge DL, Case DA, Cheatham TE, 3rd, Giudice E, Lankas F, Lavery R, Maddocks JH, Osman R, Sklenar H, et al. Molecular dynamics simulations of the 136 unique tetranucleotide sequences of DNA oligonucleotides. II: sequence context effects on the dynamical structures of the 10 unique dinucleotide steps. Biophys. J. 2005;89:3721–3740. doi: 10.1529/biophysj.105.067397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fujii S, Kono H, Takenaka S, Go N, Sarai A. Sequence-dependent DNA deformability studied using molecular dynamics simulations. Nucleic Acids Res. 2007;35:6063–6074. doi: 10.1093/nar/gkm627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kannan S, Kohlhoff K, Zacharias M. B-DNA under stress: over- and untwisting of DNA during molecular dynamics simulations. Biophys. J. 2006;91:2956–2965. doi: 10.1529/biophysj.106.087163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lankas F, Sponer J, Langowski J, Cheatham TE., 3rd DNA basepair step deformability inferred from molecular dynamics simulations. Biophys. J. 2003;85:2872–2883. doi: 10.1016/S0006-3495(03)74710-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lankas F, Sponer J, Langowski J, Cheatham TE., 3rd DNA deformability at the base pair level. J. Am. Chem. Soc. 2004;126:4124–4125. doi: 10.1021/ja0390449. [DOI] [PubMed] [Google Scholar]
- 37.Foloppe N, Nilsson L. Toward a full characterization of nucleic acid components in aqueous solution: simulations of nucleosides. J. Phys. Chem. B. 2005;109:9119–9131. doi: 10.1021/jp044513u. [DOI] [PubMed] [Google Scholar]
- 38.Isaacs RJ, Spielmann HP. Insight into G[bond]T mismatch recognition using molecular dynamics with time-averaged restraints derived from NMR spectroscopy. J. Am. Chem. Soc. 2004;126:583–590. doi: 10.1021/ja037333r. [DOI] [PubMed] [Google Scholar]
- 39.Rohs R, West SM, Liu P, Honig B. Nuance in the double-helix and its role in protein-DNA recognition. Curr. Opin. Struct. Biol. 2009;19:171–177. doi: 10.1016/j.sbi.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zuo X, Cui G, Merz KM, Jr, Zhang L, Lewis FD, Tiede DM. X-ray diffraction “fingerprinting” of DNA structure in solution for quantitative evaluation of molecular dynamics simulation. Proc. Natl Acad. Sci. USA. 2006;103:3534–3539. doi: 10.1073/pnas.0600022103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fratini AV, Kopka ML, Drew HR, Dickerson RE. Reversible bending and helix geometry in a B-DNA dodecamer: CGCGAATTBrCGCG. J. Biol. Chem. 1982;257:14686–14707. [PubMed] [Google Scholar]
- 42.Gorenstein DG. Phosphorus-31 NMR: Principles and Applications. New York: Academic Press; 1984. [Google Scholar]
- 43.Gorenstein DG. 31P NMR of DNA. Methods Enzymol. 1992;211:254–286. doi: 10.1016/0076-6879(92)11016-c. [DOI] [PubMed] [Google Scholar]
- 44.Isaacs RJ, Spielmann HP. NMR evidence for mechanical coupling of phosphate B(I)-B(II) transitions with deoxyribose conformational exchange in DNA. J. Mol. Biol. 2001;311:149–160. doi: 10.1006/jmbi.2001.4855. [DOI] [PubMed] [Google Scholar]
- 45.Schneider B, Neidle S, Berman HM. Conformations of the sugar-phosphate backbone in helical DNA crystal structures. Biopolymers. 1997;42:113–124. doi: 10.1002/(sici)1097-0282(199707)42:1<113::aid-bip10>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 46.Djuranovic D, Hartmann B. DNA fine structure and dynamics in crystals and in solution: the impact of BI/BII backbone conformations. Biopolymers. 2004;73:356–368. doi: 10.1002/bip.10528. [DOI] [PubMed] [Google Scholar]
- 47.Hartmann B, Piazzola D, Lavery R. BI-BII transitions in B-DNA. Nucleic Acids Res. 1993;21:561–568. doi: 10.1093/nar/21.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heddi B, Foloppe N, Bouchemal N, Hantz E, Hartmann B. Quantification of DNA BI/BII backbone states in solution. Implications for DNA overall structure and recognition. J. Am. Chem. Soc. 2006;128:9170–9177. doi: 10.1021/ja061686j. [DOI] [PubMed] [Google Scholar]
- 49.Lefebvre A, Fermandjian S, Hartmann B. Sensitivity of NMR internucleotide distances to B-DNA conformation: underlying mechanics. Nucleic Acids Res. 1997;25:3855–3862. doi: 10.1093/nar/25.19.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Srinivasan AR, Olson WK. Nucleic acid model building: the multiple backbone solutions associated with a given base morphology. J. Biomol. Struct. Dyn. 1987;4:895–938. doi: 10.1080/07391102.1987.10507690. [DOI] [PubMed] [Google Scholar]
- 51.van Dam L, Levitt MH. BII nucleotides in the B and C forms of natural-sequence polymeric DNA: A new model for the C form of DNA. J. Mol. Biol. 2000;304:541–561. doi: 10.1006/jmbi.2000.4194. [DOI] [PubMed] [Google Scholar]
- 52.Winger RH, Liedl KR, Pichler A, Hallbrucker A, Mayer E. Helix morphology changes in B-DNA induced by spontaneous B(I)<==>B(II) substrate interconversion. J. Biomol. Struct. Dyn. 1999;17:223–235. doi: 10.1080/07391102.1999.10508355. [DOI] [PubMed] [Google Scholar]
- 53.Bertrand H, Ha-Duong T, Fermandjian S, Hartmann B. Flexibility of the B-DNA backbone: effects of local and neighbouring sequences on pyrimidine-purine steps. Nucleic Acids Res. 1998;26:1261–1267. doi: 10.1093/nar/26.5.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Heddi B, Foloppe N, Hantz E, Hartmann B. The DNA structure responds differently to physiological concentrations of K(+) or Na(+) J. Mol. Biol. 2007;368:1403–1411. doi: 10.1016/j.jmb.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 55.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 56.Thastrom A, Bingham LM, Widom J. Nucleosomal locations of dominant DNA sequence motifs for histone-DNA interactions and nucleosome positioning. J. Mol. Biol. 2004;338:695–709. doi: 10.1016/j.jmb.2004.03.032. [DOI] [PubMed] [Google Scholar]
- 57.Thastrom A, Lowary PT, Widlund HR, Cao H, Kubista M, Widom J. Sequence motifs and free energies of selected natural and non-natural nucleosome positioning DNA sequences. J. Mol. Biol. 1999;288:213–229. doi: 10.1006/jmbi.1999.2686. [DOI] [PubMed] [Google Scholar]
- 58.Harp JM, Hanson BL, Timm DE, Bunick GJ. Asymmetries in the nucleosome core particle at 2.5 A resolution. Acta Crystallograph. 2000;56:1513–1534. doi: 10.1107/s0907444900011847. [DOI] [PubMed] [Google Scholar]
- 59.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J. Mol. Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 60.Suto RK, Edayathumangalam RS, White CL, Melander C, Gottesfeld JM, Dervan PB, Luger K. Crystal structures of nucleosome core particles in complex with minor groove DNA-binding ligands. J. Mol. Biol. 2003;326:371–380. doi: 10.1016/s0022-2836(02)01407-9. [DOI] [PubMed] [Google Scholar]
- 61.Tsunaka Y, Kajimura N, Tate S, Morikawa K. Alteration of the nucleosomal DNA path in the crystal structure of a human nucleosome core particle. Nucleic Acids Res. 2005;33:3424–3434. doi: 10.1093/nar/gki663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lavery R, Sklenar H. The definition of generalized helicoidal parameters and of axis curvature for irregular nucleic acids. J. Biomol. Struct. Dyn. 1988;6:63–91. doi: 10.1080/07391102.1988.10506483. [DOI] [PubMed] [Google Scholar]
- 63.Gorenstein DG. Conformation and dynamics of DNA and protein-DNA complexes by 31P NMR. Chemical Rev. 1994;94:1315–1338. [Google Scholar]
- 64.Yanagi K, Prive GG, Dickerson RE. Analysis of local helix geometry in three B-DNA decamers and eight dodecamers. J. Mol. Biol. 1991;217:201–214. doi: 10.1016/0022-2836(91)90620-l. [DOI] [PubMed] [Google Scholar]
- 65.Lefebvre A, Mauffret O, Hartmann B, Lescot E, Fermandjian S. Structural behavior of the CpG step in two related oligonucleotides reflects its malleability in solution. Biochemistry. 1995;34:12019–12028. doi: 10.1021/bi00037a045. [DOI] [PubMed] [Google Scholar]
- 66.Lefebvre A, Mauffret O, Lescot E, Hartmann B, Fermandjian S. Solution structure of the CpG containing d(CTTCGAAG)2 oligonucleotide: NMR data and energy calculations are compatible with a BI/BII equilibrium at CpG. Biochemistry. 1996;35:12560–12569. doi: 10.1021/bi9606298. [DOI] [PubMed] [Google Scholar]
- 67.Subirana JA, Faria T. Influence of sequence on the conformation of the B-DNA helix. Biophys. J. 1997;73:333–338. doi: 10.1016/S0006-3495(97)78073-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tisne C, Delepierre M, Hartmann B. How NF-kappaB can be attracted by its cognate DNA. J. Mol. Biol. 1999;293:139–150. doi: 10.1006/jmbi.1999.3157. [DOI] [PubMed] [Google Scholar]
- 69.Gardiner EJ, Hunter CA, Packer MJ, Palmer DS, Willett P. Sequence-dependent DNA structure: a database of octamer structural parameters. J. Mol. Biol. 2003;332:1025–1035. doi: 10.1016/j.jmb.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 70.Drew HR, Travers AA. DNA bending and its relation to nucleosome positioning. J. Mol. Biol. 1985;186:773–790. doi: 10.1016/0022-2836(85)90396-1. [DOI] [PubMed] [Google Scholar]
- 71.Satchwell SC, Drew HR, Travers AA. Sequence periodicities in chicken nucleosome core DNA. J. Mol. Biol. 1986;191:659–675. doi: 10.1016/0022-2836(86)90452-3. [DOI] [PubMed] [Google Scholar]
- 72.Chung HR, Vingron M. Sequence-dependent Nucleosome Positioning. J. Mol. Biol. 2009;386:1411–1422. doi: 10.1016/j.jmb.2008.11.049. [DOI] [PubMed] [Google Scholar]
- 73.Field Y, Kaplan N, Fondufe-Mittendorf Y, Moore IK, Sharon E, Lubling Y, Widom J, Segal E. Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLoS Comput. Biol. 2008;4:e1000216. doi: 10.1371/journal.pcbi.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ioshikhes IP, Albert I, Zanton SJ, Pugh BF. Nucleosome positions predicted through comparative genomics. Nature Genet. 2006;38:1210–1215. doi: 10.1038/ng1878. [DOI] [PubMed] [Google Scholar]
- 75.Liu H, Wu J, Xie J, Yang X, Lu Z, Sun X. Characteristics of nucleosome core DNA and their applications in predicting nucleosome positions. Biophys. J. 2008;94:4597–4604. doi: 10.1529/biophysj.107.117028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Peckham HE, Thurman RE, Fu Y, Stamatoyannopoulos JA, Noble WS, Struhl K, Weng Z. Nucleosome positioning signals in genomic DNA. Genome Res. 2007;17:1170–1177. doi: 10.1101/gr.6101007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Segal E, Fondufe-Mittendorf Y, Chen L, Thastrom A, Field Y, Moore IK, Wang JP, Widom J. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang JP, Widom J. Improved alignment of nucleosome DNA sequences using a mixture model. Nucleic Acids Res. 2005;33:6743–6755. doi: 10.1093/nar/gki977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Richmond TJ, Davey CA. The structure of DNA in the nucleosome core. Nature. 2003;423:145–150. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
- 80.Prunell A. A topological approach to nucleosome structure and dynamics: the linking number paradox and other issues. Biophys. J. 1998;74:2531–2544. doi: 10.1016/S0006-3495(98)77961-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ho PS. The non-B-DNA structure of d(CA/TG)n does not differ from that of Z-DNA. Proc. Natl Acad. Sci. USA. 1994;91:9549–9553. doi: 10.1073/pnas.91.20.9549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Johnston BH, Ohara W, Rich A. Stochastic distribution of a short region of Z-DNA within a long repeated sequence in negatively supercoiled plasmids. J. Biol. Chem. 1988;263:4512–4515. [PubMed] [Google Scholar]
- 83.Johnston BH, Rich A. Chemical probes of DNA conformation: detection of Z-DNA at nucleotide resolution. Cell. 1985;42:713–724. doi: 10.1016/0092-8674(85)90268-5. [DOI] [PubMed] [Google Scholar]
- 84.Foloppe N, MacKerell AD., Jr Intrinsic conformational properties of deoxyribonucleosides: implicated role for cytosine in the equilibrium among the A, B, and Z forms of DNA. Biophys. J. 1999;76:3206–3218. doi: 10.1016/S0006-3495(99)77472-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hays FA, Teegarden A, Jones ZJ, Harms M, Raup D, Watson J, Cavaliere E, Ho PS. How sequence defines structure: a crystallographic map of DNA structure and conformation. Proc. Natl Acad. Sci. USA. 2005;102:7157–7162. doi: 10.1073/pnas.0409455102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Peticolas WL, Wang Y, Thomas GA. Some rules for predicting the base-sequence dependence of DNA conformation. Proc. Natl Acad. Sci. USA. 1988;85:2579–2583. doi: 10.1073/pnas.85.8.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Virstedt J, Berge T, Henderson RM, Waring MJ, Travers AA. The influence of DNA stiffness upon nucleosome formation. J. Struct. Biol. 2004;148:66–85. doi: 10.1016/j.jsb.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 88.Segal E, Widom J. Poly(dA:dT) tracts: major determinants of nucleosome organization. Curr. Opin. Struct. Biol. 2009;19:65–71. doi: 10.1016/j.sbi.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Suter B, Schnappauf G, Thoma F. Poly(dA.dT) sequences exist as rigid DNA structures in nucleosome-free yeast promoters in vivo. Nucleic Acids Res. 2000;28:4083–4089. doi: 10.1093/nar/28.21.4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Anselmi C, Bocchinfuso G, De Santis P, Savino M, Scipioni A. A theoretical model for the prediction of sequence-dependent nucleosome thermodynamic stability. Biophys. J. 2000;79:601–613. doi: 10.1016/S0006-3495(00)76319-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Travers AA. The structural basis of DNA flexibility. Philos. Transact. A Math. Phys. Eng. Sci. 2004;362:1423–1438. doi: 10.1098/rsta.2004.1390. [DOI] [PubMed] [Google Scholar]
- 92.Ridgway P, Almouzni G. CAF-1 and the inheritance of chromatin states: at the crossroads of DNA replication and repair. J. Cell Sci. 2000;113(Pt 15):2647–2658. doi: 10.1242/jcs.113.15.2647. [DOI] [PubMed] [Google Scholar]
- 93.Widlund HR, Vitolo JM, Thiriet C, Hayes JJ. DNA sequence-dependent contributions of core histone tails to nucleosome stability: differential effects of acetylation and proteolytic tail removal. Biochemistry. 2000;39:3835–3841. doi: 10.1021/bi991957l. [DOI] [PubMed] [Google Scholar]
- 94.Miele V, Vaillant C, d'A;ubenton-Carafa Y, Thermes C, Grange T. DNA physical properties determine nucleosome occupancy from yeast to fly. Nucleic Acids Res. 2008;36:3746–3756. doi: 10.1093/nar/gkn262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, Leproust EM, Hughes TR, Lieb JD, Widom J. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.