Abstract
Expression of the seed plant mitochondrial nad5 gene involves two trans-splicing events that remove fragmented group II introns and join the small, central exon c to exons b and d. We show that in both monocot and eudicot plants, extensive mis-splicing of the bi-partite intron 2 takes place, resulting in the formation of aberrantly spliced products in which exon c is joined to various sites within exon b. These mis-spliced products accumulate to levels comparable to or greater than that of the correctly spliced mRNA. We suggest that mis-splicing may result from folding constraints imposed on intron 2 by base-pairing between exon a and a portion of the bi-partite intron 3 downstream of exon c. Consistent with this hypothesis, we find that mis-splicing does not occur in Oenothera mitochondria, where intron 3 is further fragmented such that the predicted base-pairing region is not covalently linked to exon c. Our findings suggest that intron fragmentation may lead to mis-splicing, which may be corrected by further intron fragmentation.
INTRODUCTION
In most angiosperms, mRNA of the mitochondrial nad5 gene, encoding subunit V of respiratory complex I, is derived from three different transcription units encompassing 5 exons (Figure 1A; 1,2). Assembly of the nad5 message requires two cis-splicing events, one to join exons a and b, the other to join exons d and e, and two trans-splicing events, through which the small (22 nt) central exon c is joined to exons b and d.
Figure 1.
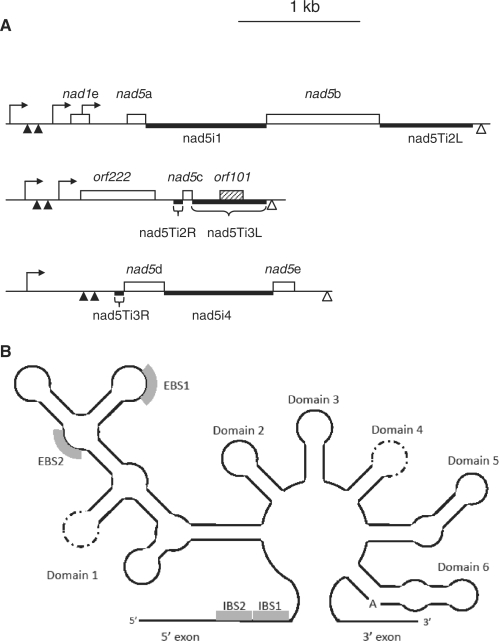
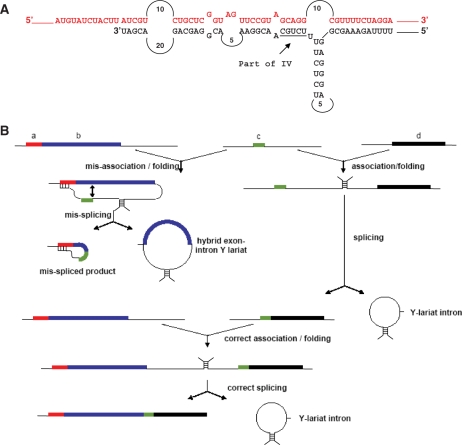
Organization and expression of B. napus nad5 exons and introns. (A) Introns are designated according to (20). Open boxes indicate exons, the shaded box the open reading frame within nad5Ti3L. Thick lines indicate predicted functional domains of introns. Transcription initiation sites are indicated as hooked arrows, 5′ processing sites as filled triangles and 3′ termini as open triangles. The L and R designations are used to refer to the 5′ and 3′ portions, respectively, of the trans-splicing introns nad5Ti2 and nad5Ti3. (B) Simplified schematic of a generalized group II intron, illustrating the various structural domains. EBS and IBS stand for exon and intron binding sites, respectively, as patterned on (42). The perforated lines indicate the domain loop sites at which the RNA is disjoined in trans-plicing introns of all flowering plants (Domain 4) and further disjoined in the Oenothera nad5 Ti3L intron (Domain 1). Thus, what is a continuous RNA in cis-splicing introns is discontinuous in trans-splicing introns; in the latter case the domain stem is thought to form through base-paring interactions between two distinct RNAs.
Figure 2.
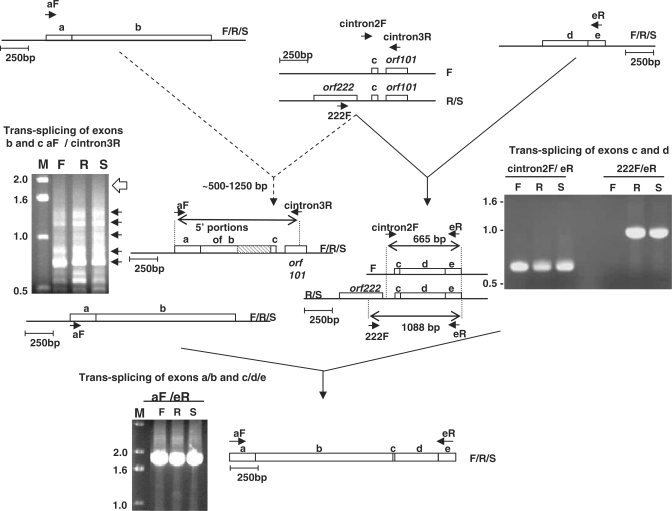
Trans-splicing and mis-splicing of Brassica nad5 transcripts. Top left: RT–PCR products generated using primers from exon a (aF) and intron nad5Ti3L/orf101 (cintron3R). Blunt ends of arrows indicate complementary sites within the corresponding transcripts. Dotted arrows on pathway 1 indicate mis-splicing. F, R and S indicate fertile, sterile and restored lines. The open arrow to the right of the gel photograph on the left indicates the position expected of an intact exon b correctly spliced to exon c. The arrows to the right of the gel image indicate the locations of individually excised, cloned and sequenced bands. The diagram to the right of the gel depicts the general structure of the mis-spliced products; the hatched box is used indicate the sections of exon b that are not present in the mis-spliced products. Top right: RT–PCR products generated using primers from exon e (eR) and intron nad5Ti2R (cintron2F) or orf222 (222F). The diagram to the right of the gel photograph indicates the sizes and structures of the products determined by excision, cloning and sequencing. Bottom: product generated from fully trans-spliced nad5 mRNA using primers from exons a and c.
The cis- and trans-spliced nad5 introns, like most plant mitochondrial introns, are group II introns, a family of intervening sequences found in organelle and prokaryotic genomes. Typically, group II intron splicing, like splicing in eukaryotic nuclei, occurs through two successive trans-esterification reactions, the first involving cleavage at the 5′ splice site with formation of a lariat intron, and the second the formation of the splice junction and release of the intron (3–5). For splicing to occur properly, the intron must fold into a well-conserved structure that juxtaposes the 5′ splice and branch sites and brings the two exons into close proximity (3,5,6). The finding that some group II introns splice autocatalytically indicates that their splicing is RNA catalyzed, although all group II introns are thought to require accessory proteins for in vivo splicing. Many group II introns encode a maturase protein that is required both for splicing and retrotransposition into related genomic sites (5).
As depicted in Figure 1B, group II introns encompass six secondary structural domains. Domain 6 (D6) is positioned within a few nucleotides of the downstream exon, and generally consists of a stem–loop with a bulged adenosine that provides the 2′ OH group for branch formation. D5, located just upstream of D6, contains an essential, highly conserved ‘catalytic triad’ motif and interacts with D1 through several long-range interactions to form the intron catalytic core (3,6,7). In trans-splicing plant mitochondrial introns, the complementary sequences that make up the D4 stem are present on distinct transcripts; it has been suggested that base-pairing between these sequences forms the D4 stem thereby allowing specific association of independently transcribed portions of the intron and assembly of a splicing-competent structure (3,8). Discontinuities within other trans-splicing group II introns have been found to occur in D3 as well as within D1 (8,9). D1 is the most complex domain, comprising multiple subdomains as well as the exon binding sites, EBS1 and EBS2, which define the 5′ splice site via Watson–Crick base pairing with the complementary intron binding sites, IBS1 and IBS2 located at the extreme 3′-end of the 5′ exon (Figure 1B).
The snRNAs of the nuclear spliceosome share certain structural features with group II introns. The U6 snRNA folds into a stem–loop configuration in which the catalytically important AGC triad is present in the same base pairing configuration as exists in group II D5 (10). In addition, nuclear U5 snRNA can functionally substitute for group II domain 1D3 in in vitro trans-splicing reactions (11) and base pairing between the U2 snRNA and sequences surrounding the intron branch site generate a structure with a bulged adenosine similar to that of group II intron D6 (12). The recently derived 3D structure of a group II intron (13) has confirmed many of the structural features deduced from comparative and biochemical studies and provided additional structural analogies with the spliceosome.
The similarities in structure and splicing mechanism, together with the finding that group II intron domains can catalyze splicing in trans (14,15), suggest that the snRNA component of the nuclear splicing apparatus may have been derived from a group II intron of an ancestral eukaryote. According to this view, snRNAs have evolved from a fragmented group II intron from which individual domains began to function in trans to catalyze splicing of introns missing corresponding structural elements (16). It is not apparent, however, how this fragmentation occurred or what types of selective pressures may have driven it. In this respect, the trans-spliced introns of plant mitochondria and chloroplasts (8) are of particular interest, in that they may resemble an early stage in the evolution from an all cis-acting to an all trans-acting splicing machinery. Despite the potential importance of trans-spliced introns in plant mitochondria, relatively little information is available about the processes associated with their splicing.
The nap form of cytoplasmic male sterility (CMS) in the canola plant Brassica napus is associated with expression of a chimeric mitochondrial gene, orf222, that is co-transcribed with the trans-spliced exon c of the nad5 gene (17 Figure 1). During a study initially aimed at determining if orf222 affected nad5 trans-splicing, we discovered that regardless of the presence or absence of an upstream ORF, trans-splicing of exon c follows an ordered pathway in which it must be joined to exon d before proper splicing with exon b can occur. Mis-ordered trans-splicing results in the generation of novel, mis-spliced products that accumulate to levels comparable to or exceeding that of the mature, properly spliced mRNA. The expression of nad5 in three other plants with the same intron organization as B. napus, the eudicot sugar beet and the monocots wheat and maize, is also associated with mis-splicing, but neither ordered splicing nor mis-splicing is associated with nad5 expression in Oenothera tetragona, a eudicot plant in which the one of the trans-splicing introns is further fragmented into a tri-partite intron. We suggest a mechanism for mis-splicing and discuss the influence that mis-splicing may have played in the evolution of plant nad5 gene structure as well as potential broader implications of our findings.
MATERIALS AND METHODS
Plant material
The B. napus lines and nomenclature used were described by L’Homme et al. (17). Plants of the cultivar Bronowski (cam, rfn/rfn) were used as a pollen parent to propagate the nap CMS line (nap, rfn/rfn); these are referred to as Fertile (F) and Sterile (S) or CMS plants, respectively. Westar (nap, Rfn/Rfn), a CMS restorer line (R), are referred to as fertility-restored or restored plants. Plants were grown as described (17). Wheat seedling mtRNA was the gift of Dr Linda Bonen, University of Ottawa. Floral buds of O. tetragona (evening primrose or ‘sundrop’) were taken from plants growing in a garden in Montreal.
Isolation and analysis of mitochondrial RNA
Brassica napus mitochondrial RNA (mtRNA) was isolated as described (17). In the case of Oenothera, RNA was extracted from isolated mitochondria using the plant and fungal protocol of the RNeasy mini-kit (Qiagen, Inc., Mississauga, ON, USA). Northern analysis of mtRNA was carried out according to Menassa et al. (18).
Reverse-Transcriptase Polymerase Chain Reaction amplification of RNA
MtRNA was initially treated with 2 U of amplification grade DNase I (Invitrogen, Carlsbad, CA, USA). Reverse-Transcriptase Polymerase Chain Reaction (RT–PCR) analysis was performed using with the SuperscriptTM One-Step RT–PCR with Platinum Taq system (Invitrogen) as recommended by the manufacturer. A 5 µg of mtRNA was reverse transcribed for 1 h at 42°C followed by 40 cycles of cDNA amplification for 1 min at 94°C, 45 s at 62°C and 4 min at 72°C. A total of 250 ng of each primer were used in each RT–PCR amplification. Amplification products were analyzed by electrophoresis through 2% agarose gels. Products were excised, purified using spin columns (Qiagen) and ligated into pCR2.1 (Invitrogen). Ligation products were transformed into Escherischia coli IVNf and clones sequenced at the commercial sequencing facility of DNA LandMarks, Inc., St Jean-sur-Richelieu, Quebec, Canada. The sequences of the primers employed in RT–PCR experiments are provided in Supplementary Table S1. To characterize Y- and lariat-shaped splicing products, the method of Vogel and Börner (19) was employed, using reagents from the Superscript III one-step RT–PCR kit from Invitrogen.
Mapping of transcript termini
We employed circular RT–PCR (cRT–PCR; 20) to identify 5′ and 3′ transcript termini (see ‘Methods, Supplementary Data). To specifically map 5′ primary transcript ends, we first removed monophosphates from 5′-ends by treatment with calf intestinal phosphatase (CIP). RNA was then treated with tobacco acid pyrophosphatase (TAP) to specifically convert initiator tri-phosphate termini into 5′ monophosphates, which could then be joined to the terminal 3′ hydroxyl group through the action of RNA ligase (21). The identity of the 5′ primary transcript terminus and 3′-end were determined from the sequence of RT–PCR products spanning the junction of the circularized RNA templates (20). The 5′-termini similarly identified through cRT–PCR of mtRNA that had not been treated with either CIP or TAP were taken to represent ends generated through nuclease processing. The identities of all 5′-termini were confirmed by 5′ RACE methodology (Methods, Supplementary Data).
RESULTS
Characterization of pre-spliced B. napus nad5 transcripts
The organization and expression of the B. napus nad5 gene is illustrated in Figure 1. Exon c is located in a transcriptional unit with two ORFs, the upstream, CMS-associated orf222 and the downstream orf101 (previously designated orf139; 17); orf101 is not well-conserved, and unlikely to have protein-coding function. We use the nomenclature of Malek and Knoop (22) to designate the different nad5 introns: the cis-splicing nad5i1 (or intron 1) separates exons a and b and cis-splicing nad5i4 (intron 4) separates exons d and e. nad5Ti2 (intron 2) is trans-splicing intron removed during the joining of exons b and c; the portion co-transcribed with exon b is designated nad5Ti2L, and that co-transcribed with exon c as nad5Ti2R. Similarly, nad5Ti3 (intron 3) is removed during c to d joining; nad5Ti3L is co-transcribed with exon c and nad5Ti3R is co-transcribed with exon d (Figure 1). A more general nomenclature designates the four angiosperm nad5 introns as nad5i230, nad5i1455, nad5i1477 and nad5i1872 (23,24).
The overall secondary structure of the different plant nad5 group II introns, including the domains and key elements of tertiary structure for the trans-splicing nad5Ti2 and nad5Ti3 introns, have been predicted for Arabidopsis thaliana and Oenothera berteriana (1,9). Since the primary structures of the B. napus exons and introns are virtually identical to those of Arabidopsis, it is likely that the structural domains of the introns are identical in these two species. After preliminary results indicated the possible occurrence of nad5 mis-splicing, we considered it important to ensure that all these domains are included in pre-spliced nad5 transcripts. We therefore used RT–PCR technology to identify transcription initiation, nuclease processing sites and 3′-termini for the three different B. napus nad5 precursor transcripts.
The results of this analysis are illustrated in Figure 1, with supporting experimental results provided in Supplementary Figures S1 and S2 and Supplementary Table S1. Importantly, all of the characterized transcripts contain the complete predicted intronic domains for trans-splicing. The fifth exon of nad1 is located 214 nucleotides upstream of the 5′-end of nad5a. Although joining nad1e to exon d is thought to occur via cis-splicing in Brassica, the independent transcription of exon e suggests that it may also occur in trans, as it does in several other plant groups (25).
nad5 cis-splicing
RT–PCR experiments performed to characterize the nad5 cis-splicing events involving nad5i1 and nad5i4 are illustrated in Supplementary Figure S3. For both introns, we analyzed transcripts generated prior to trans-splicing by designing one primer of each amplifying pair to prime within the adjacent trans-splicing intron. In most mitochondrial protein-coding transcripts, nucleotides encoded as C in DNA undergo post-transcriptional conversion to U via RNA editing; the presence of edited nucleotides in an RT–PCR product thus indicates that it is derived from RNA and not contaminating DNA. Sequence analysis of cloned cDNAs for each product confirmed the expected mRNA junction sites and also indicated the presence of editing sites in both the precursor and product forms (Supplementary Figure S4).
Ordered trans-splicing and mis-splicing
We similarly investigated trans-splicing by RT–PCR using primers designed to amplify cDNAs of transcripts that had undergone one, but not both, trans-splicing events. Two primer pairs were used to amplify products in which exon c had undergone splicing to exon d but not to exon b (Figure 2, top right). When an oligonucleotide that primes within exon e (primer eR, Figure 2) was used in conjunction with primer cintron2F, which corresponds to a sequence in nad5Ti2L, a single, major product was obtained from the three plant lines. The products of the different lines were cloned and eight clones from each line were sequenced. In all cases, the products consisted of the c/d/e exons, joined at the expected junctions. All sites were fully edited in all clones, demonstrating that the products were generated from RNA and not contaminating DNA and suggesting that editing is completed by the time trans-splicing occurs. Similar results were obtained when a primer corresponding to a site at the 3′-end of exon d was used in combination with cintron2F. When a primer matching a sequence within orf222 was used in conjunction with primer eR, a product was obtained only from plants with the nap cytoplasm (lanes R and S, Figure 2, right). The absence of an amplification product in plants with fertile (cam) cytoplasm (lane F in Figure 2, right) is expected since orf222 is not present in this mitochondrial genome (17).
Trans-splicing between exons a/b and c was analyzed by generating RT-PCR products using an oligonucleotide that primes near the 3′-end of orf101 (cintron3R, Figure 2) together with primers matching sequences at the 5′-ends of either exon a (e.g. aF, Figure 2) or exon b (not shown). Surprisingly, for all primer pairs and for all lines analyzed, no products of the size expected of a properly spliced transcript were obtained. Instead, a heterogeneous set of products, ranging in size, in the case of primer aF, from ∼500 to ∼1300 bp was obtained (Figure 2, top left). These products were individually excised, cloned and sequenced; five clones of each product, from each plant line, were analyzed. Surprisingly, all were found to correspond to mis-spliced RNAs in which the correct splice acceptor site of exon c was joined to various cryptic donor splice sites in exon b.
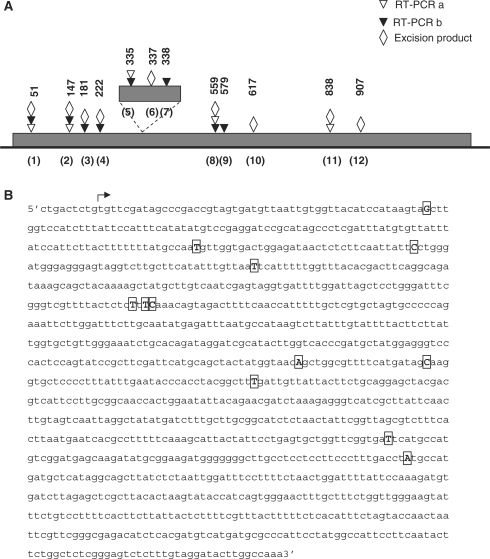
The positions of the cryptic 5′ splice sites are illustrated in Figure 3. The cryptic sites identified were located throughout exon b, but were concentrated towards its 5′-end. All the mis-spliced products were fully edited and no major differences in the abundance of the different products among the different plant lines were evident. As shown in Figure 3, five and eight cryptic sites were identified using exon a or b primers, respectively, each in combination with primer cintron3R (Figure 2); four of these sites were detected using both exon a and b primers. Because several of the RT–PCR products corresponding to mis-spliced RNAs were obtained regardless of the primers employed, and because in all cases the products corresponded to RNAs with the correct 3′ splice site, it seemed likely that they were generated from mis-spliced transcripts produced in vivo and were not produced artifactually. Moreover, because mis-splicing was found to occur in all three plant lines, it was not related to the presence of the CMS-associated orf222 upstream of nad5c, but rather appeared to be an intrinsic characteristic of nad5 expression.
Figure 3.
Structure of nad5 b to c mis-spliced products. (A) Schematic illustrating sites of mis-splicing. Triangles indicate the mis-splicing sites identified by RT–PCR using exon a (open) or b (filled) primers in combination with primer cintron3R. Diamonds indicate the location of mis-splicing sites detected in excision products. Numbers above symbols indicate the distance of the site, in nucleotides, from the 5′-end of exon b. Sites are numbered, in parentheses, according to their position from the 5′-end of exon b. (B) Sequence of nad5b; the start of the coding sequence is indicated by the arrow, boxed nucleotides are those attached to the 5′ nucleotide of exon c in mis-spliced products or adjacent to the branch site in excision products.
Importantly, only a single product is generated by RT–PCR amplification using oligonucleotides that prime within exons a and e (Figure 2, bottom). When this product was cloned and sequenced it was found to correspond to transcripts in which all exons were properly spliced and completely edited. Thus, although transcripts resulting from b to c mis-splicing accumulate, proper splicing of exons b and c also occurs, but only for transcripts in which exon c is first joined to exon d (solid lines, Figure 2). This indicates that the production of a functional nad5 transcript is contingent upon the trans-splicing events occurring in a specific sequence: splicing of exons c and d must precede the splicing of exons b and c. Moreover, since we find no evidence that b to c mis-spliced transcripts are joined to exons d and e, the mis-spliced RNAs apparently represent a ‘dead-end’ in nad5 transcript processing.
Structure of excised trans-spliced introns: novel products and confirmation of mis-splicing
Although most group II introns splice via a two-step trans-esterification pathway that leads to the formation of lariat intron products, branch-point mutants of yeast mitochondrial introns (26), as well certain chloroplast (27) and plant mitochondrial introns (28) that lack a bulged adenosine in D6, splice via a first step hydrolysis pathway that results in the formation of linear intron products. In addition, unusual plant mitochondrial excision products, containing non-encoded nucleotides, have recently been reported (28). To further characterize trans-splicing and to confirm the occurrence of mis-splicing of nad5Ti2 at the cryptic sites indicated in Figure 3, we determined the structure of excised nad5Ti2 and nad5Ti3.
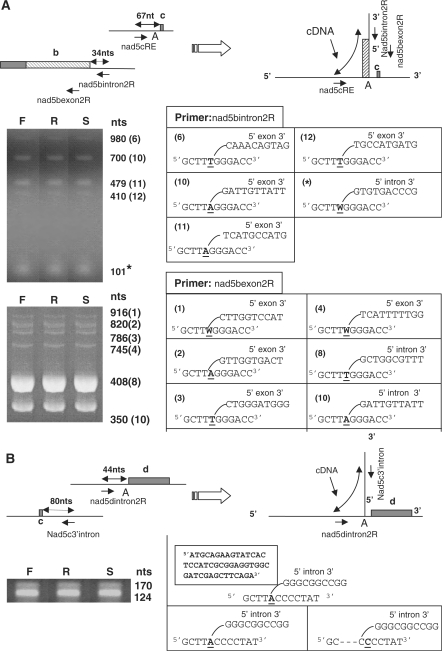
If the first step of nad5Ti2 mis-splicing proceeds through trans-esterification, a population of Y-shaped excised introns should be generated in which the branch point adenosine is linked via a 2′-5′ phosphodiester bond to the 5′-end of the intron or, in the case of nad5Ti2 mis-splicing, to a nucleotide within exon b. We used the approach of Vogel et al. (19), which relies on the capacity of certain reverse transcriptases to read through branch sites, to detect and characterize such Y-shaped molecules. Oligonucleotides designed to prime downstream of the 5′ nad5Ti2 splice site (Figure 4A, nad5bintron2R) and upstream of the potential branch point nucleotide (Figure 4A, nad5cRE) were used for RT–PCR amplification of mtRNA. For each analyzed line, five bands, ranging in size from ∼100 to ∼1200 nucleotides, were obtained (Figure 4A), each of which was excised, cloned and sequenced; 10–12 clones per band per plant line were analyzed.
Figure 4.
Excision products of trans-splicing. (A) RT–PCR analysis of nad5Ti2 trans-splicing and mis-splicing. Above: a generalized depiction of the portions of exon b incorporated into the excision products is indicated by the hatched box. nad5bintron2R, nad5bexon2R and nad5cRE are primers used in RT–PCR amplification across branch junctions; the arrows depict their annealing sites and 5′ to 3′ orientation. The branch nucleotide is indicated as A. Below: sequences of products obtained after excision and cloning of PCR products are indicated as the corresponding branched structures shown to the right of the photograph. The nucleotide of the products corresponding to the branch point, which can be copied as either A or T by the reverse transcriptase, is underlined. Numbers in parentheses indicate the corresponding mis-splicing site in (B). The asterisk indicates the excision product from correct trans-splicing. (B) RT–PCR analysis of nad5Ti3 trans-splicing. Above: nad53′cintron and nad5dintron2R are primers used in RT–PCR amplification across branch junctions. The boxed structure indicates a 46 nt, non-mtDNA encoded insertion found in 2 of 8 characterized excision products. Other aspects are as in (A).
The smallest product was found to correspond to the excised intron generated from correct b to c splicing. This 101 nucleotide cDNA begins at the nad5bintron2R primer annealing site and extends across the correct 5′ splice site, through the branch point and into the adjoining intron sequence upstream of exon c. The larger products correspond to cDNAs that arise from mis-splicing. They begin at nad5bintron2R and extend beyond the normal 5′ splice site to various sites within exon b, then further extend across the branch point into the adjoining intron sequence. In most cases, adenosine was mis-incorporated opposite the branch point, as reported previously (19,27). Although this allowed excision products of mis-splicing to be identified, most corresponded to sites of mis-splicing located closer to the 3′-end of exon b than those identified previously, and only one, at site 11 (Figure 3A) matched a mis-splicing site found earlier.
To identify excision products arising from mis-splicing at sites located closer to the 5′-end of exon b, we performed similar RT–PCR amplifications using a reverse primer located within exon b (nad5bexon2R) together with nad5cRE (Figure 4A). This allowed five additional excision products to be identified, of which four corresponded to mis-spliced RNAs identified previously (sites 1 through 4, Figure 3A). In total, excision products corresponding to nine sites where a nucleotide within exon b was joined to the branch nucleotide at the 3′-end of the nad5Ti2R intron were identified; of these, six corresponded to mis-splicing events detected using the experimental approach outlined in Figure 2. These results thus confirm the occurrence of mis-splicing at the sites illustrated in Figure 3 and provide direct evidence that mis-splicing as well as correct b to c splicing occurs through the trans-esterification pathway.
The products of trans-splicing between exons c and d were similarly examined using primers corresponding to nad5Ti3 sequences upstream of the expected branch point and downstream of exon c. As shown in Figure 4B, two amplification products similar in size to the predicted products of correct trans-splicing were obtained. When the smaller, more abundant 124 nt product was cloned and sequenced, it was found to comprise two cDNA types. In the more common type, the 5′-terminus of the intron was joined to the branch point adenosine, as expected. In the second type, the 5′ intron terminus was joined to a site three nucleotides upstream of the predicted branch point; these could represent products generated through use of an alternative branch nucleotide or the skipping of reverse transcriptase as it traversed the branch site (27). The larger PCR product represented a third, most unusual type of cDNA that possessed a 46 nt insertion situated between the 5′-end of the intron and the branch site. The inserted sequence is not present in the B. napus mitochondrial genome but possesses near-perfect similarity with a nuclear Arabidopsis sequence within predicted gene At4g02940, and encodes amino acids 105 to 121 of a 2-oxoglutarate dependent, iron dependent oxygenase (2OG-Fe[II] oxygenase). Although the presence of this sequence within the mitochondria seems extraordinary, several plant mitochondrial tRNAs are imported from the cytoplasm and it has been reported that chloroplasts import the nuclear-encoded eIF4E mRNA (29). Interestingly, no RNA of this type was found in analysis of the excision products of c to d splicing from wheat or O. berteriana mtRNA (see below). While it is likely that the incorporation of this insertion was generated artifactually during the RT–PCR amplification process, no clear source of such an artifact, such as extensive sequence complementarity between At4g02940 and the excised intron, is apparent. Collectively, the results indicate that both correct and incorrect trans-splicing of the nad5Ti2 and nad5Ti3 introns involve the formation of lariat intermediates and take place via trans-esterification events. In the case of nad5Ti3, it is conceivable that trans-splicing can occasionally result in the integration of a relatively large, non-mtDNA encoded oligonucleotide.
As shown in the upper panel of Figure 4A, the amounts of RT–PCR products corresponding to the excision products of correct splicing and mis-splicing appeared comparable, suggesting that mis-spliced RNAs might accumulate to a level that is similar to or even exceeds that of the properly spliced message. The results of two additional experiments are consistent with this possibility. Exon a–b probes were found to detect a heterogenous set of discrete transcripts in RNA gel-blot experiments that are shorter than any anticipated correctly spliced transcript, including the full-length properly spliced nad5 message or a cis-spliced a–b transcript that might accumulate prior to trans-splicing (Supplementary Figure S5). The sizes of these smaller transcripts fall in the size range of RNAs in which mis-splicing at sites indicated in Figure 3 had taken place; gel-blot analysis of the same mtRNA preparation with orf222 sequences detects only a single major product, indicating that the analyzed smaller RNAs did not result from non-specific degradation of the RNA. In addition, an exon c reverse primer used in combination with an exon a primer amplifies both mis-spliced and properly trans-spliced RNAs in RT–PCR amplifications, but the levels of the mis-spliced products exceed that of full-length product (Supplementary Figure S5). While neither of these experiments are strictly quantitative, they both suggest that level of mis-spliced products to accumulate to a degree rivaling or exceeding that of the properly spliced mRNA.
Mis-splicing may result from mis-folding of nad5a/b/c precursor transcripts
The above results can be explained by postulating that nad5Ti2 intron is unable to trans-splice correctly when the portion of nad5Ti3 that is positioned downstream of exon c (nad5Ti3L, Figure 1), remains attached to this small, 22 bp, exon. This suggests that when nad5Ti3L remains attached to exon c, it interferes with the capacity of nad5Ti2 to fold in a manner that allows recognition of its correct 5′ splice site. The folding of trans-splicing group II intron domains 1–3 into a splicing competent structure involves intra-molecular secondary and tertiary structural interactions that are expected to occur rapidly after transcription. The D4 sequences, however, remain unpaired until they encounter complementary intron sequences on a separate transcript, an inter-molecular process expected to occur considerably slower than the initial domain folding. Should nad5Ti2 assemble via D4 base-pairing prior to the assembly of nad5Ti3, unpaired nad5Ti3L D4 sequences would have an opportunity to interact with the upstream exons following the folding of D1-3. Because it seemed that such interactions might underlie mis-splicing, we searched for potential secondary structural interactions that could occur between the nad5Ti3L and the nad5a/b/Ti2L transcripts.
Analysis of the folding of an RNA that could arise from the association of nad5a/b and nad5c precursor transcripts using the on-line resource mfold (30; http://mfold.bioinfo.rpi.edu/cgi-bin/rna-form1.cgi; Supplementary Data, Methods section) revealed an extended potential duplex between nad5 exon a and nad5Ti3 sequences that spans over 50 nucleotides and encompasses the entire half-domain 4 sequence of nad5Ti3 (Figure 5A). This sequence is predicted to be considerably more stable (ΔG = −44.3 kcal/mol) than the normal D4 duplex, which encompasses only 6 base pairs (9). Thus, if an exon c transcript were to associate with a cis-spliced a/b transcript while nad5Ti3L was still attached to it, it seems likely that the free 3′-end of the intron could base pair with to exon a. The formation of this duplex might then position the 3′ splice site in the vicinity of the cryptic sites of exon b and prevent the juxtaposition of the normal 5′ and 3′ splice sites (Figure 5B). The small size of exon c might make nad5Ti2 particularly susceptible to such folding interference by the downstream intron. Conceivably, the presence of nad1 intron sequences upstream of nad5a might also contribute to the opportunity for mis-folding.
Figure 5.
Model for b to c mis-splicing based on constrained folding in the b/c splicing complex. (A) Potential duplex structure between Brassica exon a (red) and intron sequences downstream of exon c (black). The 3′-end of the sequence in black corresponds to the mapped 3′ terminus of the nad5Ti3L partial intron. (B) Proposed model for mis-splicing: thick lines indicate exons, thin lines indicate introns and hatch marks indicate regions of known or proposed base-pairing. If exon c (green) associates with exon b prior to its splicing to exon d, intron sequences downstream of c pair with a complementary region in exon a to constrain the folding of the intron and prevent proper positioning of the normal 5′-splice; splicing occurs to sites within exon b with release of a hybrid exon-intron Y lariat. If the exon c and d transcripts associate first, normal splicing of both exons c and d and exons c and b occurs, with the release of the introns as Y-lariat structures.
Figure 5B depicts a model for the ordered splicing and mis-splicing of nad5 transcripts. It is based on the premise that the nad5Ti2R/exon c/nad5Ti3L transcript is initially free to associate with the exons a/b or exons d/e transcripts. According to this model, if the exon c transcript associates with the a/b transcript prior to its association with d/e, Ti3L will form a duplex with exon a and the Ti2 3′ splice site will be positioned opposite cryptic splice sites within exon b. This will lead to mis-splicing and the release of a hybrid Y lariat molecule composed of both exon and intron sequences. Importantly, the nad5Ti3 intron sequences would remain annealed to exon a and be unable to participate in the joining of the mis-spliced molecules to exon d; hence these products would represent a splicing dead-end, as observed. If, however, nad5Ti3L sequences on the exon c transcript associate with their nad5Ti3R counterparts on the d/e transcript prior to the association of nad5Ti2L and nad5Ti2R, exon c would be correctly joined to exons d and e. The resulting spliced RNA would lack the interfering nad5Ti3L sequences and thus be capable of correctly splicing with exons a and b to form the observed, functional, trans-spliced mRNA.
Mis-splicing is eliminated when the nad5Ti3 intron is further fragmented
Because plant mitochondria are not amenable to transformation, it is not possible to test the hypothesis for mis-splicing outlined above by direct experimentation. Natural variation in the structure of the nad5 gene, however, does afford a means to test the hypothesis less directly. In the eudicot plant, O. berteriana, nad5Ti3 intron is fragmented into three instead of two segments (9). In this plant, only the portion of D1 encompassing subdomains 1A, 1B and 1C is located immediately downstream of exon c (Figure 1B). The central portion of the intron is encoded by a distinct locus, designated tix, that encompasses D1 subdomain 1D, as well as D2, D3 and the portion of D4 present in nad5Ti3L in other plant mtDNAs (9). The formation of functional nad5Ti3 in Oenothera therefore involves the assembly of three distinct transcripts; the tix RNA is thought to associate with the portion of the intron co-transcribed with exon c through base-pairing interactions involving D1 and, as in other plant mitochondria, with the nad5Ti3R half-intron through base-pairing in D4. If the model for mis-splicing articulated above is correct, we would not expect to observe mis-splicing in Oenothera because the region predicted to base-pair with exon a sequences is not co-transcribed with exon c. Moreover, because of the high sequence similarity between the Oenothera and Brassica (or Arabidopsis) nad5 genes and because the nad5 transcripts are predicted to fold in the same manner (1,9), we would anticipate that major differences in nad5 splicing between the two species would likely result from the further fragmentation of the Oenothera nad5Ti3L half-intron rather than other structural differences.
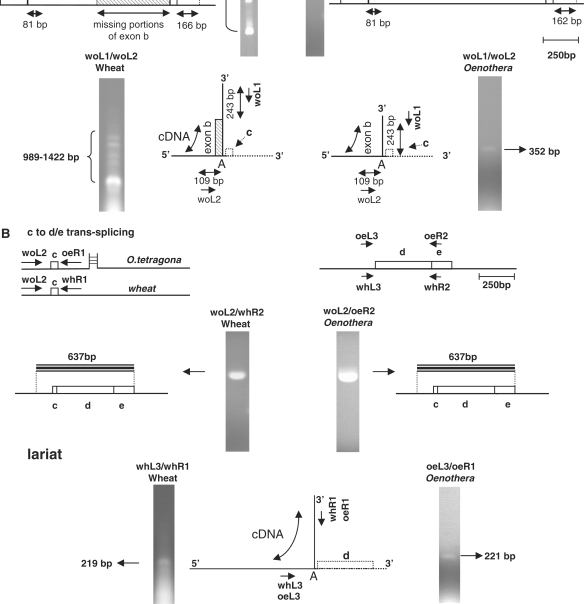
RNA isolated from floral buds of Oenothera tetragona mitochondria was subjected to RT–PCR analyses similar to those described above. Preliminary PCR analyses on mtDNA from this Oenothera species indicated that, as expected, sub-domain 1A is not coded on a DNA segment contiguous with that encoding subdomain 1D (not shown). When a primer located at the 5′-end of exon b was used in RT–PCR amplifications with a primer annealing to the portion nad5Ti3R immediately downstream of exon c (and not located within the tix transcript), only a single band, of the size expected for a correctly spliced RNA, was obtained; sequencing of the cloned product confirmed the expected composition (Figure 6A, right). As expected, RT-PCR analysis of Oenothera mtRNA using primers that anneal to nad5Ti2R and exon e yielded only products corresponding to RNAs in which exon d was correctly spliced to exon c. In both cases, the analysis of intron excision products was consistent with the absence of mis-splicing. Thus, in contrast to what was observed for B. napus, we find no evidence for nad5 b to c mis-splicing, or ordered splicing, in Oenothera mitochondria. These findings are consistent with the view that the pairing of sequences at the 3′-end of the Brassica nad5Ti3L transcript with nad5a transcripts is a key factor in mis-splicing.
Figure 6.
Mis-splicing of exons b and c occurs in wheat but not Oenothera. (A) Upper: products of b to c splicing in wheat (left) and Oenothera (right) as analyzed by RT–PCR using primers that anneal downstream of exon c and within exon b. Primers specific to wheat mtRNA are indicated with the ‘wh’ prefix, those specific to Oenothera with the ‘oe’ prefix and primers suitable for use on both templates are indicated with the ‘wo’ prefix. All products were individually cloned and sequenced to determine the identity of the sequences within them. Regions of exon b that are missing in one or more of the analyzed products are indicated by hatched lines. Only a single product, consisting of exon b correctly spliced to exon c was obtained from Oenothera. Mis-spliced products predominated in wheat mtRNA; mis-splicing occurred at sites corresponding to those of Brassica. The smallest wheat excision product, corresponding to the properly spliced intron, is not shown. Lower: excision products of b to c mis-splicing (wheat) splicing in (Oenothera). Multiple products, corresponding to Y-shaped excision products derived from mis-splicing, are observed in wheat mitochondria (left), whereas only a single product, corresponding to the excised intron, is observed in Oenothera mitochondria (right). (B) Upper: splicing of exons c and d in wheat and Oenothera; as in Brassica, only a single RT–PCR product, corresponding to a properly spliced mRNA, is observed. Lower: only a single excision product, corresponding to the correctly spliced intron is observed in both wheat and Oenthera.
To determine if the absence of mis-splicing in Oenothera mitochondria might be accounted for by factors other than the fragmentation of the nad5Ti3L half-intron, we similarly analyzed mtRNA isolated from wheat seedlings. The structure of the nad5 gene in wheat mtDNA is identical to that found in flowering plants other than Oenothera: introns 2 and 3 are located in the same positions with respect to the protein coding sequence and are similarly fragmented into half-introns that undergo trans-splicing (2). The nucleotide sequence of the gene in this monocot, however, is significantly more diverged from the Brassica gene than Oenothera nad5 (4.7% versus 1.4% dissimilarity in exons). Despite this, the 3′-end of wheat nad5Ti3L, like Brassica nad5Ti3L, is potentially capable of base-pairing with exon a (not shown), and would therefore be expected to mis-splice according to the model of Figure 5B.
As shown in the left portion of Figure 6A, when primers corresponding to those used to analyze the trans-splicing of Oenothera exons b and c were used in RT–PCR amplifications with wheat mtRNA, a heterogenous set of products, similar to that observed with Brassica, was obtained. When these products were individually excised from the gel, cloned and sequenced they were found, as with the Brassica mis-spliced products, to correspond to transcripts in which the precise 5′-end of exon c was joined to various cryptic splice sites within exon b. Of the six sites of mis-splicing represented in these products, four were found to coincide precisely with corresponding cryptic sites in Brassica nad5b. Similar experiments conducted using mtRNA from another eudicot plant, sugar beet (Beta vulgaris) as well as the monocot maize have similarly revealed the occurrence of mis-splicing between nad5 exons b and c (Supplementary Figure S6). The occurrence of mis-splicing in wheat and its absence in Oenothera was confirmed by analysis of the Y-shaped intron excision products (Figure 6A). Thus, mis-splicing is a general characteristic of nad5 expression in angiosperm plants: Oenothera is sole observed exception. These experiments suggest that the absence of mis-splicing in Oenothera results from the additional fragmentation of the nad5Ti3L intron and provide support for the view that mis-splicing of nad5Ti2 results from the pairing of sequences at the 3′-end of the nad5c transcript with sequences in exon a.
DISCUSSION
Excision products of trans-splicing
This investigation is the first to characterize excision products of trans-splicing plant mitochondrial introns. In most cases, the products identified are consistent with a splicing mechanism involving successive trans-esterification reactions, as has been generally inferred for most plant mitochondrial introns (4). We also identify a minor excision product of Brassica nad5Ti3 splicing, however, in which 46 non-encoded nucleotides are inserted at the branch point junction. It is unclear how such molecules could have been generated. Li-Pook-Than and Bonen (28) recently characterized a number of unusual excision products from several plant mitochondrial group II introns that are also inconsistent with a simple trans-esterification model. It is conceivable that following the initial excision of the branched intron it may have undergone additional reaction(s), possibly involving reverse splicing, with another RNA in the mitochondria.
Cryptic splice-site selection
In self-splicing group II introns, pairing between two sequence elements at the 3′-end of the upstream exon, intron-binding sites 1 and 2 (IBS1, 2), with corresponding exon binding sites in D1 (EBS1, 2) has been shown to be important for defining the 5′ cleavage site, which normally occurs at the nucleotide immediately adjacent to IBS1 (31). Studies of in vitro autocatalytic splicing have shown that deletion of the IBS2 sequence can result in the enhancement of splicing at cryptic 5′ sites, which can base pair with native EBS1 (32). The sequences immediately adjacent to the cryptic splice sites in the B. napus nad5 intron do not, in general, appear capable of forming base-paired structures with the EBS elements identified by Knoop et al. (1) however, and share little evident sequence similarity among one another. Nevertheless, mis-splicing clearly occurs at a select group of sites, and these sites are the same in Brassica and wheat nad5 transcripts.
Other experiments have indicated that the 5′ splice site is recognized as the 3′-terminal nucleotide at the junction between single and double-stranded residues formed between the intron and EBS1 (33). Even with the flexibility allowed by this model, however, it is difficult to explain the observed selection of cryptic sites by nad5Ti2, although their general location within the exon is consistent with the positional constraints imposed by the pairing of nad5Ti3L with exon a, as proposed. This difficulty is compounded by lack of certainty regarding the identity of EBS1 itself. The nad5i2 intron, like several other plant mitochondrial introns, has proven difficult to fold into secondary structures that satisfy canonical group II intron features, and potential EBS1 elements differ among different published secondary structures (1,34). It is clear, however, that mis-splicing proceeds through the conventional trans-esterification pathway and it seems likely therefore that the bulged adenosine in nad5Ti2 D6, although situated in an unusual context (28), functions as a promiscuous nucleophile in the first splicing step.
Mis-splicing and evolution of the plant nad5 gene
The modern angiosperm nad5 gene likely evolved from an ancestor that had the same intron complement but in which nad5i2 and nad5i3 existed in a cis- rather than trans-splicing form (22). The additional fragmentation of nad5Ti3 into a tripartite structure has thus far only been observed in plants of the genus Oenothera. Of the plants analyzed here, Oenothera (order Myrtales), according to the DNA-based phylogeny of Wikström et al. (35), is most closely related to B. napus (order Brassicales). Thus, conversion of the intron from a bipartite to a tripartite structure has taken place since the divergence of these two plant lineages an estimated 110–120 Myr ago (35).
Although transcript mis-splicing occurs frequently in most eukaryotic cells, the cytoplasmic mRNAs so generated normally contain premature termination codons and are subjected to nonsense-mediated decay. We find that nad5 b to c mis-spliced RNAs appear to accumulate to levels comparable to, or exceeding that of the mature, correctly spliced message. We are not aware of any example of mis-splicing that results in the accumulation aberrant mRNAs on the scale observed here. This mis-splicing is inherently wasteful, since its products represent a dead-end in nad5 transcript processing. Moreover, it is possible that aberrant, mis-spliced transcripts might be translated to produce a protein that contains only the N-terminus of the NAD5 protein, and could compete with native NAD5 and thereby have a negative affect mitochondrial function. Nevertheless, cells in most plant lineages have apparently tolerated nad5 mis-splicing since fragmentation of the nad5i2 and nad5i3 introns prior to the monocot–dicot divergence over 150 Myr ago (35). In this respect, it should be noted that many plants, including B. napus, can tolerate the expression of aberrant, chimeric mitochondrial genes associated with the trait of cytoplasmic male sterility (36).
General implications of mis-splicing and group II intron fragmentation
The frequent rearrangements that plant mitochondrial genomes undergo has resulted in the separation of some introns into two independently transcribed parts which retain functionality due to the capacity of the D4 stem to assemble in trans. Trans-splicing introns are also known to occur in mitochondria of the green alga, Mesostigma viride (37) as well as in chloroplasts of land plants and the green alga Chlamydomonas reinhardtii (8,38). We show here that when angiosperm nad5 introns 2 and 3 both exist in a bipartite, trans-splicing form, mis-splicing of intron 2 occurs. We suggest that mis-splicing is the result of intron 2 mis-folding due to base-pairing between the 3′-end of the nad5Ti3L half-intron and exon a. We hypothesize that this leads to a mis-positioning of the 3′ splice site in the mis-folded intron such that it is located near the cryptic sites in exon b. In support of this view, we find that when the nad5Ti3 intron is further fragmented into a tripartite form such that the sequences predicted to pair with exon a are no longer attached to exon c and the nad5Ti2R half-intron, as in Oenothera mitochondria, mis-splicing does not occur. Mis-splicing does occur, however, in all other plants examined in which the nad5Ti2 and nad5Ti3 introns are both organized in bipartite form.
While it is conceivable that other factors, such as the evolution of a novel splicing protein unique to the Oenothera lineage, could also account for the absence of mis-splicing in this plant, we deem it more likely that the correlation of correct splicing and additional intron fragmentation in this plant is not coincidental. The fragmentation of nad5Ti3 into a tripartite form may have been selectively favored since it could have eliminated the wasteful and potentially detrimental consequences of mis-splicing. The finding that at least one other trans-splicing intron, intron 1 of the psaA gene in C. reinhardtii (38), has undergone fragmentation into tripartite form, is consistent with the view that additional fragmentation of group II introns may, in some circumstances, be selectively advantageous.
The notion that further fragmentation of trans-splicing group II introns may, in the appropriate circumstance, be selectively favored, has interesting evolutionary implications. Group II introns are found in many different bacterial taxa, including the α- proteobacteria from which the symbiotic precursor to present day mitochondria is thought to have been derived (39,40). Many bacterial group II introns appear to function as both introns and retroelements, with their retrotransposition into related genomic sites being catalyzed by the maturase encoded within domain 4 (5,40). This, together with increased understanding of the similarities between group II and splicesomal introns, has led to the plausible hypothesis that a group II retrotransposon/intron, possibly acquired from the original mitochondrial endosymbiont, proliferated within the genome of an ancestral ‘protoeukaryote’ (41). The fragmentation of such an intron into separate domains capable of assembling in trans could then have given rise to a primitive splicing apparatus that would have allowed for the loss of functional domains in other introns of the same primordial nuclear genome. The recent finding that many locations within a group II intron can serve as sites for fragmentation that allow for retention of splicing functionality (42) supports this view.
The conversion of the angiosperm nad5Ti3 intron from a bi- to a tripartite structure created an intron in which one segment, the tix transcript, functions in a purely trans capacity. It seems possible that selective forces similar to those that we suggest may have aided nad5Ti2 fragmentation also acted to favor the conversion of related, two-segmented group II introns in the protoeukaryote into multiple, trans-acting parts. Although it is not presently possible to experimentally evaluate the mis-folding hypothesis for plant mitochondrial nad5 mis-splicing, it may be possible to devise a test for a more general case, using, for example, in vitro group II intron trans-splicing systems (6,14,15,33,43). This, in turn, could provide an important conceptual link between trans-splicing group II introns and the nuclear spliceosome. In any case, the high degree of nad5 transcript mis-splicing reported here represents an unexpected and novel feature of plant mitochondria that may open several avenues for further investigation in organelle gene expression and evolution.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Discovery Grants from the Natural Sciences and Engineering Research Council (NSERC) of Canada; Scholarships from NSERC; Fonds québécois de recherche sur la nature et les technologies (FQRNT) to H.E. Funding for open access charge: NSERC Discovery Grant.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We extend thanks Dr Linda Bonen of the University of Ottawa for the gift of wheat mtRNA, and to her and members of her laboratory for helpful discussions. Special thanks to the late Sheila Sullivan Brown for access to Oenothera tetragona in her garden.
REFERENCES
- 1.Knoop V, Schuster W, Wissinger B, Brennicke A. Trans splicing integrates an exon of 22 nucleotides into the nad5 mRNA in higher plant mitochondria. EMBO J. 1991;10:3483–3493. doi: 10.1002/j.1460-2075.1991.tb04912.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pereira de Souza A, Jubier MF, Delcher E, Lancelin D, Lejeune B. A trans-splicing model for the expression of the tripartite nad5 gene in wheat and maize mitochondria. Plant Cell. 1991;3:1363–1378. doi: 10.1105/tpc.3.12.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Michel F, Ferat JL. Structure and activities of group II introns. Annu. Rev. Biochem. 1995;64:435–436. doi: 10.1146/annurev.bi.64.070195.002251. [DOI] [PubMed] [Google Scholar]
- 4.Bonen L, Vogel J. The ins and outs of group II introns. Trends Genet. 2001;17:322–331. doi: 10.1016/s0168-9525(01)02324-1. [DOI] [PubMed] [Google Scholar]
- 5.Lambowitz AM, Zimmerly S. Mobile group II introns.Annu. Rev. Genet. 2004;38:1–35. doi: 10.1146/annurev.genet.38.072902.091600. [DOI] [PubMed] [Google Scholar]
- 6.Qin PZ, Pyle AM. The architectural organization and mechanistic function of group II intron structural elements. Curr. Opin. Struct. Biol. 1998;8:301–308. doi: 10.1016/s0959-440x(98)80062-6. [DOI] [PubMed] [Google Scholar]
- 7.Fedorova O, Zingler N. Group II introns: structure, folding and splicing mechanism. Biol. Chem. 2007;388:665–678. doi: 10.1515/BC.2007.090. [DOI] [PubMed] [Google Scholar]
- 8.Bonen L. Trans-splicing of pre-mRNA in plants, animals, and protists. FASEB J. 1993;7:40–46. doi: 10.1096/fasebj.7.1.8422973. [DOI] [PubMed] [Google Scholar]
- 9.Knoop V, Altwasser M, Brennicke A. A tripartite group II intron in mitochondria of an angiosperm plant. Mol. Gen. Genet. 1997;255:269–276. doi: 10.1007/s004380050497. [DOI] [PubMed] [Google Scholar]
- 10.Sashital DG, Cornilescu G, McManus CJ, Brow DA, Butcher SE. U2-U6 RNA folding reveals a group II intron-like domain and a four-helix junction. Nat. Struct. Mol. Biol. 2004;11:1237–1242. doi: 10.1038/nsmb863. [DOI] [PubMed] [Google Scholar]
- 11.Hetzer M, Wuzer G, Schweyen R, Mueller MW. Trans-activation of group II intron splicing by nuclear U5 snRNA. Nature. 1997;386:417–420. doi: 10.1038/386417a0. [DOI] [PubMed] [Google Scholar]
- 12.Villa T, Pleiss JA, Guthrie C. Spliceosomal snRNAs: Mg2+-dependent chemistry at the catalytic core? Cell. 2002;109:149–152. doi: 10.1016/s0092-8674(02)00726-2. [DOI] [PubMed] [Google Scholar]
- 13.Toor N, Keating KS, Taylor SD, Pyle AM. Crystal structure of a self-spliced group II intron. Science. 2008;320:77–82. doi: 10.1126/science.1153803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jarrell KA, Dietrich RC, Perlman PS. Group II intron domain 5 facilitates a trans-splicing reaction. Mol. Cell Biol. 1988;8:2361–2366. doi: 10.1128/mcb.8.6.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chin K, Pyle AM. Branch-point attack in group II introns is a highly reversible transesterification, providing a potential proofreading mechanism for 5′-splice site selection. RNA. 1995;1:391–406. [PMC free article] [PubMed] [Google Scholar]
- 16.Sharp PA. “Five easy pieces”. Science. 1991;254:663. doi: 10.1126/science.1948046. [DOI] [PubMed] [Google Scholar]
- 17.L’Homme Y, Stahl RJ, Li XQ, Hameed A, Brown GG. Brassica nap cytoplasmic male sterility is associated with expression of a mtDNA region containing a chimeric gene similar to the pol CMS-associated orf224 gene. Curr. Genet. 1997;31:325–335. doi: 10.1007/s002940050212. [DOI] [PubMed] [Google Scholar]
- 18.Menassa R, L’Homme Y, Brown GG. Post-transcriptional and developmental regulation of a CMS-associated mitochondrial gene region by a nuclear restorer gene. Plant J. 1999;17:491–499. doi: 10.1046/j.1365-313x.1999.00397.x. [DOI] [PubMed] [Google Scholar]
- 19.Vogel J, Hess WR, Börner T. Precise branch point mapping and quantification of splicing intermediates. Nucleic Acids Res. 1997;25:2030–2031. doi: 10.1093/nar/25.10.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuhn J, Binder S. RT-PCR analysis of 5′ to 3′-end-ligated mRNAs identifies the extremities of cox2 transcripts in pea mitochondria. Nucleic Acids Res. 2002;30:439–446. doi: 10.1093/nar/30.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kühn K, Weihe A, Börner T. Multiple promoters are a common feature of mitochondrial genes in Arabidopsis. Nucleic Acids Res. 2005;33:337–346. doi: 10.1093/nar/gki179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malek O, Knoop V. Trans-splicing group II introns in plant mitochondria: the complete set of cis-arranged homologs in ferns, fern allies, and a hornwort. RNA. 1998;4:1599–1609. doi: 10.1017/s1355838298981262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dombrovska O, Qiu YL. Distribution of introns in the mitochondrial gene nad1 in land plants: phylogenetic and molecular evolutionary implications. Mol. Phylogenet. Evol. 2004;32:246–263. doi: 10.1016/j.ympev.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 24.Groth-Malonek M, Pruchner D, Grewe F, Knoop V. Ancestors of trans-splicing mitochondrial introns support serial sister group relationships of hornworts and mosses with vascular plants. Mol. Biol. Evol. 2005;22:117–125. doi: 10.1093/molbev/msh259. [DOI] [PubMed] [Google Scholar]
- 25.Qiu YL, Palmer JD. Many independent origins of trans splicing of a plant mitochondrial group II intron. J. Mol. Evol. 2004;59:80–89. doi: 10.1007/s00239-004-2606-y. [DOI] [PubMed] [Google Scholar]
- 26.Podar M, Chu VT, Pyle AM, Perlman PS. Group II intron splicing in vivo by first-step hydrolysis. Nature. 1998;391:915–918. doi: 10.1038/36142. [DOI] [PubMed] [Google Scholar]
- 27.Vogel J, Börner T. Lariat formation and a hydrolytic pathway in plant chloroplast group II intron splicing. EMBO J. 2002;21:3794–3803. doi: 10.1093/emboj/cdf359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li-Pook-Than J, Bonen L. Multiple physical forms of excised group II intron RNAs in wheat mitochondria. Nucleic Acids Res. 2006;34:2782–2790. doi: 10.1093/nar/gkl328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nicolai M, Duprat A, Sormanji R, Rodriguez C, Roncato MA, Rolland N, Robaglia C. Higher plant chloroplasts import the mRNA coding for the eukaryotic translation initiation factor 4E. FEBS Lett. 2007;581:3921–3926. doi: 10.1016/j.febslet.2007.07.017. [DOI] [PubMed] [Google Scholar]
- 30.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jacquier A, Michel F. Multiple exon binding sites in class II self-splicing introns. Cell. 1987;50:17–29. doi: 10.1016/0092-8674(87)90658-1. [DOI] [PubMed] [Google Scholar]
- 32.Müller MW, Schweyen RJ, Schmelzer C. Selection of cryptic 5’ splice sites by group II intron RNAs in vitro. Nucleic Acids Res. 1988;16:7383–7395. doi: 10.1093/nar/16.15.7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Su LJ, Qin PZ, Michels WJ, Pyle AM. Guiding ribozyme cleavage through motif recognition: the mechanism of cleavage site selection by a group II intron ribozyme. J. Mol. Biol. 2001;306:655–668. doi: 10.1006/jmbi.2000.4323. [DOI] [PubMed] [Google Scholar]
- 34.Malek O, Brennicke A, Knoop V. Evolution of trans-splicing mitochondrial introns in pre-Permian times. Proc. Natl Acad. Sci. USA. 1997;94:553–558. doi: 10.1073/pnas.94.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wikström N, Savolainen V, Chase MW. Evolution of the angiosperms: calibrating the family tree. Proc. Biol. Sci. 2001;268:2211–2220. doi: 10.1098/rspb.2001.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hanson MR, Bentolila S. Interactions of mitochondrial and nuclear genes that affect male gametophyte development. Plant Cell. 2004;16:S154–S169. doi: 10.1105/tpc.015966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Turmel M, Otis C, Lemieux C. The complete mitochondrial DNA sequence of Mesostigma viride identifies this green alga as the earliest green plant divergence and predicts a highly compact mitochondrial genome in the ancestor of all green plants. Mol. Biol. Evol. 2002;19:24–38. doi: 10.1093/oxfordjournals.molbev.a003979. [DOI] [PubMed] [Google Scholar]
- 38.Goldschmidt-Clermont M, Choquet Y, Giriard-Bascou J, Michel F, Schirmer-Rahire M, Rochaix JD. A small chloroplast RNA may be required for trans-splicing in Chlamydomonas reinhardtii. Cell. 1991;65:473–482. doi: 10.1016/0092-8674(91)90415-u. [DOI] [PubMed] [Google Scholar]
- 39.Ferat JL, Michel F. Group II self-splicing introns in bacteria. Nature. 1993;364:358–361. doi: 10.1038/364358a0. [DOI] [PubMed] [Google Scholar]
- 40.Dai L, Zimmerly S. Compilation and analysis of group II intron insertions in bacterial genomes: evidence for retroelement behavior. Nucleic Acids Res. 2002;30:1091–1102. doi: 10.1093/nar/30.5.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koonin EV. The origin of introns and their role in eukaryogenesis: a compromise solution to the introns-early vs introns-late debate? Biol. Direct. 2006;1:22. doi: 10.1186/1745-6150-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Belhocine K, Mak AB, Cousineau B. Trans-splicing versatility of the L1.LtrB group II intron. RNA. 2008;14:1782–1790. doi: 10.1261/rna.1083508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fedorova O, Su LJ, Pyle AM. Group II introns: highly specific nucleases with modular structures and diverse catalytic functions. Methods. 2002;28:323–335. doi: 10.1016/s1046-2023(02)00239-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.