Abstract
Synonymous codon usage of protein coding genes of thirty two completely sequenced mycobacteriophage genomes was studied using multivariate statistical analysis. One of the major factors influencing codon usage is identified to be compositional bias. Codons ending with either C or G are preferred in highly expressed genes among which C ending codons are highly preferred over G ending codons. A strong negative correlation between effective number of codons (Nc) and GC3s content was also observed, showing that the codon usage was effected by gene nucleotide composition. Translational selection is also identified to play a role in shaping the codon usage operative at the level of translational accuracy. High level of heterogeneity is seen among and between the genomes. Length of genes is also identified to influence the codon usage in 11 out of 32 phage genomes. Mycobacteriophage Cooper is identified to be the highly biased genome with better translation efficiency comparing well with the host specific tRNA genes.
1. Introduction
The genetic code uses 64 codons to represent the 20 standard amino acids and the translation termination signal. Each codon is recognized by a subset of a cell's transfer ribonucleotide acid molecules (tRNAs), and with the exception of a few codons that have been reassigned in some lineages [1, 2] the genetic code is remarkably conserved, although it is still in a state of evolution [3].
Codons can be grouped into 20 disjoint families and each family in the universal genetic code contains between 1 and 6 codons. The usage of alternate synonymous codons in an organism is understood to be nonrandom. Grantham et al. [4, 5] proposed that each genome has a particular codon usage signature that reflects particular evolutionary forces acting within that genome. The synonymous codons and amino acids are not used at equal frequencies both within and between organisms [5–7]; the patterns of codon usage vary considerably among organisms, and also among genes from the same genome [8].
Several factors such as directional mutational bias [9–11], translational selection [11–15], secondary structure of proteins [16–21], replicational and transcriptional selection [22, 23], and environmental factors [24, 25] have been reported to influence the codon usage in various organisms. In contrast, amino acid usage has been shown to be influenced by factors such as hydrophobicity, aromaticity, cysteine residue (Cys) content, and mean molecular weight (MMW) [24, 26–30]. Compositional constraints and translational selection are thought to be the two main factors for the codon usage variation among the genes in and across genomes. Compositional bias shapes the codon usage variation among the genes in the extremely AT or GC rich unicellular organisms [31–33]. Analysis of codon usage pattern can provide a basis for understanding the relevant mechanism for biased usage of synonymous codons [34] and has both practical and theoretical importance in understanding the basics of molecular biology [7, 35–39].
Bacteriophages generally use the translational machinery of their hosts to synthesize both their structural and regulatory proteins. This indicates that the amount of codon usage in the protein coding genes in the phages and their bacterial hosts should be similar.
Mycobacteriophages have the potential to be used in diagnosis of tuberculosis and as molecular tools to study mycobacteria. Understanding the codon usage pattern of these phages should guide in the selection of appropriate ones for such purposes. The present venture is to study and understand codon usage patterns of all the mycobacteriophages so far sequenced.
Codon usage analysis was previously done for fourteen phages by Sau et al. [40]. Eighteen more mycobacteriophage genomes were subsequently sequenced and became available in Genbank. In the present work we have analysed all these 32 phage genomes to study and compare their codon usage pattern. Other codon usage indices that affect the genomes of these phages are also studied.
2. Materials and Methods
2.1. Sequences
The complete genome sequences of 32 mycobacteriophages were downloaded from GenBank. Genes having more than 100 codons with proper start and stop codons and without any intermediate stop codon were selected for the current study.
2.2. Analysis
Numbers of codons (Ncs), Relative Synonymous Codon Usage (RSCU), and GC composition at every position of codons were calculated for each gene. The analysis was carried out by GCUA [41], CODONW 1.4.2 (http://codonw.sourceforge.net/).
Nc, the “effective number of codons” used in a gene measures the bias away from equal usage of codons within synonymous groups [19]. Nc can take values from 20 to 61, when only one codon or all synonyms in equal frequencies were used per amino acid, respectively. Nc appears to be a good measure of general codon usage bias [19, 42]. The sequences in which Nc values are <30 are highly expressed while those with >55 are poorly expressed genes [12, 43].
Relative Synonymous Codon Usage. Relative synonymous codon usage (RSCU) is defined as the ratio of the observed frequency of codons to the expected frequency if all the synonymous codons for those amino acids are used equally [21]. RSCU is used to observe the synonymous codon usage variation among the genes.
Base composition. The frequency of A, T, G, C, and GC at first, second, and third positions of synonymously variable sense codons which can potentially vary from 0 to 1 was calculated. The variation of GC3s among genes was characterized by its standard deviation.
2.3. Statistical Methods
Correspondence analysis (CA) is used to study the codon usage variation between genes in different organisms in which the data are plotted in a multidimensional space of 59 axes excluding those of Met, Trp, and stop codons [19]. For understanding the codon usage variation of mycobacteriophages chosen for the current study, RSCU values are used for CA in order to minimize the amino acid composition. To investigate the difference between high and low expressed genes, we have compared the codon usage variation between 10% of the genes located at the extreme right of axis 1 and 10% of the genes located at the extreme left of the axis 1 produced by CA using RSCU. To estimate the codon usage variation between these two sets of genes we have performed Chi square tests taking P < .01 as significant criterion.
The Pearson correlation coefficient and linear regression were calculated to identify the indices that influence the codon usage variation in mycobacteriophages using SPSS version 10.0. The levels of statistical significance were defined as P < .01 or P < .05.
3. Results and Discussion
3.1. Overall Codon Usage Analysis in Mycobacteriophages
The RSCU values of 32 mycobacteriophage genomes show that G- and/or C-ending codons are predominantly used (Table 1), in which 13 are C-ending and 6 are G-ending codons. This was expected, as these phages have a high genomic content. However, from the overall RSCU values, it can be assumed that compositional constraint is the only factor responsible for shaping the codon usage variation among the genes in these genomes. Although the overall RSCU values could unveil the codon usage pattern for the genomes, it may hide the codon usage variation among different genes in a genome.
Table 1.
Relative synonymous codon usage for all the genes of 32 mycobacteriophages was calculated. AA and N are the amino acid and number of codons, respectively.
| AA | Condon | N | RSCU | AA | Condon | N | RSCU |
|---|---|---|---|---|---|---|---|
| Phe | UUU | 1795 | (0.20) | Ser | UCU | 2127 | (0.40) |
| UUC | 15765 | (1.80) | UCC | 7032 | (1.32) | ||
| Leu | UUA | 195 | 0.03 | UCA | 1889 | (0.35) | |
| UUG | 5037 | (0.66) | UCG | 10888 | (2.04) | ||
| Tyr | UAU | 2447 | (0.31) | Cys | UGU | 874 | (0.33) |
| UAC | 13288 | (1.69) | UGC | 4496 | (1.67) | ||
| ter | UAA | 393 | (0.00) | Ter | UGA | 1365 | (0.00) |
| ter | UAG | 388 | (0.00) | Trp | UGG | 11727 | (1.00) |
| Leu | CUU | 2805 | (0.37) | Pro | CCU | 3503 | (0.42) |
| CUC | 13867 | (1.82) | CCC | 10146 | (1.21) | ||
| CUA | 1400 | (0.18) | CCA | 2630 | (0.31) | ||
| CUG | 22302 | (2.93) | CCG | 17369 | (2.06) | ||
| His | CAU | 2427 | (0.40) | Arg | CGU | 5190 | (0.83) |
| CAC | 9611 | (1.60) | CGC | 16237 | (2.60) | ||
| Gln | CAA | 3076 | (0.30) | CGA | 3195 | (0.51) | |
| CAG | 17766 | (1.70) | CGG | 9987 | (1.60) | ||
| Ile | AUU | 3524 | (0.40) | Thr | ACU | 2990 | (0.32) |
| AUC | 21992 | (2.52) | ACC | 20215 | (2.20) | ||
| AUA | 636 | (0.07) | ACA | 2910 | (0.32) | ||
| Met | AUG | 11923 | (1.00) | ACG | 10698 | (1.16) | |
| Asn | AAU | 2886 | (0.29) | Ser | AGU | 1693 | (0.32) |
| AAC | 16738 | (1.71) | AGC | 8424 | (1.58) | ||
| Lys | AAA | 3185 | (0.29) | Arg | AGA | 692 | (0.11) |
| AAG | 18997 | (1.71) | AGG | 2110 | (0.34) | ||
| Val | GUU | 4218 | (0.40) | Ala | GCU | 7738 | (0.50) |
| GUC | 18361 | (1.73) | GCC | 26730 | (1.72) | ||
| GUA | 1855 | (0.17) | GCA | 6591 | (0.42) | ||
| GUG | 17981 | (1.70) | GCG | 21142 | (1.36) | ||
| Asp | GAU | 8082 | (0.42) | Gly | GGU | 10057 | (0.78) |
| GAC | 30303 | (1.58) | GGC | 27920 | (2.17) | ||
| Glu | GAA | 8983 | (0.50) | GGA | 5241 | (0.41) | |
| GAG | 26657 | (1.50) | GGG | 8345 | (0.65) |
3.2. Codon Usage Variation in 32 Mycobacteriophages
The codon usage bias in the coding regions of 32 completely sequenced mycobacteriophages of varying G + C content has been investigated. The average values of the effective numbers of codon (Nc) in different mycobacteriophages varied from 31.44 to 47.96 in mycobacteriophage Cooper and mycobacteriophage Barnyard, respectively. Nucleotide usage pattern in third codon position of all the mycobacteriophages showed high codon usage variation (Table 2). The average GC3s values for individual genomes varied from 65.84 to 89.35 in mycobacteriophage Barnyard and mycobacteriophage Cooper, respectively. In addition, there are marked intragenomic variations in Nc and GC3s values with standard deviation of >3.5 in both the indices. There seems to be a considerable heterogeneity in compositional bias and codon usage pattern within and among the genome of these phages. Of the 32 mycobacteriophages, the genome of Cooper is identified to have the lowest Nc and the highest GC3s values while Barnyard has the highest Nc and the lowest GC3s values indicating that highly GC rich genomes are more biased than poor GC rich genomes.
Table 2.
Nc and GC3s values for 32 mycobacteriophages with standard deviation within brackets.
| Phages | Nc | GC3s |
|---|---|---|
| 244 | 38.98 (6.96) | 80.65 (7.87) |
| Bxb1 | 40.17 (7.28) | 80.05 (7.334) |
| Bxz2 | 37.93 (5.70) | 82.72 (6.46) |
| Che9c | 39.48 (6.96) | 81.01 (7.842) |
| Rosebush | 32.53 (4.04) | 88.44 (4.27) |
| Omega | 43.91 (6.54) | 75.49 (7.01) |
| Halo | 37.87 (5.24) | 82.46 (5.59) |
| Barnyard | 47.96 (7.24) | 65.84 (8.87) |
| Bxz1 | 35.80 (5.64) | 85.09 (6.48) |
| Cjw1 | 38.80 (6.40) | 80.64 (7.83) |
| Corndog | 37.87 (6.69) | 82.65 (7.23) |
| Orion | 37.79 (5.55) | 82.85 (5.17) |
| Plot | 44.28 (5.69) | 73.35 (6.59) |
| Llij | 43.52 (6.18) | 72.23 (5.51) |
| Pipefish | 35.12 (3.55) | 87.35 (3.53) |
| PMC | 42.62 (6.28) | 71.95 (5.30) |
| Qyrzula | 32.45 (3.93) | 88.56 (4.37) |
| Wildcat | 47.62 (6.63) | 66.37 (6.29) |
| D29 | 37.83 (5.28) | 82.58 (5.59) |
| L5 | 40.67 (5.70) | 80.79 (5.86) |
| PBI1 | 44.07 (5.67) | 73.71 (6.63) |
| PG1 | 37.59 (5.20) | 82.7 (4.90) |
| Cooper | 31.44 (4.65) | 89.35 (5.90) |
| Che12 | 39.06 (4.83) | 81.94 (5.75) |
| Catera | 35.44 (5.66) | 85.53 (6.24) |
| TM4 | 34.07 (3.60) | 88.19 (4.64) |
| Che8 | 43.62 (6.33) | 70.57 (5.37) |
| Tweety | 42.76 (5.81) | 72.07 (5.51) |
| U2 | 39.58 (7.03) | 81.10 (7.26) |
| Bethlehem | 40.22 (6.98) | 80.35 (6.57) |
| Giles | 36.48 (5.04) | 83.38 (5.24) |
| Che9d | 44.10 (7.13) | 71.6 (5.8) |
In unicellular organisms, a strong correlation between gene expressivity and the extent of codon usage bias is reported for Escherichia coli and Saccharomyces cerevisiae and phages of Staphycoccus aureus and mycobacteria [13, 40, 44–48]. Our analysis reveals that the genome of mycobacteriophage Cooper is highly biased than other 31 mycobacteriophage genomes. Based on the comparison of the highly represented codons of cooper and the copy number of host specific tRNA, the data indicate that the putatively highly expressed genes of this phage have better translational efficiency comparatively and may be expressed rapidly by the host translation machinery.
Table 3 represents the base composition for the 32 completely sequenced mycobacteriophages. GC composition at the third codon position is always higher than the second and first + second codon positions observed in other GC-rich genomes [9–11]. It is also identified that there is major variation in GC3s content among the genomes studied with no major variation in GC1s and GC2s. This suggests that GC3s has a major role to play than GC1s and GC2s and is tightly associated with the codon usage bias of these genomes.
Table 3.
GC % and base composition in the third codon position for 32 mycobacteriophage genomes.
| Virus | G + C | GC1s | GC2s | GC (1st + 2nd) | GC3s | C3s | T3s | A3s | G3s |
|---|---|---|---|---|---|---|---|---|---|
| 244 | 63.26 | 63.3 | 44.7 | 54 | 81 | 51.14 | 10.58 | 8.88 | 30.01 |
| Bxb1 | 63.43 | 63.4 | 45.7 | 54.55 | 80.4 | 44.89 | 11.53 | 8.53 | 35.63 |
| Bxz2 | 64.41 | 64 | 45.6 | 54.8 | 83 | 48.62 | 9.12 | 8.25 | 34.56 |
| Che9C | 65.66 | 66 | 48.9 | 57.45 | 81.4 | 47.27 | 10.17 | 8.9 | 34.16 |
| Rosebush | 69.16 | 68.3 | 49.8 | 59.05 | 88.5 | 51.21 | 6.26 | 5.36 | 37.73 |
| Omega | 61.48 | 62.4 | 45.1 | 53.75 | 76.1 | 45.43 | 13.88 | 10.78 | 30.52 |
| Halo | 63.79 | 63.4 | 44.5 | 53.95 | 82.7 | 47.4 | 9.57 | 8.06 | 35.5 |
| Barnyard | 58.05 | 60.9 | 45.5 | 53.2 | 67 | 38.55 | 18.39 | 15.97 | 27.68 |
| Bxz1 | 65.04 | 64.1 | 44.9 | 54.5 | 85.3 | 51.84 | 9.61 | 5.39 | 33.72 |
| Cjw1 | 63.52 | 63.7 | 45.1 | 54.4 | 81 | 51.39 | 10.2 | 9.27 | 29.76 |
| Corndog | 65.64 | 64.8 | 48.4 | 56.6 | 83 | 46.49 | 9.76 | 7.7 | 36.64 |
| Orion | 66.9 | 66.6 | 50.2 | 58.4 | 83.17 | 50.46 | 10.19 | 7.04 | 32.85 |
| Plot | 60.22 | 61.6 | 44.3 | 52.95 | 74.1 | 42.4 | 15.73 | 11.05 | 31.34 |
| Llij | 61.66 | 62.4 | 48.9 | 55.65 | 73.1 | 39.73 | 16.12 | 11.79 | 32.87 |
| Pipefish | 67.64 | 65.4 | 49.3 | 57.35 | 87.5 | 49.1 | 7.19 | 5.51 | 38.72 |
| PMC | 61.46 | 62.8 | 48.2 | 55.5 | 72.8 | 39.67 | 15.81 | 12.38 | 32.66 |
| Qyrzula | 69.14 | 68.1 | 49.8 | 58.95 | 88.6 | 51.07 | 6.08 | 5.41 | 38 |
| Wildcat | 57.03 | 59.8 | 42.8 | 51.3 | 67.7 | 35.37 | 22.27 | 11.57 | 31.45 |
| D29 | 63.78 | 63.4 | 44.4 | 53.9 | 82.9 | 47.52 | 9.51 | 8 | 35.49 |
| L5 | 62.49 | 62.1 | 43.5 | 52.8 | 81.2 | 47.45 | 10.01 | 9.3 | 33.79 |
| PBI1 | 60.28 | 61.4 | 44.3 | 52.85 | 74.5 | 42.71 | 15.41 | 10.99 | 31.37 |
| PG1 | 66.83 | 66.5 | 50.2 | 58.35 | 83.12 | 50.46 | 10.24 | 7.07 | 32.79 |
| Cooper | 69.26 | 67.6 | 50 | 58.8 | 89.47 | 52.65 | 6.32 | 4.38 | 37.19 |
| Che12 | 63.26 | 62.5 | 44.4 | 53.45 | 82.25 | 48.61 | 9.86 | 8.3 | 33.79 |
| Catera | 65.17 | 64.1 | 44.9 | 54.5 | 85.8 | 52.29 | 9.44 | 5.1 | 33.71 |
| Che8 | 61.41 | 63.71 | 48.6 | 56.16 | 70.57 | 37.08 | 16.05 | 12.52 | 34.32 |
| TM4 | 68.7 | 68.3 | 48.9 | 58.6 | 88.19 | 47.26 | 6.66 | 5.08 | 40.99 |
| Che9d | 61.4 | 63.02 | 48.15 | 55.59 | 71.6 | 37.07 | 15.54 | 11.94 | 35.41 |
| Bethlehem | 63.29 | 63.37 | 45.22 | 54.30 | 80.35 | 42.56 | 11.32 | 7.93 | 38.15 |
| Giles | 67.89 | 66.55 | 52.72 | 59.64 | 83.38 | 48.11 | 6.26 | 10.06 | 35.55 |
| Tweety | 61.83 | 63.12 | 48.89 | 56.01 | 72.07 | 38.77 | 14.85 | 12.15 | 34.2 |
| U2 | 63.87 | 63.61 | 45.92 | 54.77 | 81.1 | 43.53 | 10.68 | 7.83 | 37.94 |
3.3. Synonymous Codon Usage in Different Mycobacteriophages
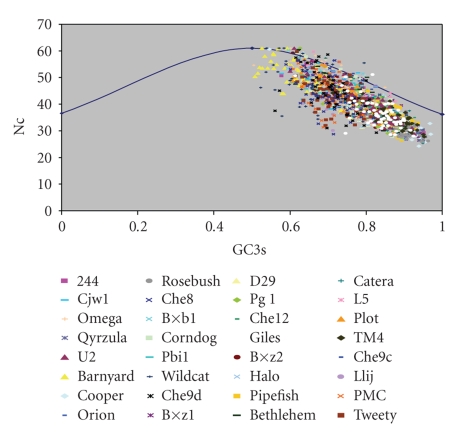
A plot of Nc versus GC3s (Nc plot) has been widely used to study the codon usage variation among genes in different organisms [19]. It was demonstrated that the comparison of the actual distribution of the genes, with the expected distribution under no selection, indicates that the codon usage bias of the genes has influences other than the compositional bias. In contrast, if GC3s were the only determinants of the codon usage variation among the genes, then the values of Nc would fall on the continuous curve between Nc and GC3s.
It is evident from Nc plot for the mycobacteriophages studied that most of the genes fall within a restricted cloud, at GC3s between 0.65 and 0.93, and Nc values 28 and 47 (Figure 1). Nc values for these genes lie just below the expected curve, indicating that these genes have additional codon usage bias apart from compositional bias. The rest of the genes have higher Nc values and lower GC3s values, mostly lying on and close to the expected curve. Consequently, the Nc values of these genes are substantially higher relative to expected values. However, strong influence of compositional constraints on codon usages bias in all the phages analyzed could be understood from the presence of significant negative correlation between GC3s and Nc (r = −0.969; P < .01).
Figure 1.
Nc plot of thirty two mycobacteriophages. The genes for individual phages are represented by different colors.
3.4. Differential Base Usage in Third Codon Position
The correlation of the frequencies of four bases in the third position against Nc values of different genes of these 32 mycobacteriophages has been estimated (Table 4). As there is no information of gene expression level of mycobacteriophages so far, we have considered the highly biased genes having low Nc values as those highly expressed and vice versa.
Table 4.
Correlation coefficient of Nc with C3s, T3s, A3s, and G3s base composition.
| NC | ||||
|---|---|---|---|---|
| Phage name | C3s | T3s | A3s | G3s |
| 244 | −.814** | .816** | .655** | −0.126NS |
| Bxb1 | −.509** | .727** | .867** | −.532** |
| Bxz2 | −.631** | .636** | .829** | −.315* |
| Che9C | −.705** | .829** | .831** | −0.19NS |
| Rosebush | −.599** | .688** | .609** | −0.063NS |
| Omega | −.714** | .635** | .618** | −0.141NS |
| Halo | −.564** | .394** | .690** | −0.129NS |
| Barnyard | −.641** | .694** | .796** | −.621** |
| Bxz1 | −.742** | .725** | .777** | −0.123NS |
| Cjw1 | −.808** | .774** | .629** | −0.104NS |
| Corndog | −.484** | .788** | .621** | −.320** |
| Orion | −.817** | .726** | .673** | 0.147NS |
| Plot | −.666** | .709** | .717** | −.401** |
| Llij | −.278* | .348** | 0.212NS | −0.041NS |
| Pipefish | −.372** | .527** | .567** | −0.085NS |
| PMC | −.442** | .409** | −0.047 | 0.247NS |
| Qyrzula | −.657** | .743** | .665** | −0.048NS |
| Wildcat | −0.216NS | 0.069NS | .487** | −.274* |
| D29 | −.564** | .391** | .690** | −0.13NS |
| L5 | −.642** | .668** | .735** | −0.182NS |
| PBI1 | −.662** | .718** | .702** | −.387** |
| PG1 | −.815** | .735** | .616** | 0.214NS |
| Cooper | −.672** | .806** | .844** | −0.165NS |
| Che12 | −.711** | .708** | .628** | −0.115NS |
| Catera | −.716** | .768** | .702** | −0.125NS |
| TM4 | −.575** | .635** | .601** | −0.021NS |
| Bethlehem | −.583** | .700** | .758** | −.468** |
| Che8 | −.404** | .410** | 0.036NS | 0.109NS |
| Tweety | −.359** | .353** | 0.106NS | 0.088NS |
| Giles | −.535** | .502** | .406** | 0.158NS |
| U2 | −.514** | .783** | .880** | −.544** |
| Che9d | −0.102NS | 0.085 | .401** | −0.194NS |
Notable significant relationships are marked by **P < .01 or *P < .05, NSNonsignificant.
In all mycobacteriophages analyzed, except Che9d, the frequency of C at the third codon position increases with decreasing Nc values, whereas frequencies of T and A increase with Nc. However, the frequency of G is not influenced in the third codon position excluding for few phages such as Bxb1, Barnyard, Corndog, Plot, PBI1, and Bethelhem. Thus the influence of mutational bias of these phages is reflected in the choice of bases in the third position. However, this is expected since the optimal codons are, in general, chosen in accordance with the mutational bias of these phages. In other words, it is due to the translational selection that the mutational bias appears to be more prominent in the third codon position of highly expressed genes [18].
3.5. Effect of Translational Selection on the Synonymous Codon Usage in Mycobacteriophage Genomes
Some of the earlier reports showed that synonymous codon usage in the highly expressed genes of diverse array of organisms is influenced by cellular tRNA abundance [44, 49–52]. Kanaya et al. [52, 53] have reported that the cellular tRNA abundance in several organisms are directly proportional to their tRNA copy number.
Of the 32 mycobacteriophages analyzed, 10 phages (244 (2 tRNA genes), Bxz1 (26 tRNA genes), Omega (2 tRNA genes), Cjw1 (1 tRNA gene), Wildcat (21 tRNA genes), D29 (5 tRNA genes), L5 (3 tRNA genes), Che12 (3 tRNA genes), Catera (26 tRNA genes)) encode tRNA genes for few of the amino acids. Both Bxz1 and Catera are identified to encode large number of tRNA genes (26 tRNA genes) and Wildcat encoding for 21 tRNA genes. Of the 10 phage genomes encoding tRNA genes, excluding 244 and Omega, eight carry tRNA genes for the overrepresented codons in highly and lowly expressed genes.
To see whether the synonymous codon usage of putatively highly expressed genes of these mycobacteriophage genomes is positively correlated with the host tRNA abundance, the number of over represented synonymous codons in such genes was determined by comparing with that of the putatively lowly expressed genes. It was found that among the 22 overrepresented synonymous codons in highly expressed genes, 21 codons are recognized by the M.tuberculosis specific tRNAs (data not shown). Based on the above analysis, the data indicate that the putatively highly expressed genes of these phages have translational efficiency.
3.6. Relationship between Codon Bias and Gene Length
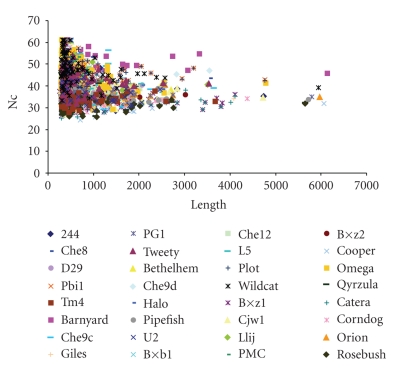
Selection for translational accuracy is predicted to have a positive correlation between codon bias and gene length [20]. Previously, the relationship between gene length and synonymous codon usage bias has been reported for Drosophila melanogaster, Escherichia coli, Saccharomyces cerevisiae, Pseudomonas aeruginosa, and Yersinia pestis [11, 15]. From the plot drawn with gene length against Nc (Figure 2), it is understood that shorter genes have a much wider variance in Nc values, and vice versa for longer genes. Lower Nc values in longer genes may be due to the direct effect of translation time on fitness or to the extra energy cost of proofreading associated with longer translating time. A significant correlation was identified in 11 phages revealing that gene length influences codon usage of these genomes (Table 5). Similar results were also reported for S. pneumoniae, P. aeruginosa and SARS coronavirus [11, 16]. Eyre-Walker [20] has reported that the selection for fidelity in protein translation is likely to be greater in longer genes because the cost of producing a protein is proportional to its length. Therefore selection of translational accuracy predicts a positive correlation between codon usage bias and gene length. And this selection may be stronger at constrained codons coding for evolutionarily conserved amino acids than the nonconserved amino acid. As the codon bias is lower in longer genes than shorter ones, further analysis in finding these constrained codons will help us in understanding whether it is the same in all genes irrespective of their length.
Figure 2.
Plot of Nc versus Gene length for all mycobacteriophage genomes.
Table 5.
Correlation coefficient of gene length with Nc and GC3s values.
| Length | ||
|---|---|---|
| Phage name | Nc | GC3s |
| Bxb1 | −.440** | .445** |
| Bxz2 | −.328* | .414** |
| Omega | −.223* | .243** |
| Bxz1 | −.241** | .205* |
| Cjw1 | −.227* | 0.213NS |
| Corndog | −.331** | .336** |
| Llij | −.342** | .332** |
| Wildcat | −.280* | 0.026NS |
| Catera | −.200* | .168* |
| U2 | −.475** | .427** |
| Bethlehem | −.472** | .445** |
Notable significant relationships are marked by **P < .01 or *P < .05, NSNonsignificant.
3.7. Correspondence Analysis Using RSCU Values
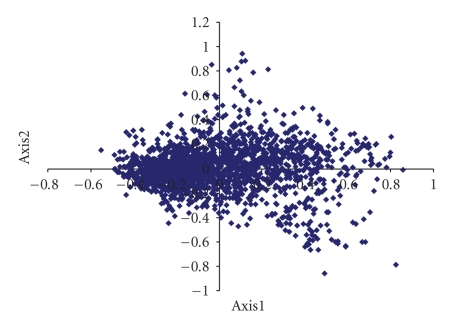
In order to determine the factors that influence variations in codon usage among the genes of mycobacteriophage genomes, correspondence analysis was conducted on the RSCU values of its genes. Only the distributions of the genes along the first two major axes were shown, as these accounted for 13.63% and 6.89% of the total variation (Figure 3).
Figure 3.
Correspondence analysis of Relative Synonymous Codon Usage values of mycobacteriophages (32 genomes).
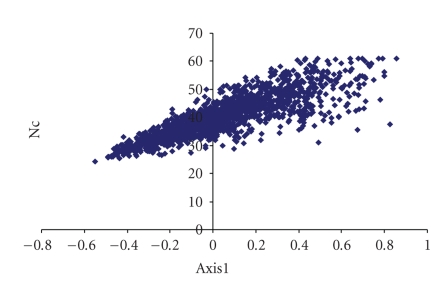
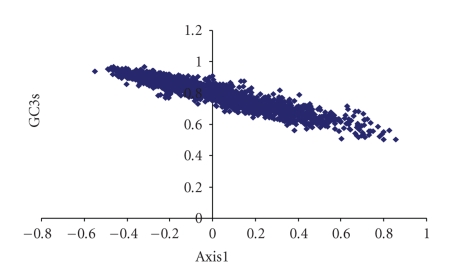
The first major axis is negatively correlated with G3s (r = −.235, P < .01) and C3s (r = −.778, P < .01) but correlated positively with A3s (r = .687, P < .01) and T3s (r = .827, P < .01). Interestingly, high degree of positive correlation exists between position of genes along the first axis with Nc (r = .863, P < .01) (Figure 4) and high degree of negative correlation with GC3s (r = −.934, P < .01) (Figure 5). These findings suggest that highly biased genes, those with G- and C-ending codons, are clustered on the negative side, whereas the codons ending in A and T predominate on the positive side of the first major axis.
Figure 4.
Scatter plot of mycobacteriophages and Nc values.
Figure 5.
Scatter plot of mycobacteriophages and GC3s values.
Additionally, significant negative correlation is observed with Nc against GC3s and GC. Highly expressed genes tend to use “C” or “G” at the synonymous positions compared with lowly expressed genes. It is also studied that C-ending codons are preferred over G-ending codons in highly expressed genes. Preference of C-ending codons in the highly expressed genes might be related to the translational efficiency of the genes as it has been reported that RNY (R-purine, N- any nucleotide base, and Y-pyrimidine) codons are more advantageous for translation [54]. Thus, compositional mutation bias possibly plays an important role in shaping the genome of these phages.
The genomes of Llij, PMC, Wildcat, TM4, Che8, Tweety, and Che9d phages showed no significant correlation between first major axis and GC3s. Whereas, phages such as 244, Bxb1, Bxz2, Che9c, Rosebush, Omega, Halo, Barnyard, Bxz1, Cjw1, Corndog, Orion, Plot, Qyrzula, and Giles show strong negative correlation with GC3s. The primary trend in codon usage variation in these phages can be attributed to the presence of putatively foreign genes acquired through horizontal gene transfer with unusually A + T rich codon usage. However, in phages such as D29, L5, PBI1, PG1, Cooper, Che12, Catera, Bethelhem, and U2, axis 1 coordinates are significantly positively correlated with GC3s values (Table 6). Moreover, when G3s and C3s are considered separately, the correlation coefficient exhibited by the positions of genes along the first axis with C3s is significantly larger than that with G3s (Table 6), indicating that the contribution of C3s to the interspecies variation in overall GC3s content is greater than that of G3s.
Table 6.
Correlation of Axis1 with other codon usage indices.
| Axis 1 | |||||||
|---|---|---|---|---|---|---|---|
| Virus | GC3s | A3s | T3s | G3s | C3s | Gravy | Aromaticity |
| 244 | −.930** | .729** | .829** | −.230* | −.798** | −0.206NS | 0.178NS |
| Bxb1 | −.891** | .871** | .689** | −.417** | −.603** | −0.222NS | 0.078NS |
| Bxz2 | −.904** | .865** | .676** | −.503** | −.502** | −.323* | −0.16NS |
| Che9C | −.921** | .854** | .848** | −0.201NS | −.717** | −0.002NS | −0.053NS |
| Rosebush | −.689** | .323** | .743** | 0.108NS | −.638** | 0.151NS | 0.028NS |
| Omega | −.897** | .650** | .771** | −.262** | −.736** | 0.004NS | −0.055NS |
| Halo | −.746** | .720** | .498** | −0.125NS | −.641** | −0.091NS | 0.013NS |
| Barnyard | −.933** | .843** | .792** | −.647** | −.730** | 0.02NS | 0.023NS |
| Bxz1 | −.901** | .859** | .729** | −.221** | −.718** | 0.005NS | −0.048NS |
| Cjw1 | −.929** | .796** | .742** | −.362** | −.712** | −.259* | 0.153NS |
| Corndog | −.904** | .647** | .838** | −0.175NS | −.652** | −0.184NS | −0.002NS |
| Orion | −.763** | .641** | .644** | 0.099NS | −.729** | −.362** | −0.169NS |
| Plot | −.826** | .820** | .594** | −.427** | −.631** | 0.172NS | 0.229NS |
| Llij | 0.15NS | .763** | −.846** | −.912** | .896** | −0.186NS | −0.166NS |
| Pipefish | .370** | −0.205NS | −.348** | −0.036NS | .263* | −0.105NS | 0.185NS |
| PMC | 0.097NS | .853** | −.919** | −.944** | .908** | −0.229NS | −0.078NS |
| Qyrzula | −.641** | .323** | .731** | 0.09NS | −.588** | 0.061NS | −0.076NS |
| Wildcat | 0.14NS | −.788** | .556** | .411** | −0.208NS | 0.017NS | −0.036NS |
| D29 | .740** | −.721** | −.479** | 0.126NS | .634** | 0.241NS | −0.041NS |
| L5 | .862** | −.775** | −.670** | .384** | .485** | .296* | −0.071NS |
| PBI1 | .845** | −.806** | −.664** | .365** | .705** | −0.131NS | −.269* |
| PG1 | .740** | −0.524NS | −.678** | −0.219NS | .753** | .277** | 0.118NS |
| Cooper | .782** | −.782** | −.609** | 0.153NS | .563** | 0.2NS | −0.001NS |
| Che12 | .803** | −.752** | −.596** | 0.201NS | .650** | 0.214NS | −0.249NS |
| Catera | .914** | −.840** | −.766** | .233** | .702** | 0.041NS | 0.002NS |
| TM4 | 0.085NS | ‒.313* | 0.258NS | 0.182NS | −0.094NS | 0.003NS | 0.008NS |
| Bethelhem | .882** | −.881** | −.625** | .505** | .602** | 0.229NS | .294* |
| Che8 | 0.124NS | .816** | −.883** | −.917** | .899** | −0.242NS | −0.189NS |
| Tweety | −0.113NS | −.797** | .891** | .934** | −.902** | 0.231NS | 0.098NS |
| Giles | −.352** | .730** | −0.447NS | −.887** | .691** | −.444** | −0.102NS |
| U2 | .922** | −.918** | −.737** | .522** | .537** | 0.066NS | 0.01NS |
| Che9d | 0.16NS | .778** | −.831** | −.857** | .862** | −0.027NS | −0.231NS |
Notable significant relationships are marked by **P < .01 or *P < .05, NSNonsignificant.
Table 7 shows RSCU values for each codon for the two groups of genes. The asterisk represents the codons whose occurrences are significantly higher in the genes situated on the extreme left side of axis 1, compared to the genes present on the extreme right of the first major axis. It is important to note that out of 22 codons that are statistically overrepresented in genes located on the extreme left side of axis 1 there is 16 C-ending codons and 5-G ending codons. UGA is the most frequent stop codon among highly and lowly expressed genes. Similar pattern is also seen in Mycobacterium tuberculosis genome, where the highly expressed genes prefer codons ending with “C” and “G” [18].
Table 7.
Relative Synonymous Codon Usage for the highly and lowly expressed genes.
| AA | Codon | RSCUa | Na | RSCUb | Nb | AA | Codon | RSCUa | Na | RSCUb | Nb |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Phe | UUU | 0.08 | (34) | 0.57 | (185) | Ser | UCU | 0.11 | (27) | 0.94 | (178) |
| UUC* | 1.92 | (839) | 1.43 | (466) | UCC* | 1.20 | (284) | 0.88 | (166) | ||
| Leu | UUA | 0.00 | (0) | 0.16 | (41) | UCA | 0.10 | (23) | 0.84 | (160) | |
| UUG | 0.09 | (33) | 1.55 | (390) | UCG | 1.76 | (417) | 1.67 | (317) | ||
| CUU | 0.07 | (26) | 0.93 | (235) | Pro | CCU | 0.16 | (60) | 0.85 | (249) | |
| CUC* | 1.38 | (518) | 1.16 | (292) | CCC* | 1.49 | (545) | 0.88 | (257) | ||
| CUA | 0.01 | (5) | 0.49 | (123) | CCA | 0.12 | (43) | 0.71 | (209) | ||
| CUG* | 4.45 | (1676) | 1.70 | (429) | CCG* | 2.23 | (813) | 1.56 | (459) | ||
| Ile | AUU | 0.19 | (78) | 1.01 | (288) | Thr | ACU | 0.09 | (47) | 0.78 | (217) |
| AUC* | 2.81 | (1181) | 1.74 | (495) | ACC* | 3.11 | (1573) | 1.20 | (333) | ||
| AUA | 0.00 | (1) | 0.25 | (72) | ACA | 0.07 | (33) | 0.70 | (193) | ||
| Met | AUG | 1.00 | (648) | 1.00 | (427) | ACG | 0.73 | (367) | 1.32 | (364) | |
| Val | GUU | 0.15 | (88) | 0.92 | (332) | Ala | GCU | 0.22 | (183) | 0.91 | (428) |
| GUC* | 1.97 | (1131) | 1.04 | (373) | GCC* | 2.50 | (2078) | 1.03 | (483) | ||
| GUA | 0.08 | (48) | 0.34 | (123) | GCA | 0.22 | (181) | 0.76 | (355) | ||
| GUG | 1.79 | (1025) | 1.69 | (608) | GCG | 1.06 | (882) | 1.30 | (611) | ||
| Tyr | UAU | 0.10 | (41) | 0.69 | (185) | Cys | UGU | 0.06 | (7) | 0.83 | (123) |
| UAC* | 1.90 | (767) | 1.31 | (350) | UGC* | 1.94 | (212) | 1.17 | (175) | ||
| TER | UAA | 0.56 | (20) | 0.79 | (28) | TER | UGA | 1.77 | (63) | 1.63 | (58) |
| UAG | 0.67 | (24) | 0.59 | (21) | Trp | UGG | 1.00 | (531) | 1.00 | (468) | |
| His | CAU | 0.12 | (36) | 0.89 | (236) | Arg | CGU | 0.52 | (164) | 1.37 | (364) |
| CAC* | 1.88 | (570) | 1.11 | (296) | CGC* | 3.79 | (1189) | 1.50 | (399) | ||
| Gln | CAA | 0.06 | (29) | 0.63 | (221) | CGA | 0.18 | (55) | 0.85 | (227) | |
| CAG* | 1.94 | (1021) | 1.37 | (480) | CGG | 1.41 | (442) | 1.23 | (328) | ||
| Asn | AAU | 1.10 | (50) | 0.75 | (265) | Ser | AGU | 0.11 | (27) | 0.73 | (139) |
| AAC* | 1.90 | (926) | 1.25 | (446) | AGC* | 2.71 | (642) | 0.94 | (178) | ||
| Lys | AAA | 0.06 | (33) | 0.60 | (229) | Arg | AGA | 0.00 | (1) | 0.41 | (109) |
| AAG* | 1.94 | (1046) | 1.40 | (539) | AGG | 0.11 | (33) | 0.64 | (170) | ||
| Asp | GAU | 0.15 | (151) | 0.89 | (564) | Gly | GGU | 0.54 | (305) | 1.04 | (414) |
| GAC* | 1.85 | (1868) | 1.11 | (706) | GGC* | 2.90 | (1646) | 1.19 | (471) | ||
| Glu | GAA | 0.29 | (270) | 0.84 | (513) | GGA | 0.14 | (81) | 0.91 | (363) | |
| GAG* | 1.71 | (1609) | 1.16 | (704) | GGG | 0.41 | (235) | 0.86 | (341) |
*Codons whose occurrences are significantly higher (P < .01) in the extreme left side of axis 1 than the genes present on the extreme right of the first major axis. Each group contains 10% of sequences at either extreme of the major axis generated by correspondence analysis. AA: amino acid; N: number of codon; agenes on extreme left of axis 1; bgenes on extreme right of axis 1.
4. Conclusion
Compositional bias and translational forces had been reported to play a major role in shaping the codon usage of 14 mycobacteriophages. Our observations corroborate with the earlier report with respect to all the 32 mycobacteriophages. Gene length has a minor role in the selection of codon usage of 11 out of 32 mycobacteriophage genes analyzed. High level of heterogeneity is seen within and among the mycobacteriophage genomes. Cooper is identified to be the highly biased genome with better translation efficiency comparing well with the host specific tRNA genes.
Acknowledgments
S. Hassan acknowledges ICMR—Biomedical Informatics Project and Tuberculosis Research Centre for the funding provided. The authors acknowledge the Department of Biotechnology, New Delhi, for their financial support of this initiative under the collaborative study titled “Establishment of National Database on Tuberculosis.” They thank Dr. P. Venkatesan, Gomathi Sivaramakrishnan, Azger Dusthackeer, and Balaji Subramanyam of Tuberculosis Research Centre for their help in preparation of the manuscript.
References
- 1.Osawa S, Jukes TH. Codon reassignment (codon capture) in evolution. Journal of Molecular Evolution. 1989;28(4):271–278. doi: 10.1007/BF02103422. [DOI] [PubMed] [Google Scholar]
- 2.Osawa S, Muto A, Jukes TH, Ohama T. Evolutionary changes in the genetic code. Proceedings of the Royal Society B. 1990;241(1300):19–28. doi: 10.1098/rspb.1990.0060. [DOI] [PubMed] [Google Scholar]
- 3.Osawa S, Jukes TH, Watanabe K, Muto A. Recent evidence for evolution of the genetic code. Microbiological Reviews. 1992;56(1):229–264. doi: 10.1128/mr.56.1.229-264.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grantham R, Gautier C, Gouy M, Mercier R, Pavé A. Codon catalog usage and the genome hypothesis. Nucleic Acids Research. 1980;8(1):r49–r62. doi: 10.1093/nar/8.1.197-c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grantham R, Gautier C, Gouy M. Codon frequencies in 119 individual genes confirm consistent choices of degenerate bases according to genome type. Nucleic Acids Research. 1980;8(9):1893–1912. doi: 10.1093/nar/8.9.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin CE, Scheinbach S. Expression of proteins encoded by foreign genes in Saccharomyces cerevisiae . Biotechnology Advances. 1989;7(2):155–185. doi: 10.1016/0734-9750(89)90357-1. [DOI] [PubMed] [Google Scholar]
- 7.Lloyd AT, Sharp PM. Evolution of codon usage patterns: the extent and nature of divergence between Candida albicans and Saccharomyces cerevisiae . Nucleic Acids Research. 1992;20(20):5289–5295. doi: 10.1093/nar/20.20.5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grocock RJ, Sharp PM. Synonymous codon usage in Cryptosporidium parvum: identification of two distinct trends among genes. International Journal for Parasitology. 2001;31(4):402–412. doi: 10.1016/s0020-7519(01)00129-1. [DOI] [PubMed] [Google Scholar]
- 9.D’Onofrio G, Bernardi G. A universal compositional correlation among codon positions. Gene. 1992;110(1):81–88. doi: 10.1016/0378-1119(92)90447-w. [DOI] [PubMed] [Google Scholar]
- 10.Majumdar S, Gupta SK, Sundararajan VS, Ghosh TC. Compositional correlation studies among the three different codon positions in 12 bacterial genomes. Biochemical and Biophysical Research Communications. 1999;266(1):66–71. doi: 10.1006/bbrc.1999.1774. [DOI] [PubMed] [Google Scholar]
- 11.Gupta SK, Ghosh TC. Gene expressivity is the main factor in dictating the codon usage variation among the genes in Pseudomonas aeruginosa . Gene. 2001;273(1):63–70. doi: 10.1016/s0378-1119(01)00576-5. [DOI] [PubMed] [Google Scholar]
- 12.Sharp PM, Cowe E. Synonymous codon usage in Saccharomyces cerevisiae . Yeast. 1991;7(7):657–678. doi: 10.1002/yea.320070702. [DOI] [PubMed] [Google Scholar]
- 13.Alvarez F, Robello C, Vignali M. Evolution of codon usage and base contents in kinetoplastid protozoans. Molecular Biology and Evolution. 1994;11(5):790–802. doi: 10.1093/oxfordjournals.molbev.a040159. [DOI] [PubMed] [Google Scholar]
- 14.Moriyama EN, Powell JR. Gene length and codon usage bias in Drosophila melanogaster, Saccharomyces cerevisiae and Escherichia coli . Nucleic Acids Research. 1998;26(13):3188–3193. doi: 10.1093/nar/26.13.3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hou Z-C, Yang N. Factors affecting codon usage in Yersinia pestis . Sheng Wu Hua Xue Yu Sheng Wu Wu Li Xue Bao Shanghai. 2003;35(6):580–586. [PubMed] [Google Scholar]
- 16.Gu W, Zhou T, Ma J, Sun X, Lu Z. Analysis of synonymous codon usage in SARS Coronavirus and other viruses in the Nidovirales. Virus Research. 2004;101(2):155–161. doi: 10.1016/j.virusres.2004.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hou Z-C, Yang N. Analysis of factors shaping S. pneumoniae codon usage. Yi Chuan Xue Bao. 2002;29(8):747–752. [PubMed] [Google Scholar]
- 18.Pan A, Dutta C, Das J. Codon usage in highly expressed genes of Haemophillus influenzae and Mycobacterium tuberculosis: translational selection versus mutational bias. Gene. 1998;215:405–413. doi: 10.1016/s0378-1119(98)00257-1. [DOI] [PubMed] [Google Scholar]
- 19.Wright F. The ‘effective number of codons’ used in a gene. Gene. 1990;87(1):23–29. doi: 10.1016/0378-1119(90)90491-9. [DOI] [PubMed] [Google Scholar]
- 20.Eyre-Walker A. Synonymous codon bias is related to gene length in Escherichia coli: selection for translational accuracy? Molecular Biology and Evolution. 1996;13(6):864–872. doi: 10.1093/oxfordjournals.molbev.a025646. [DOI] [PubMed] [Google Scholar]
- 21.Sharp PM, Li WH. The codon Adaptation Index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Research. 1987;15(3):1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ranjan A, Vidyarthi AS, Poddar R. Evaluation of codon bias perspectives in phage therapy of Mycobacterium tuberculosis by multivariate analysis. In Silico Biology. 2007;7(4-5, article 0030):423–431. [PubMed] [Google Scholar]
- 23.Gupta SK, Bhattacharyya TK, Ghosh TC. Synonymous codon usage in Lactococcus lactis: mutational bias versus translational selection. Journal of Biomolecular Structure & Dynamics. 2004;21(4):527–536. doi: 10.1080/07391102.2004.10506946. [DOI] [PubMed] [Google Scholar]
- 24.Basak S, Banerjee T, Gupta SK, Ghosh TC. Investigation on the causes of codon and amino acid usages variation between thermophilic Aquifex aeolicus and mesophilic Bacillus subtilis . Journal of Biomolecular Structure & Dynamics. 2004;22(2):205–214. doi: 10.1080/07391102.2004.10506996. [DOI] [PubMed] [Google Scholar]
- 25.Lynn DJ, Singer GAC, Hickey DA. Synonymous codon usage is subject to selection in thermophilic bacteria. Nucleic Acids Research. 2002;30(19):4272–4277. doi: 10.1093/nar/gkf546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lobry JR, Gautier C. Hydrophobicity, expressivity and aromaticity are the major trends of amino-acid usage in 999 Escherichia coli chromosome-encoded genes. Nucleic Acids Research. 1994;22(15):3174–3180. doi: 10.1093/nar/22.15.3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garat B, Musto H. Trends of amino acid usage in the proteins from the unicellular parasite Giardia lamblia . Biochemical and Biophysical Research Communications. 2000;279(3):996–1000. doi: 10.1006/bbrc.2000.4051. [DOI] [PubMed] [Google Scholar]
- 28.Zavala A, Naya H, Romero H, Musto H. Trends in codon and amino acid usage in Thermotoga maritima . Journal of Molecular Evolution. 2002;54(5):563–568. doi: 10.1007/s00239-001-0040-y. [DOI] [PubMed] [Google Scholar]
- 29.Banerjee T, Basak S, Gupta SK, Ghosh TC. Evolutionary forces in shaping the codon and amino acid usages in Bochmannia floridanus . Journal of Biomolecular Structure & Dynamics. 2004;22(1):13–23. doi: 10.1080/07391102.2004.10506976. [DOI] [PubMed] [Google Scholar]
- 30.Naya H, Zavala A, Romero H, Rodríguez-Maseda H, Musto H. Correspondence analysis of amino acid usage within the family Bacillaceae . Biochemical and Biophysical Research Communications. 2004;325(4):1252–1257. doi: 10.1016/j.bbrc.2004.10.170. [DOI] [PubMed] [Google Scholar]
- 31.Ohkubo S, Muto A, Kawauchi Y, Yamao F, Osawa S. The ribosomal protein gene cluster of Mycoplasma capricolum . Molecular & General Genetics. 1987;210(2):314–322. doi: 10.1007/BF00325700. [DOI] [PubMed] [Google Scholar]
- 32.Wright F, Bibb MJ. Codon usage in the G+C-rich Streptomyces genome. Gene. 1992;113(1):55–65. doi: 10.1016/0378-1119(92)90669-g. [DOI] [PubMed] [Google Scholar]
- 33.Gupta SK, Bhattacharyya TK, Ghosh TC. Compositional correlation and codon usage studies in Buchnera aphidicola . Indian Journal of Biochemistry & Biophysics. 2002;39(1):35–48. [PubMed] [Google Scholar]
- 34.Lü H, Zhao W-M, Zheng Y, Wang H, Qi M, Yu X-P. Analysis of synonymous codon usage bias in Chlamydia . Acta Biochimica et Biophysica Sinica. 2005;37(1):1–10. doi: 10.1111/j.1745-7270.2005.00009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fickett JW. Recognition of protein coding regions in DNA sequences. Nucleic Acids Research. 1982;10(17):5303–5318. doi: 10.1093/nar/10.17.5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chiapello H, Ollivier E, Landes-Devauchelle C, Nitschke P, Risler J-L. Codon usage as a tool to predict the cellular location of eukaryotic ribosomal proteins and aminoacyl-tRNA synthetases. Nucleic Acids Research. 1999;27(14):2848–2851. doi: 10.1093/nar/27.14.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martín A, Bertranpetit J, Oliver JL, Medina JR. Variation in G+C-content and codon choice: differences among synonymous codon groups in vertebrate genes. Nucleic Acids Research. 1989;17(15):6181–6189. doi: 10.1093/nar/17.15.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moriyama EN, Hartl DL. Codon usage bias and base composition of nuclear genes in Drosophila. Genetics. 1993;134(3):847–858. doi: 10.1093/genetics/134.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McInerney JO. Replicational and transcriptional selection on codon usage in Borrelia burgdorferi . Proceedings of the National Academy of Sciences of the United States of America. 1998;95(18):10698–10703. doi: 10.1073/pnas.95.18.10698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sahu K, Gupta SK, Sau S, Ghosh TC. Comparative analysis of the base composition and codon usages in fourteen mycobacteriophage genomes. Journal of Biomolecular Structure & Dynamics. 2005;23(1):63–71. doi: 10.1080/07391102.2005.10507047. [DOI] [PubMed] [Google Scholar]
- 41.McInerney JO. GCUA: general codon usage analysis. Bioinformatics. 1998;14(4):372–373. doi: 10.1093/bioinformatics/14.4.372. [DOI] [PubMed] [Google Scholar]
- 42.Comeron JM, Aguadé M. An evaluation of measures of synonymous codon usage bias. Journal of Molecular Evolution. 1998;47(3):268–274. doi: 10.1007/pl00006384. [DOI] [PubMed] [Google Scholar]
- 43.Sau K, Gupta SK, Sau S, Ghosh TC. Synonymous codon usage bias in 16 Staphylococcus aureus phages: implication in phage therapy. Virus Research. 2005;113(2):123–131. doi: 10.1016/j.virusres.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 44.Grantham R, Gautier C, Gouy M, Jacobzone M, Mercier R. Codon catalog usage is a genome strategy modulated for gene expressivity. Nucleic Acids Research. 1981;9(1):r43–r74. doi: 10.1093/nar/9.1.213-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bennetzen JL, Hall BD. Codon selection in yeast. Journal of Biological Chemistry. 1982;257(6):3026–3031. [PubMed] [Google Scholar]
- 46.Gouy M, Gautier C. Codon usage in bacteria: correlation with gene expressivity. Nucleic Acids Research. 1982;10(22):7055–7074. doi: 10.1093/nar/10.22.7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sharp PM, Li W-H. An evolutionary perspective on synonymous codon usage in unicellular organisms. Journal of Molecular Evolution. 1986;24(1-2):28–38. doi: 10.1007/BF02099948. [DOI] [PubMed] [Google Scholar]
- 48.Sharp PM, Devine KM. Codon usage and gene expression level in Dictyostelium discoideum: highly expressed genes do ‘prefer’ optimal codons. Nucleic Acids Research. 1989;17(13):5029–5039. doi: 10.1093/nar/17.13.5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sharp PM, Rogers MS, McConnell DJ. Selection pressures on codon usage in the complete genome of bacteriophage T7. Journal of Molecular Evolution. 1985;21(2):150–160. doi: 10.1007/BF02100089. [DOI] [PubMed] [Google Scholar]
- 50.Ikemura T. Transfer RNA in Protein Synthesis. London, UK: CRC Press; 1992. [Google Scholar]
- 51.Zhou J, Liu WJ, Peng SW, Sun XY, Frazer I. Papillomavirus capsid protein expression level depends on the match between codon usage and tRNA availability. Journal of Virology. 1999;73(6):4972–4982. doi: 10.1128/jvi.73.6.4972-4982.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kanaya S, Yamada Y, Kudo Y, Ikemura T. Studies of codon usage and tRNA genes of 18 unicellular organisms and quantification of Bacillus subtilis tRNAs: gene expression level and species-specific diversity of codon usage based on multivariate analysis. Gene. 1999;238(1):143–155. doi: 10.1016/s0378-1119(99)00225-5. [DOI] [PubMed] [Google Scholar]
- 53.Kanaya S, Yamada Y, Kinouchi M, Kudo Y, Ikemura T. Codon usage and tRNA genes in eukaryotes: correlation of codon usage diversity with translation efficiency and with CG-dinucleotide usage as assessed by multivariate analysis. Journal of Molecular Evolution. 2001;53(4-5):290–298. doi: 10.1007/s002390010219. [DOI] [PubMed] [Google Scholar]
- 54.Shepherd JC. Method to determine the reading frame of a protein from the purine/pyrimidine genome sequence and its possible evolutionary justification. Proceedings of the National Academy of Sciences of the United States of America. 1981;78(3):1596–1600. doi: 10.1073/pnas.78.3.1596. [DOI] [PMC free article] [PubMed] [Google Scholar]