Abstract
The MHC class I-related chain (MIC) A and MICB ligands for the activating receptor NKG2D can be shed from tumor cells, and the presence of these soluble molecules in sera is related with compromised immune response and progression of disease. Recently, thiol disulphide isomerases and members of the ADAM (a disintegrin and metalloproteinase) gene family were identified as key enzymes in mediating MICA/B shedding from cells. Here, we report shedding of the most frequently expressed MICA allele in human populations (MICA*008) into exosomes, small membrane vesicles that are secreted upon fusion with the plasma membrane. Although similar to other MICA/B molecules in the extracellular domain, the predicted transmembrane and cytoplasmic domains of MICA*008 are quite different, and this difference seemed to be critical for the mode of release from tumor cells. Treatment of natural killer (NK) cells with exosomes containing MICA*008 molecules not only triggered downregulation of NKG2D from the cell surface but also provoked a marked reduction in NK cytotoxicity that is independent of NKG2D ligand expression by the target cell. Our findings reveal a mechanism of NK suppression in cancer that may facilitate immune escape and progression.
Keywords: MICA*008, NKG2D, exosomes, inhibition
Introduction
NKG2D is a C-type lectin-like activating receptor expressed on all NK cells, as well as TCRγδ+ and CD8+ TCRαβ+ T cells (1) where it can act to costimulate the activation of naïve T cells (2) and even trigger cytotoxicity in the absence of TCR ligation (3). In humans, NKG2D binds to MHC class I-related chain (MIC) A, MICB, and UL16-binding proteins (ULBPs) whose expression is restricted or absent on normal tissues, but is induced in situations of stress and disease (4). The molecular basis of this altered expression is not well understood, although it has been described that a variety of stresses can upregulate NKG2D ligand (NKG2D-L) expression (5-7). The expression of NKG2D-L has been described in multiple types of tumours (4, 8) and evidence is accumulating that strongly suggests that activation of the immune system, mediated by engagement of NKG2D with its ligands, plays an important role in immunosurveillance of cancer (9, 10). This strong selection pressure appears to have led tumour cells to evolve mechanisms to minimise or avoid the response mediated by NKG2D by shedding NKG2D-L from the cell surface. The shedding of soluble MICA by human tumours not only hinders recognition of the MICA-expressing tumor cells, but also leads to down-regulation of NKG2D expression on circulating CD8 T cells, NK cells, and γδT cells, and so the anti-tumour immune response is impaired (11). Indeed high levels of soluble MIC molecules in sera correlate strongly with poor clinical outcome in patients suffering from various types of cancer, including colon (12) and prostate cancers (13). The mechanisms of shedding of MICA/B molecules have been studied intensively. Proteolytic cleavage of MICA has been shown to depend on the thiol isomerase ERp5 (14) and metalloproteases(15), specifically the ADAM (A Disintegrin And Metalloproteinase) family members, ADAM10 and ADAM17 (also known as TACE, TNF-Alpha-Converting Enzyme) (16, 17). However, no reduction in soluble MICA shedding was observed from HeLa and A375 cells after treatment with metalloproteinase inhibitors (14). One explanation for this discrepancy could be that the extensive polymorphism of MIC molecules affects the shedding process. Here it is interesting to note that the allele of MICA expressed by Hela cells is MICA*008 which is by far the most frequently expressed allele of MICA, with an allele frequency of 21% to 47% in diverse populations (18-22). Strikingly, while MICA*008 is very similar to other MIC molecules in the extracellular domain, the predicted transmembrane (TM) and cytoplasmic domains of this allele are very different. This sequence difference is accompanied by functional differences. Specifically, whilst surface expression of many MICA molecules is blocked in cells infected by Human Cytomegalovirus (HCMV) or Kaposi Sarcoma associated Herpesvirus Virus (KSHV), MICA*008 appears to be resistant to downregulation by these viruses (23, 24).
The transmembrane and cytoplasmic regions of proteins usually play an important role in their trafficking within the cell. Thus, the different MICA alleles might follow different pathways that could affect their mechanism of shedding. We have recently described that some alleles of MICB can be endocytosed, and are found intracellularly in the trans golgi network (TGN) and the endosomal system (25). Late endosomes derive from early endosomes through a maturation process that involves gradual change in content, as well as the incorporation of material from the plasma membrane and TGN-derived transport vesicles. During this process, late endosomes can accumulate up to hundreds of internal vesicles which, after fusion of the organelle with the plasma membrane, can be released to the surrounding media as exosomes [for review (26)]. Here, we describe that full length, and therefore membrane resident, MICA*008 proteins can be released from cells in exosomes. In contrast, the MICA*019 allele, which is almost identical to MICA*008 in the extracellular domains but has transmembrane and cytoplasmic tail domains typical of most MICA molecules, was only found in cell culture supernatant as a soluble truncated molecule. Despite this difference in mechanism of shedding, both soluble MICA*019 and exosomal MICA*008 molecules are able to trigger downregulation of NKG2D from the NK cell surface. In addition, exposure of NK cells to MICA*008 containing exosomes triggers a marked loss of cytotoxic function. Thus, exosome release can be an alternative pathway to metalloprotease-mediated shedding of MICA/B molecules for immune evasion.
Materials and methods
Cells and antibodies
The cell lines Hela (MICA*008/008)(18), HepG2 (MICA*002/009)(24) and MelJuSo were cultured in DMEM supplemented with 10% FCS, L-glutamine and sodium pyruvate. HCT116 cells (MICA*001/009)(27) were cultured in RPMI 1640 medium with the same supplements. CHO cells were cultured in Hams F12 medium supplemented with 10% FCS, L-Gln and pyruvate.
The isolation and culture of human primary polyclonal NK cells, as well as culture of the cell lines 721.221, RPMI-8866 and Daudi were as previously described (28). Influenza specific T-cell lines were derived from peripheral blood lymphocytes of an HLA-A2 positive donor by growth on irradiated autologous B lymphoblasts pulsed with influenza A matrix peptide 58-66 (29). Goat polyclonal antibodies and mouse monoclonal antibody (mAb) specific for MICA/B were purchased from R&D systems, mAb anti-NKG2D from Santa Cruz Biotechnology), mAb MEM-259 (specific for CD63) from Abcam and rabbit anti-caveolin antibody from Transduction labs.
Molecular biology, plasmids and transfection
The MICA*019 expression construct has been described previously (30). The MICA*008 expression plasmid was constructed by subcloning cDNA of full-length MICA*008, a gift from Dr Paul Lehner, into pcDNA3. The 019EC/008TMT chimaera was constructed by excising the extracellular domain of the 008 allele by digestion with Hind III and Dra III and introducing the extracellular domain of 019 (also as a Hind III-Dra III fragment). Each MICA expression plasmid was mixed (9:1 ratio) with a vector conferring resistance to puromycin (31), then CHO cells were transfected with this mixture using Lipofectamine 2000. Stable transfectants were generated by culture of transfected CHO cells in selective medium (8μg/ml puromycin, Calbiochem) and, where necessary, by cell sorting.
Exosome isolation and characterization
Exosomes were isolated by differential centrifugation as described previously (32). Briefly, adherent monolayers were washed extensively with PBS and recultured in fresh medium (no serum) for 24 hours. Cell culture media were centrifuged twice for 10 minutes at 300 xg to remove cell debris, and then centrifuged for 30 minutes at 10,000 xg and 2 hours at 100,000 xg sequentially. Soluble proteins were recovered from the 100,000 xg supernatant by TCA precipitation. The pellets were solubilized in reducing SDS sample buffer and analyzed by Western blot. In some experiments, further purification of exosomes by flotation on a sucrose gradient was performed as described previously (33). Briefly, exosome samples (500 μL) were mixed with 2.5 vol. of 85% (w/v) sucrose in 20 mm Tris/HCl (pH 7.5) containing 150 mm NaCl and 5 mm EDTA (TNE), and placed in centrifuge tubes. The mixtures were layered successively with 4 mL of 60% (w/v), 3 mL of 30% (w/v) and 1 mL of 5% (w/v) sucrose in TNE, and centrifuged at 200 000 xg for 18 h at 4 °C. 1ml fractions were collected from the top to the bottom of the tube. Aliquots of these fractions were analyzed by Western blot.
DRM fractionation
Detergent resistant and detergent soluble membrane fractions were prepared as previously described (16). Western blot was performed using antibodies specific for MICA/B and caveolin. Quantitative analysis of the western blot data was done using Image J software (Rasband, W.S., ImageJ, U. S. National Institutes of Health, Bethesda, Maryland, USA, http://rsb.info.nih.gov/ij/, 1997-2008).
Western blot
Cell lysates were prepared by incubation in TNE buffer containing 1% NP-40 and protease inhibitors. After centrifugation to remove nuclei and cell debris, samples were run on 10% or 12% SDS-PAGE gels (for experiments visualizing CD63 the samples were run under non-reducing conditions) and transferred to Immobilon-P (Millipore). The membrane was blocked using 5% non-fat dry milk in PBS-0.1% Tween-20 (PBS-T), and then specific antigens were detected by incubating the membrane with the indicated first antibody followed by HRP conjugated secondary antibodies. Proteins were visualized using the ECL system (Amersham Pharmacia). In some experiments, samples were treated with peptide N-glycosidase F (PNGase F) (New England Biolabs, Ipswich, MA), according to the manufacturer’s instructions.
Electron microscopy
Electron microscope examination of exosomes was carried out by floating a carbon-coated 400-mesh Formvar EM grid on top of one drop of freshly prepared exosomes (60ug/ml in PBS) for around 1 minute. The grid was then briefly washed with deionised water and floated on a drop of 2% uranyl acetate. Samples were examined using a Philips CM100 operating at 60 or 80 keV.
NKG2D downregulation
1 × 105 IL-2 activated human NK cells, 3 days after stimulation with IL-2 (50U/ml, R&D Systems), were incubated, in 96 well flat-bottomed plates, for 24 hours with supernatants of untransfected CHO cells or CHO cells transfected with either MICA*019 or MICA*008. When using exosome fractions, 40-100 ng of total protein was added to the NK cells. NKG2D surface expression was monitored by staining with mAb specific for NKG2D (clone 1D11, 1μg/105 cells) and flow cytometry using a FACSCan cytometer running Cellquest software (BD Biosciences).
Cytotoxicity assay
Cytotoxicity assays were carried out using a one-step fluorimetric assay based on the use of AlamarBlue (Invitrogen) (34). Effector cells alone, target cells alone and mixes of effectors and target cells at the indicated E:T ratios were incubated with AlamarBlue in 96 well flat-bottomed plates at 37°C in a humidified 5% CO2 incubator overnight. Following the incubation, the fluorescence of the AlamarBlue was read on a Synergy HT plate reader (Biotek) with excitation at 530nm and emission at 590nm at 37°C. The percentage specific lysis was calculated using the following formula:
where AF = absolute fluorescence units.
Results
Biochemical analysis of MIC proteins shed from tumour cell lines
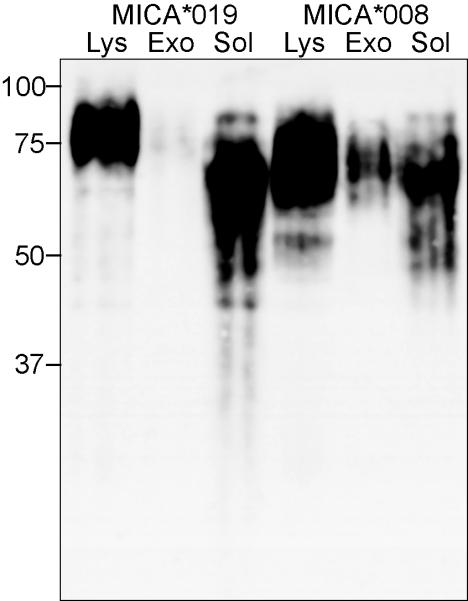
In previous experiments, biochemical analysis of MICA/B molecules shed from transfectants showed that the shed MICB was a truncated species that remained soluble after centrifugation at 100,000 xg (16). We extended this analysis to study the MICA/B molecules shed from a panel of tumour cell lines that express endogenous MICA and MICB molecules (Supplementary Figure 1). It is not possible to distinguish between MICA and MICB in Western Blot experiments due to their high homology, but we could see that soluble MICA/B molecules shed from the colon carcinoma cell line HCT116 had a molecular weight similar to the soluble MIC shed from transfectants (16, 17), corresponding to MIC protein cleaved in the stalk region that connects the α3 and transmembrane domains (Figure 1a). In contrast, analysis of MICA/B molecules released from Hela cells showed that these proteins were not soluble as they pelleted on centrifugation at 100,000 xg. Moreover, their size corresponded to the full length MICA/B protein present in the Hela cell lysate (Figures 1a and b). Hela cells express the MICA*008 allele (18) and very little MICB (Supp. Fig. 1)(25), moroever MICA*008 is distinct from other alleles of MICA due to the insertion of a guanine at position 952 in the transmembrane (TM) region, which results in expression of a protein with a truncated TM region and a very short cytoplasmic tail (35). For this reason, MICA*008 can be easily distinguished from other MICA and MICB molecules by its molecular weight. Analysis of other tumour cell lines showed that while some of the MICA/B molecules released by the cell lines HepG2 and MelJuso were soluble and comparable in size to those shed by the HCT116 cell line, a proportion of the MICA/B molecules shed from the melanoma cell line MelJuSo were present in the 100,000 xg pellet with a molecular weight of 35kDa after N-Glycanase digestion (Fig 1c). Cloning and sequencing experiments confirmed that this cell line expressed MICA*008 (not shown), thus these results demonstrate that different MICA/B molecules can be released as biochemically different species even in the same cell line. In addition, the data show that MICA*008 is preferentially released as a full-length, membrane anchored molecule. The simplest interpretation of these data is that MICA*008 was released in exosomes rather than by proteolysis.
Figure 1. MICA molecules released from Hela pellet at 100,000 xg.
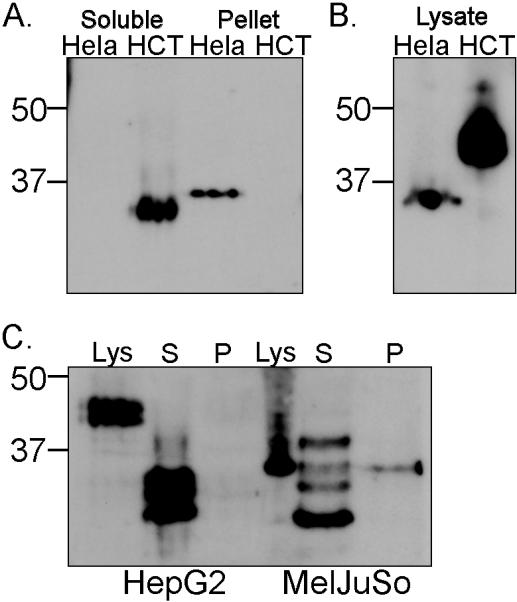
(A.) Supernatants were collected from cells that had been cultured for 16 hours centrifuged to eliminate large membrane fragments and organelles and then fractionated to separate proteins that pellet at 100,000 xg, from soluble proteins (recovered by TCA precipitation). Samples were digested with PNGase F before western blot analysis. (B.) Total cell lysates were analysed for comparison. (C.) Analysis of MICA/B released by the tumour cell lines HepG2 and MelJuSo. Lys – lysate, S – soluble, P – pellet. Data are representative of three experiments. Positions of Mr weight markers (kDa) are indicated.
MICA*008 protein is released from Hela cells in exosomes
Exosomes are bioactive vesicles formed through the fusion of multivesicular bodies (MVBs) with the plasma membrane, leading to the release of 30- to 100-nm lipid bilayer vesicles (36). Physically, exosomes are equivalent to cytoplasm enclosed in a lipid bilayer, with the external domains of transmembrane proteins exposed to the extracellular environment. Although exosome composition varies depending on the cell type of origin, one of the common characteristic features of these vesicles is the presence of tetraspanins such as CD63 (36, 37). The MICA*008 containing pellet obtained by centrifugation at 100,000 xg, but not the supernatant, was enriched for CD63 (Fig 2a). Previous studies (36) have shown that exosomes display equilibrium buoyant densities between 1.10-1.19 g/ml on linear sucrose gradients. So, to confirm that tetraspan proteins and Hela cell MICA*008 localize to membranes with the same characteristics, exosomes isolated from the culture medium by differential centrifugation were floated into a linear sucrose gradient. The distribution of MICA and CD63 in the gradient was analyzed by Western blotting and shown to overlap substantially (Fig. 2B), providing further evidence that MICA*008 was shed from Hela cells in exosomes. This conclusion was strengthened by electron microscopy analysis which showed that the MICA containing 100,000 xg pellet obtained from HeLa culture supernatant was composed of small membrane vesicles with diameters of 50 to 100 nm (Fig. 2C), typical of exosomes.
Figure 2. Exosome preparations from Hela contain MICA and are enriched in the tetraspanin CD63.
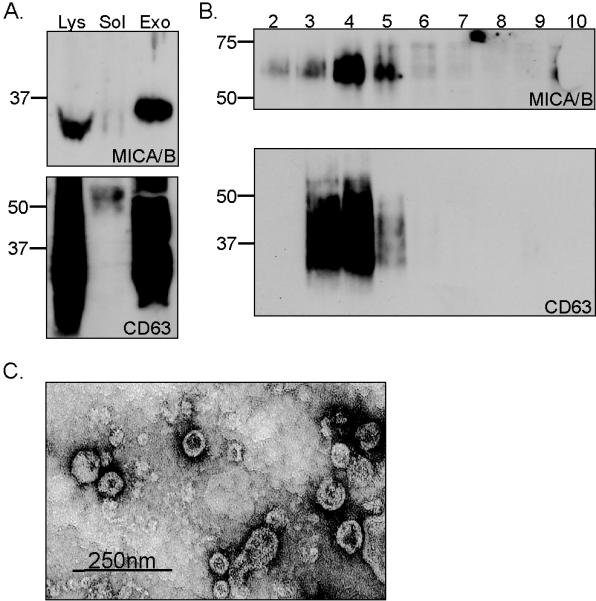
A. Cells were cultured for 16 hours in serum free medium and exosomes were isolated. The fractions corresponding to soluble protein (sol), exosomes (exo) and total cell lysate (lys) were analysed by western blot, probing with either anti-MICA antibody or anti-CD63, as indicated. Samples were digested with PNGase F before western blot analysis for MICA. B. Exosomes were further fractionated on a step sucrose gradient and fractions analysed for MICA and CD63. C. Exosomes were visualised by electron microscopy. Data are representative of two experiments.
Exosome shedding of MICA*008 is an intrinsic property of this allele
Analysis of MICA shedding in CHO cells stably transfected with either the allele MICA*019 or MICA*008 confirmed that whilst MICA*019 was found solely as a truncated molecule in the 100,000 xg supernatant, a considerable proportion of MICA*008 molecules was shed in exosomes (Fig. 3).
Figure 3. MICA*008, but not MICA*019, is released in exosomes.
Exosomes and soluble proteins were isolated from cell free culture supernatants of CHO cells stably transfected with either MICA*019 or MICA*008. The preparations of exosomes and soluble proteins were analysed by western blot (3 experiments).
These data indicated that the incorporation of MICA*008 into exosomes was an intrinsic property of this allele, and not of the cell type in which it is expressed. Nevertheless, some soluble MICA*008 was released from the CHO transfectants. Since soluble MICA*008 is not detected on preparations from Hela cells, which express the protein endogenously, our hypothesis is that detection of the truncated protein is related to the very high level MICA*008 expression by the transfected cells (Supplementary Figure 2a), that is much higher than the levels of expression reported for MICA expression on cell lines and in vivo (38).
MICA*008 is found in membrane microdomains
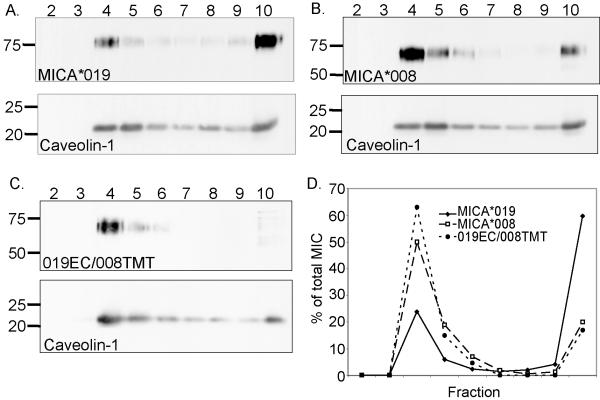
The best characterised mechanism underlying recruitment of proteins to MVBs is monoubiquitination and interaction with the ESCRT machinery (40). However, this route cannot be relevant for incorporation of MICA*008 to exosomes since this protein does not have lysine residues in its cytoplasmic tail. Another route for cargo recruitment to exosomes depends on the intrinsic physical properties of the protein and preferential segregation into raft-like microdomains (41). Thus, we analysed whether MICA*008 was present in detergent-resistant membrane microdomains. In agreement with observations made with MICB (16), approximately 25% of MICA*019 molecules are found in low density detergent insoluble fractions whilst more than 60% of this protein is present in the detergent soluble fractions (Fig. 4A). In contrast, the majority of MICA*008 molecules are present in low density detergent resistant fractions, while only 20% are found at the bottom of the gradient (Fig. 4B). MICA*019 and MICA*008 vary by only one amino-acid in the extracellular domain, in a region distant from the NKG2D binding site (39), but are very different in the TM region and cytoplasmic tail (35)(Supplementary Fig. 2b). This suggested that the TM/cytoplasmic tail of MICA*008 controlled the localisation of MICA*008 to DRMs. This hypothesis was confirmed by the preferential localization in DRMs of a chimaeric protein comprising the extracellular domain of MICA*019 and the transmembrane and cytoplasmic tail of the MICA*008 molecule (Fig. 4C and D). Analysis of culture supernatants of CHO cell transfectants expressing the chimaeric 019EC/008TMT molecule revealed that this molecule was also shed in exosomes (Supplementary Fig. 3).
Figure 4. The majority of MICA*008 is present in detergent resistant membranes (DRMs).
CHO cells transfected with MICA*019 (A), MICA*008 (B), or a chimaeric protein comprising the extracellular domain of MICA*019 fused to the transmembrane and cytoplasmic tail of MICA*008 (019EC/008TMT) (C) were lysed and fractionated by centrifugation on sucrose gradients. Equal volumes of these fractions were analysed, by western blot, for the presence of MICA. Data are representative of at least three experiments. (D) Quantitative analysis to determine the proportion of MICA protein present in each fraction was carried out using Image J software.
Supernatants containing either MICA*019 or MICA*008 can downregulate surface expression of the NKG2D receptor
As well as promoting tumor evasion by reducing the cell surface expression of NKG2D ligands on tumors, the release of soluble MICA has also been reported to trigger a systemic down-regulation of the NKG2D receptor on NK cells and CD8 T cells (11-13). In this context, it was of interest to compare the effect of culture supernatants containing MICA*008 (released in exosomes) or MICA*019 (soluble) on cell surface expression of NKG2D on primary human NK cells. To avoid the confounding effects of factors such as exosomal TGF-β on lymphocyte proliferation and NKG2D expression (42-44), we chose to compare supernatants of MICA*019 or MICA*008 transfectants with supernatants of untransfected CHO cells. A representative example of such an experiment is shown in Figure 5A. These data clearly indicate that both MICA*019 and MICA*008 containing supernatants can decrease cell surface NKG2D expression. Analysis of multiple experiments shows that these effects are statistically significant (Fig. 5B). Interestingly, although the supernatants from CHO cells expressing MICA*008 shed less MICA than MICA*019 expressing cells, the MICA*008 culture supernatant consistently triggered more downregulation of NKG2D (5/5 experiments).
Figure 5. Both soluble and exosomal MICA downmodulate surface expression of NKG2D.
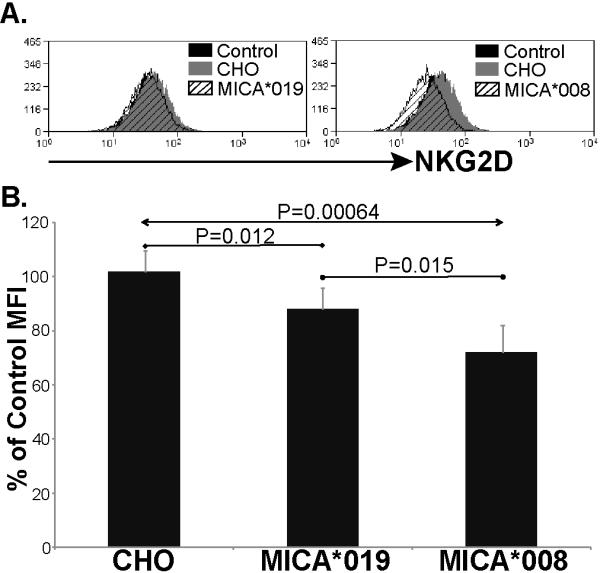
Culture supernatants were collected from CHO cells transfected with MICA*019 or *008. Supernatant from untransfected CHO cells was used as control. A. Supernatant from CHO cells and MICA transfected CHO cells were incubated with IL-2 activated human NK cells for 24 hours. The amount of NKG2D on the surface of the NK cells was then analysed by flow cytometry. The result shown is representative of 5 experiments. B. Downmodulation of NKG2D is significant (Student’s T Test) after treatment with either MICA*019 or *008 containing supernatants. Data are expressed as a percentage of the NKG2D expression observed on NK cells incubated in medium alone.
Exosomal MICA*008 can downmodulate surface expression of NKG2D and compromise NK cell cytotoxicity
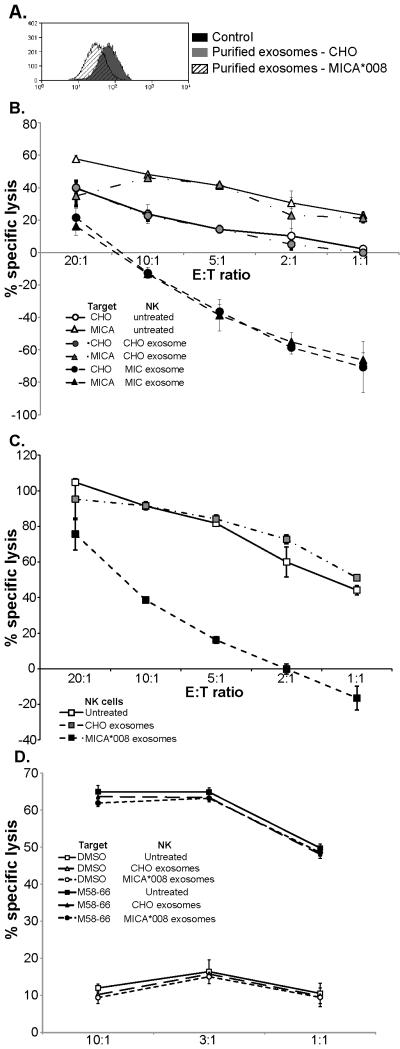
Unlike Hela cells, the culture supernatant of the CHO cells overexpressing MICA*008 contains some soluble MICA*008 protein. Thus, it was important to test whether purified exosomes containing MICA*008 molecules affected cell surface expression of NKG2D on NK cells. Figure 6A shows that exposure to exosomes purified from MICA*008 transfectants, but not from untransfected cells, trigger a marked reduction in cell surface NKG2D on NK cells. Importantly, while pretreatment of NK cells with exosomes prepared from untransfected CHO cells had essentially no effect on the ability of these NK cells to lyse CHO and MICA*019 transfectants, treatment with exosomes containing MICA*008 molecules was associated with a marked reduction in cytotoxicity against both of these target cells (Figure 6B). These observations suggested that treatment with MICA*008 exosomes might trigger a general deficit in NK cell cytotoxic function. To test this idea, the effect of exosome treatment on the ability of NK cells to lyse 721.221 cells was tested, since recognition of this cell line by NK cells is mediated by multiple activating receptors including 2B4 (45), NKp44 and NKp46 (46). Strikingly, pretreatment with MICA*008 exosomes markedly impaired NK cell mediated lysis of 721.221 cells. In the light of these observations, we investigated whether treatment with exosomes would also inhibit immune recognition by antigen-specific CTL expressing NKG2D. While exposure to MICA*008 containing exosomes, but not CHO exosomes, provoked downregulation of CTL surface NKG2D (Supplementary Table 1), this pretreatment had no effect on the ability of these CTL to recognize and specifically lyse peptide-pulsed target cells (Fig 6D).
Figure 6. Exosomal MICA*008 can trigger both NKG2D downmodulation and compromise NK cell cytotoxicity.
Incubation of NK cells with purified exosomes isolated from CHO cells transfected with MICA*008, but not untransfected CHO cells, leads to a marked reduction in NKG2D cell surface expression (A) (4 experiments) and compromised NK cell mediated lysis of parental and MICA transfected CHO cells (B), as well as unrelated “third-party” cells such as 721.221 (C). Note that AlamarBlue measures cell metabolism, thus negative values for percentage specific lysis indicate target cell proliferation. Pre-incubation of CTL specific for the influenza matrix peptide M58-66 bound to HLA-A2, with either control or MICA*08 containing exosomes has no effect on specific cytotoxicity (D) (2 experiments).
Discussion
MICA*008 is the most frequently expressed allele in nearly all populations studied (18-22). Although similar to other MICA molecules in the extracellular domain, MICA*008 is distinct from other alleles of MICA due to a single nucleotide insertion in the TM region, which gives rise to a truncated TM region and cytoplasmic tail (35). Here we show that the particular C-terminus of MICA*008 is associated with a different mechanism of release from the cell surface, and an altered distribution of the protein to microdomains within the plasma membrane. In contrast to other MICA alleles that are shed as truncated soluble species after proteolysis by ADAM17/TACE (16, 17), MICA*008 accumulates in the supernatant as a full-length molecule present in the membrane of exosomes. This observation probably explains why metalloproteinase inhibitors do not inhibit the release of MICA molecules from HeLa cells (14). It is interesting to note that MICA008 could potentially be released as a soluble protein, since CHO cell transfectants with high levels of expression of MICA*008 do release some soluble MIC. However, soluble MICA*008 was not observed in our experiments studying Hela cells suggesting that shedding of soluble MICA*008 is an artifact of the approximately 100-fold higher levels of expression of MICA*008 in the CHO transfectant compared to Hela cells.
The trafficking of MICA*008 into the MVBs (which give rise to exosomes) appears to depend on the truncated C-terminus of this protein since this region confers preferential recruitment of MICA*008 into the particular lipid environment associated with the detergent resistant membranes enriched in exosomes (41). Further studies are underway to characterise the particular physical properties of the MICA*008 protein that are responsible for this pattern of trafficking. However, it is interesting to speculate that this differential trafficking of MICA*008 is related to the resistance of MICA*008 to the HCMV and KSHV proteins that downregulate surface expression of many other MICA alleles (23, 24). These data also emphasize that sequence variation between different MICA and MICB alleles can be associated with marked differences in their function, and confirm that trafficking of MICA/B molecules can have a profound effect on the mechanism of shedding of these molecules (16).
The release of MICA/B molecules from tumour cells is associated with a marked downregulation in cell surface expression of NKG2D on circulating NK and CD8+ T cells. The current data show that incubation with either soluble MICA*019 or exosomal MICA*008 molecules leads to downregulation of cell surface NKG2D expression, suggesting that both species are able to bind the receptor. However, although the culture supernatant of MICA*008 transfectants contains less MICA protein than that of cells transfected with MICA*019 (Figure 3), incubation of NK cells with the MICA*008 containing supernatant triggers significantly more NKG2D downregulation than the MICA*019 culture supernatant (Figure 5). This might be related to differences in how the two MICA species exist in the supernatant: soluble, probably monomeric MICA*019 versus multivalent MICA*008 molecules in exosome membranes. Importantly, purified exosomes containing MICA*008 protein triggered significant downregulation of NK cell surface NKG2D and compromised NK cell function. Strikingly, incubation with exosomes containing MICA*008 also impaired NK cytotoxicity known to be triggered by other activating receptors. The molecular basis of this general loss of NK function is unclear, but this phenomenon is reminiscent of previous descriptions of a global deficit in NK cell cytotoxic function after overnight incubation with NKG2D ligand expressing cells (47). In contrast, exposure of CTL to MICA*008 containing exosomes, although triggering NKG2D downregulation (Supplementary Table 1), did not significantly affect the ability of the CTL to specifically lyse target cells. CTL recognition of the HLA-A2/M58-66peptide complex is mediated by an immunodominant high affinity TCR (48). Thus, it remains possible that treatment with MICA*008 containing exosomes could affect other lower affinity CTL, but it is interesting to note that these data are consistent with previous observations where constitutive Rae-1ε transgene expression triggered local and systemic NKG2D downregulation with generalized defects in NK cell-mediated cytotoxicity, but only mild CD8+ T cell effects (49).
Elevated levels of soluble MICA have been detected in the sera of patients suffering from various types of cancer, including gastrointestinal malignancies, breast and lung tumors, melanoma, prostate cancer, pancreatic carcinomas, hepatocellular cancer and leukemia (11, 50). Moreover, the levels of soluble MICA and MICB in patient sera can be used as diagnostic markers for cancer progression, where elevated levels of these proteins often correlate with a poor prognosis for the patient (50). This relationship is likely to be related to the known effects of MICA/B shedding: a reduction in cell surface density of NKG2D ligands leading to a reduced susceptibility to NKG2D-mediated cytotoxicity and systemic down-regulation of NKG2D on NK cells and CD8 T cells in cancer patients. These considerations suggest that either blockade of MIC release, or neutralisation of shed soluble MIC would be a useful addition to immunological approaches for cancer therapy. In this context, an understanding of the mechanisms involved in the release of soluble MICA/B molecules from tumour cells is crucial for the development of effective strategies to block the shedding of these proteins. ADAM17/TACE has been identified as a key protease mediating proteolytic cleavage of both MICA and MICB molecules (16, 17). However, the data in this manuscript now show that MICA molecules can be released from a tumour cell in more than one way. The MICA*008 molecule, that is by far the most frequent allele of MICA in multiple populations (18-22), is not released from cells by proteolysis, but rather as a membrane-anchored molecule in exosomes. Thus, effective blockade of the accumulation of soluble MICA in patient sera will require different strategies depending on the MICA alleles expressed by the individual patient.
Supplementary Material
Acknowledgements
The authors thank SJ Powis (U. St Andrews), P. Roda-Navarro, Dr. F. Colucci and his group and Dr. A. Kelly for helpful discussions and Prof. J. Trowsdale for critical reading of the manuscript. We would like to thank N. Miller for assistance with cell sorting.
This work was supported by grants from the MRC and the LRF. MV-G is a recipient of a New Investigator Award Grant from the MRC; OA and PB were partially supported by The Newton Trust; SA-G was supported by Fundacion CajaMadrid and Ibercaja.
References
- 1.Bauer S, Groh V, Wu J, et al. Activation of NK cells and T cells by NKG2D, a receptor for stress- inducible MICA. Science. 1999;285:727–9. doi: 10.1126/science.285.5428.727. [DOI] [PubMed] [Google Scholar]
- 2.Maasho K, Opoku-Anane J, Marusina AI, Coligan JE, Borrego F. NKG2D is a costimulatory receptor for human naive CD8+ T cells. J Immunol. 2005;174:4480–4. doi: 10.4049/jimmunol.174.8.4480. [DOI] [PubMed] [Google Scholar]
- 3.Verneris MR, Karami M, Baker J, Jayaswal A, Negrin RS. Role of NKG2D signaling in the cytotoxicity of activated and expanded CD8+ T cells. Blood. 2004;103:3065–72. doi: 10.1182/blood-2003-06-2125. [DOI] [PubMed] [Google Scholar]
- 4.Gonzalez S, Groh V, Spies T. Immunobiology of human NKG2D and its ligands. Curr Top Microbiol Immunol. 2006;298:121–38. doi: 10.1007/3-540-27743-9_6. [DOI] [PubMed] [Google Scholar]
- 5.Groh V, Bahram S, Bauer S, Herman A, Beauchamp M, Spies T. Cell stress-regulated human major histocompatibility complex class I gene expressed in gastrointestinal epithelium. Proc Natl Acad Sci U S A. 1996;93:12445–50. doi: 10.1073/pnas.93.22.12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamamoto K, Fujiyama Y, Andoh A, Bamba T, Okabe H. Oxidative stress increases MICA and MICB gene expression in the human colon carcinoma cell line (CaCo-2) Biochim Biophys Acta. 2001;1526:10–2. doi: 10.1016/s0304-4165(01)00099-x. [DOI] [PubMed] [Google Scholar]
- 7.Gasser S, Orsulic S, Brown EJ, Raulet DH. The DNA damage pathway regulates innate immune system ligands of the NKG2D receptor. Nature. 2005;436:1186–90. doi: 10.1038/nature03884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pende D, Rivera P, Marcenaro S, et al. Major histocompatibility complex class I-related chain A and UL16-binding protein expression on tumor cell lines of different histotypes: analysis of tumor susceptibility to NKG2D-dependent natural killer cell cytotoxicity. Cancer Res. 2002;62:6178–86. [PubMed] [Google Scholar]
- 9.Smyth MJ, Swann J, Cretney E, Zerafa N, Yokoyama WM, Hayakawa Y. NKG2D function protects the host from tumor initiation. J Exp Med. 2005;202:583–8. doi: 10.1084/jem.20050994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guerra N, Tan YX, Joncker NT, et al. NKG2D-deficient mice are defective in tumor surveillance in models of spontaneous malignancy. Immunity. 2008;28:571–80. doi: 10.1016/j.immuni.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Groh V, Wu J, Yee C, Spies T. Tumour-derived soluble MIC ligands impair expression of NKG2D and T-cell activation. Nature. 2002;419:734–8. doi: 10.1038/nature01112. [DOI] [PubMed] [Google Scholar]
- 12.Doubrovina ES, Doubrovin MM, Vider E, et al. Evasion from NK cell immunity by MHC class I chain-related molecules expressing colon adenocarcinoma. J Immunol. 2003;171:6891–9. doi: 10.4049/jimmunol.171.12.6891. [DOI] [PubMed] [Google Scholar]
- 13.Wu JD, Higgins LM, Steinle A, Cosman D, Haugk K, Plymate SR. Prevalent expression of the immunostimulatory MHC class I chain-related molecule is counteracted by shedding in prostate cancer. J Clin Invest. 2004;114:560–8. doi: 10.1172/JCI22206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaiser BK, Yim D, Chow IT, et al. Disulphide-isomerase-enabled shedding of tumour-associated NKG2D ligands. Nature. 2007;447:482–6. doi: 10.1038/nature05768. [DOI] [PubMed] [Google Scholar]
- 15.Salih HR, Rammensee HG, Steinle A. Cutting edge: down-regulation of MICA on human tumors by proteolytic shedding. J Immunol. 2002;169:4098–102. doi: 10.4049/jimmunol.169.8.4098. [DOI] [PubMed] [Google Scholar]
- 16.Boutet P, Aguera-Gonzalez S, Atkinson S, et al. Cutting edge: The metalloproteinase ADAM17/TNF-alpha-converting enzyme regulates proteolytic shedding of the MHC class I-related chain B protein. J Immunol. 2009;182:49–53. doi: 10.4049/jimmunol.182.1.49. [DOI] [PubMed] [Google Scholar]
- 17.Waldhauer I, Goehlsdorf D, Gieseke F, et al. Tumor-associated MICA is shed by ADAM proteases. Cancer Res. 2008;68:6368–76. doi: 10.1158/0008-5472.CAN-07-6768. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, Lazaro AM, Lavingia B, Stastny P. Typing for all known MICA alleles by group-specific PCR and SSOP. Hum Immunol. 2001;62:620–31. doi: 10.1016/s0198-8859(01)00241-5. [DOI] [PubMed] [Google Scholar]
- 19.Marin ML, Savioli CR, Yamamoto JH, Kalil J, Goldberg AC. MICA polymorphism in a sample of the Sao Paulo population, Brazil. Eur J Immunogenet. 2004;31:63–71. doi: 10.1111/j.1365-2370.2004.00446.x. [DOI] [PubMed] [Google Scholar]
- 20.Tian W, Boggs DA, Ding WZ, Chen DF, Fraser PA. MICA genetic polymorphism and linkage disequilibrium with HLA-B in 29 African-American families. Immunogenetics. 2001;53:724–8. doi: 10.1007/s00251-001-0392-x. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y, Lazaro AM, Zou Y, et al. MICA polymorphism in South American Indians. Immunogenetics. 2002;53:900–6. doi: 10.1007/s00251-001-0426-4. [DOI] [PubMed] [Google Scholar]
- 22.Romphruk AV, Naruse TK, Romphruk A, et al. Diversity of MICA (PERB11.1) and HLA haplotypes in Northeastern Thais. Tissue Antigens. 2001;58:83–9. doi: 10.1034/j.1399-0039.2001.580203.x. [DOI] [PubMed] [Google Scholar]
- 23.Zou Y, Bresnahan W, Taylor RT, Stastny P. Effect of human cytomegalovirus on expression of MHC class I-related chains A. J Immunol. 2005;174:3098–104. doi: 10.4049/jimmunol.174.5.3098. [DOI] [PubMed] [Google Scholar]
- 24.Thomas M, Boname JM, Field S, et al. Down-regulation of NKG2D and NKp80 ligands by Kaposi’s sarcoma-associated herpesvirus K5 protects against NK cell cytotoxicity. Proc Natl Acad Sci U S A. 2008;105:1656–61. doi: 10.1073/pnas.0707883105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aguera-Gonzalez S, Boutet P, Reyburn HT, Vales-Gomez M. Brief residence at the plasma membrane of the MHC class I-related chain B is due to clathrin-mediated cholesterol-dependent endocytosis and shedding. J Immunol. 2009;182:4800–8. doi: 10.4049/jimmunol.0800713. [DOI] [PubMed] [Google Scholar]
- 26.Fevrier B, Raposo G. Exosomes: endosomal-derived vesicles shipping extracellular messages. Curr Opin Cell Biol. 2004;16:415–21. doi: 10.1016/j.ceb.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 27.Zwirner NW, Marcos CY, Mirbaha F, Zou Y, Stastny P. Identification of MICA as a new polymorphic alloantigen recognized by antibodies in sera of organ transplant recipients. Hum Immunol. 2000;61:917–24. doi: 10.1016/s0198-8859(00)00162-2. [DOI] [PubMed] [Google Scholar]
- 28.Roda-Navarro P, Vales-Gomez M, Chisholm SE, Reyburn HT. Transfer of NKG2D and MICB at the cytotoxic NK cell immune synapse correlates with a reduction in NK cell cytotoxic function. Proc Natl Acad Sci U S A. 2006;103:11258–63. doi: 10.1073/pnas.0600721103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morrison J, Elvin J, Latron F, et al. Identification of the nonamer peptide from influenza A matrix protein and the role of pockets of HLA-A2 in its recognition by cytotoxic T lymphocytes. EurJImmunol. 1992;22:903–7. doi: 10.1002/eji.1830220404. [DOI] [PubMed] [Google Scholar]
- 30.Vales-Gomez M, Browne H, Reyburn HT. Expression of the UL16 glycoprotein of Human Cytomegalovirus protects the virus-infected cell from attack by natural killer cells. BMC Immunol. 2003;4:4. doi: 10.1186/1471-2172-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de la Luna S, Ortin J. pac gene as efficient dominant marker and reporter gene in mammalian cells. Methods Enzymol. 1992;216:376–85. doi: 10.1016/0076-6879(92)16035-i. [DOI] [PubMed] [Google Scholar]
- 32.Raposo G, Nijman HW, Stoorvogel W, et al. B lymphocytes secrete antigen-presenting vesicles. J Exp Med. 1996;183:1161–72. doi: 10.1084/jem.183.3.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oshima K, Aoki N, Kato T, Kitajima K, Matsuda T. Secretion of a peripheral membrane protein, MFG-E8, as a complex with membrane vesicles. Eur J Biochem. 2002;269:1209–18. doi: 10.1046/j.1432-1033.2002.02758.x. [DOI] [PubMed] [Google Scholar]
- 34.Nociari MM, Shalev A, Benias P, Russo C. A novel one-step, highly sensitive fluorometric assay to evaluate cell-mediated cytotoxicity. J Immunol Methods. 1998;213:157–67. doi: 10.1016/s0022-1759(98)00028-3. [DOI] [PubMed] [Google Scholar]
- 35.Mizuki N, Ota M, Kimura M, et al. Triplet repeat polymorphism in the transmembrane region of the MICA gene: a strong association of six GCT repetitions with Behcet disease. Proc Natl Acad Sci U S A. 1997;94:1298–303. doi: 10.1073/pnas.94.4.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stoorvogel W, Kleijmeer MJ, Geuze HJ, Raposo G. The biogenesis and functions of exosomes. Traffic. 2002;3:321–30. doi: 10.1034/j.1600-0854.2002.30502.x. [DOI] [PubMed] [Google Scholar]
- 37.Escola JM, Kleijmeer MJ, Stoorvogel W, Griffith JM, Yoshie O, Geuze HJ. Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J Biol Chem. 1998;273:20121–7. doi: 10.1074/jbc.273.32.20121. [DOI] [PubMed] [Google Scholar]
- 38.Groh V, Rhinehart R, Secrist H, Bauer S, Grabstein KH, Spies T. Broad tumor-associated expression and recognition by tumor-derived gamma delta T cells of MICA and MICB. Proc Natl Acad Sci U S A. 1999;96:6879–84. doi: 10.1073/pnas.96.12.6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li P, Morris DL, Willcox BE, Steinle A, Spies T, Strong RK. Complex structure of the activating immunoreceptor NKG2D and its MHC class I-like ligand MICA. Nat Immunol. 2001;2:443–51. doi: 10.1038/87757. [DOI] [PubMed] [Google Scholar]
- 40.Hicke L, Dunn R. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu Rev Cell Dev Biol. 2003;19:141–72. doi: 10.1146/annurev.cellbio.19.110701.154617. [DOI] [PubMed] [Google Scholar]
- 41.de Gassart A, Geminard C, Fevrier B, Raposo G, Vidal M. Lipid raft-associated protein sorting in exosomes. Blood. 2003;102:4336–44. doi: 10.1182/blood-2003-03-0871. [DOI] [PubMed] [Google Scholar]
- 42.Castriconi R, Cantoni C, Della Chiesa M, et al. Transforming growth factor beta 1 inhibits expression of NKp30 and NKG2D receptors: consequences for the NK-mediated killing of dendritic cells. Proc Natl Acad Sci U S A. 2003;100:4120–5. doi: 10.1073/pnas.0730640100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clayton A, Mitchell JP, Court J, Mason MD, Tabi Z. Human tumor-derived exosomes selectively impair lymphocyte responses to interleukin-2. Cancer Res. 2007;67:7458–66. doi: 10.1158/0008-5472.CAN-06-3456. [DOI] [PubMed] [Google Scholar]
- 44.Clayton A, Mitchell JP, Court J, Linnane S, Mason MD, Tabi Z. Human tumor-derived exosomes down-modulate NKG2D expression. J Immunol. 2008;180:7249–58. doi: 10.4049/jimmunol.180.11.7249. [DOI] [PubMed] [Google Scholar]
- 45.Bottino C, Augugliaro R, Castriconi R, et al. Analysis of the molecular mechanism involved in 2B4-mediated NK cell activation: evidence that human 2B4 is physically and functionally associated with the linker for activation of T cells. Eur J Immunol. 2000;30:3718–22. doi: 10.1002/1521-4141(200012)30:12<3718::AID-IMMU3718>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 46.Vitale M, Bottino C, Sivori S, et al. NKp44, a novel triggering surface molecule specifically expressed by activated Natural Killer cells, is involved in non-major histocompatibility-restricted tumour cell lysis. J Exp Med. 1998;187:2065–72. doi: 10.1084/jem.187.12.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coudert JD, Scarpellino L, Gros F, Vivier E, Held W. Sustained NKG2D engagement induces cross-tolerance of multiple distinct NK cell activation pathways. Blood. 2008;111:3571–8. doi: 10.1182/blood-2007-07-100057. [DOI] [PubMed] [Google Scholar]
- 48.Moss PAH, Moots RJ, Rosenberg WMC, et al. Extensive conservation of a and b chains of the human T-cell antigen receptor recognizing HLA-A2 and influenza A matrix peptide. Proc Natl Acad Sci USA. 1991;88:8987–90. doi: 10.1073/pnas.88.20.8987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oppenheim DE, Roberts SJ, Clarke SL, et al. Sustained localized expression of ligand for the activating NKG2D receptor impairs natural cytotoxicity in vivo and reduces tumor immunosurveillance. Nat Immunol. 2005;6:928–37. doi: 10.1038/ni1239. [DOI] [PubMed] [Google Scholar]
- 50.Salih HR, Holdenrieder S, Steinle A. Soluble NKG2D ligands: prevalence, release, and functional impact. Front Biosci. 2008;13:3448–56. doi: 10.2741/2939. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.