Abstract
In 2000, Marquardt et al. (A. Marquardt, H. Stöhr, K. White, and B. H. F. Weber. 2000. cDNA cloning, genomic structure, and chromosomal localization of three members of the human fatty acid desaturase family. Genomics. 66: 176–183.) described the genomic structure of the fatty acid desaturase (FADS) cluster in humans. This cluster includes the FADS1 and FADS2 genes encoding, respectively, for the Δ5- and Δ6-desaturases involved in polyunsaturated fatty acid biosynthesis. A third gene, named FADS3, has recently been identified but no functional role has yet been attributed to the putative FADS3 protein. In this study, we investigated the FADS3 occurrence in rat tissues by using two specific polyclonal antibodies directed against the N-terminal and C-terminal ends of rat FADS3. Our results showed three potential protein isoforms of FADS3 (75 kDa, 51 kDa, and 37 kDa) present in a tissue-dependent manner. The occurrence of these FADS3 isoforms did not depend on the mRNA level determined by real-time PCR. In parallel, mouse tissues were also tested and showed the same three FADS3 isoforms but with a different tissue distribution. Finally, we reported the existence of FADS3 in human cells and tissues but different new isoforms were identified. To conclude, we showed in this study that FADS3 does exist under multiple protein isoforms depending on the mammalian tissues. These results will help further investigations to determine the physiological function of FADS3.
Keywords: protein isoform, mRNA level, rat, human
PUFAs are key components involved in a variety of physiological functions (1). Some of them, belonging to the n-6 or n-3 families, have to be fulfilled from the diet or derived from the biosynthetic pathways resulting in the conversion of essential precursors to their respective elongated polyenoic products.
The availability of PUFA in mammalian cells greatly depends on the activity of enzymes involved in FA metabolism. In animals and humans, the Δ5- and Δ6-desaturases are the pivotal enzymes introducing de novo unsaturations in the carbon chain of precursors leading to the synthesis of long-chain PUFA (LC-PUFA). These enzymes were cloned 10 years ago from mammals (2–5). In parallel, Marquardt et al. (6) described the human genomic structure of the fatty acid desaturase (FADS) cluster including the FADS1 and FADS2 genes coding, respectively, for the Δ5- and Δ6-desaturases. A third gene, named FADS3, was identified, revealing 62% and 70% nucleotide sequence identity with FADS1 and FADS2, respectively. Further studies showed a significant correlation between FADS3 polymorphism and lipid metabolism markers such as PUFA, high density- or low density-lipoprotein cholesterol, and triglyceride levels (7–10). The newly discovered gene was thereafter integrated into a serial analysis of gene expression and a DNA microarray succeeding in more physiological data. FADS3 was therefore found to be highly expressed at the implantation site of the embryo in mouse uterus (11) and downregulated during human neurogenic differentiation (12). More recently, Park et al. described, in baboon, different alternative transcripts of FADS3 generated by alternative splicing, which suggests the occurrence of multiple FADS3 gene products (13). This study also showed a different pattern of expression in response to human neuroblastoma SK-N-SH cell differentiation. All data together only concern the FADS3 gene with no description of the functional role of the putative FADS3 protein.
A protein database search identified FADS3 as a front end desaturase as well as FADS1 and FADS2. The predicted structure of FADS3 describes a membrane-bound desaturase composed of an N-terminal cytochrome b5-like domain and a C-terminal FA desaturase domain (Fig. 1A). These two domains are thought to be potentially involved in the regulation or the catalytic assignment of the desaturase as previously reported for FADS2 (14), suggesting an FA desaturase role of FADS3. It is therefore possible to assume that FADS3 exists in cells.
Fig. 1.
Structure of the putative rat FADS3 protein and peptides designed to produce specific rat FADS3 antibodies. A: Rat FADS3 displays an FA membrane-bound desaturase structure with two distinct domains. In the N-terminal end, the cytochrome b5-like domain is specific of front-end desaturases, introducing a double bond before the eighth carbon of the FA carbon chain; the HPGG motif present in that domain is a pivotal pattern for the desaturase activity. In the carboxyl-terminal end, the FA desaturase domain is characterized by three histidine boxes necessary for the catalysis; in the third box, the first amino acid is a glutamine (Q) instead of a histidine (H), which is characteristic of the front-end desaturases such as Δ5-, Δ6-, and Δ8-desaturases, as compared with the methyl-end desaturases like Δ12- and Δ15-desaturases. B: Different antibodies were produced to detect FADS3 in rat tissues. Several peptides were, thus, designed according to the putative sequence of rat FADS3. Among the eight generated antibodies, only three recognized FADS3.
A recent study reported an enhanced mRNA expression of Fads3 in liver of Fads2−/− mice (15). Deletion of the Fads2 gene modifies the enzymatic pathways of LC-PUFA biosynthesis without impairing the normal lifespan of Fads2−/− mice but causing a variety of pathologies (16). FADS2 is known to catalyze the conversion of multiple FA substrates (17–19) and thus, to take part in many physiological functions. The close relationship between Fads2 and Fads3 in these knockout mice is thought to underline the putative involvement of Fads3 in the FA metabolism. No biological assignment has yet been attributed to that latest gene; no evidence has emerged concerning the translation from the transcript into the protein. Thus, the question is, does the FADS3 protein exist? To answer this query, we reported in this study the expression of Fads3 in mammalian tissues. We first analyzed the transcript level of Fads3 in rat tissues and then we investigated the occurrence of the FADS3 protein in mammalian cells or tissues.
MATERIALS AND METHODS
Materials
DMEM and antibiotics were purchased from Eurobio (Les Ulis, France). Fetal calf serum (FCS) was obtained from Lonza (Levallois-Perret, France). African green monkey kidney Cos-7 cell lines (from ECACC), HRP conjugated mouse anti-IgG, mouse monoclonal anti-GAPDH antibody, and chemicals were provided by Sigma-Aldrich (Saint-Quentin Fallavier, France). Chemicals for PCR were from Eurogentec (Angers, France). pCMV/ myc/ cyto and pCMV•SPORT-βgal plasmids were from Invitrogen (Cergy Pontoise, France). Gene Pulser XcellTM electroporation system and Kaleidoscope prestained standard were purchased from Bio-Rad (Marnes-la-Coquette, France). Customized human normal tissue blot was from Interchim (Montluçon, France). HRP conjugated rabbit anti-IgG antibody and Immobilon detection kit were from Millipore (Guyancourt, France). Human neuroblastoma SH-SY5Y cells were kindly given by J. M. Alessandri (INRA, Jouy-en-Josas, France). HUH7 cells were a generous gift from the Laboratory of Genetics (Agrocampus Ouest, Rennes, France). BALB/c female mouse tissues were kindly given by V. Le Moigne (Agrocampus Ouest, Rennes, France).
Animals
Male and female Sprague-Dawley rats (150–200g body weight) were obtained from the Elevage Janvier breeding center (Le Genest Saint Isle, France). Animals had access to standard rodent chow (Scientific Animal Food and Engineering, Augy, France) and water ad libitum. Rats were fasted 24 h before euthanasia and bled by decapitation after anesthesia and analgesia using intraperitoneal injection of pentothal (75 mg/kg body weight). Fresh tissues were used to extract RNA and proteins. White adipose tissue (AT) was perirenal, intestine was from jejunum, and skeletal muscle corresponded to the thigh dorsal muscles. The experimental procedure was in compliance with recommendations of the 2003/65/CEE European directive for animal experimentation.
Cell culture
Cos-7, SH-SY5Y, and HUH7 cell lines were cultured in DMEM supplemented with 10% FCS and complemented with antibiotics (50 UI/ml penicillin, 50 µg/ml streptomycin, 10µg/ml gentamycin). Cells were maintained at 37°C in a humidified atmosphere of air (95%) and CO2 (5%) up to 70–80% confluence. Cell extracts were prepared in radio immuno precipitation assay (RIPA) buffer containing PBS (150 mM NaCl, 0.95 mM NaH2PO4, 4.05 mM Na2HPO4; pH7.4), 1% Nonidet P40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM EDTA, and 10 mg/ml aprotinin. Samples were also diluted in native lysis buffer (50 mM Tris-HCl pH8, 1% Nonidet P40, 150 mM NaCl) when loading in native PAGE.
Rat Fads3 cloning and plasmid construction
The human FADS3 nucleotide sequence (GenBank accession number AF084560) was used to blast-search the rat genomic database (6). Designed from the Rattus norvegicus chromosome 1 genomic contig NW_047563.2, an oligonucleotide primer (5′-CAG CAA CGA AGA GGA GCA GTG TCC CAG TGG CA-3′) was produced to screen the rat liver Marathon-ready cDNA library by Rapid Amplification cDNA Ends (RACE) reactions using the advantage-GC 2 PCR kit (Clontech, Saint-Germain-en-Laye, France). The 5′ end of the rat Fads3 cDNA containing a candidate ATG start codon was obtained, as verified by the sequencing of the resulting PCR product (SCRIBE, Rennes, France). Oligonucleotide primers were then designed to PCR amplify the full-length rat Fads3 coding sequence from the rat liver Marathon-ready cDNA library. The forward primer (5′-CAT CTC TCA GAC CTC TGC CAC GTA-3′) was deduced from the former 5′ end sequence. The reverse primer (5′-CTG GGA AGA CAT GCT ATG CTC ACC-3′) was designed from a rat EST sequence (GenBank accession number AI178762) presenting a high sequence homology with the human FADS3 3′ flanking region. After the first-round PCR, a nested PCR amplification was performed using the cloning forward primer (5′-TTG TGC ATA GCG GCC ATG GGC GGC GT-3′) including the candidate translation start codon (in bold) and a NcoI restriction site (underlined) and the cloning reverse primer (5′-ACT GCG GCC GCC TTC ACT GAT GGA GGT ACG C-3′) with the putative stop codon (in bold) and a NotI restriction site (underlined). The resulting 1350-bp PCR product was cloned into a pCMV / myc/ cyto expression vector and the in-frame orientation was confirmed by DNA sequencing. The recombinant plasmid is referred to as pCMV / Fads3. The sequence of the full-length rat Fads3 cDNA is available in the GenBank database (accession number AJ494720).
Plasmid transfection
Rat recombinant FADS1, FADS2, and FADS3 were expressed after transfection of pCMV / myc/ cyto plasmids containing the rat Fads1 (pCMV/Fads1), Fads2 (pCMV/Fads2), or Fads3 (pCMV/Fads3) gene inserts (17). Recombinant plasmids were transiently transfected into Cos-7 cells using electroporation. Briefly, 70% confluent cells were electroporated (250V-1500µF) and then cultured in DMEM with 10% FCS. After 48 h, cells were scraped off and protein extracts were diluted in RIPA buffer. The transfection efficiency was assessed by β-galactosidase colorimetric assay after cotransfection with pCMV•SPORT-βgal.
Quantification of the Fads mRNA level by real-time PCR
Total RNA was extracted from the tissues of three rats with Extract-All® (Eurobio, Les Ulis, France) and retrotranscripted in duplicates using SuperScriptTM II reverse transcriptase (Invitrogen, Cergy Pontoise, France) according to the manufacturer's instructions. Real-time PCR was performed in duplicates with the TaqMan Universal PCR Master Mix (Applied Biosystems, Courtaboeuf, France) containing 40 ng of retrotranscripted RNA, 0.5 µM of each primer, and 0.25 µM of TaqMan probe (Table 1). PCR was run using ABI Prism 7000 sequence detection system (Applied Biosystems, Courtaboeuf, France) as follows: 2 min at 50°C, 5 min at 95°C, 40 cycles of 10 s at 95°C, and 1 min at 60°C (SENAH, INRA, Saint-Gilles, France). Normalization was assessed using the Yakima Yellow®-Eclipse® DarkQuencher 18S rRNA control kit (Eurogentec, Angers, France). The PCR efficiency was estimated by calibration curves and by calculation using LinReg (20). The relative expression was evaluated as delta Cycle threshold (ΔCt = Ctfads − Ct18S).
TABLE 1.
Primers and TaqMan probes used for quantification of the Fads mRNA levels determined by real-time PCR
| Forward Primer (5′-3′) | Reverse Primer (5′-3′) | 5′-FAM/TAMRA-3′ Probe | |
|---|---|---|---|
| Fads1 | agctttgaacccaccaagaataag | cagcaggatgtgcagcagatag | ttccgggagctgcgggcc |
| Fads2 | tccacaaggaccccgacata | ttgtagggcagatatttcagcttctt | ttggagagtggcagcccctcgagta |
| Fads3 | tgctgtgggctgccagtt | caccctgacagcaacgaaga | ccccttctatggtgccactgggacac |
Rat FADS3 antibody production
Two polyclonal antibodies against Rattus norvegicus FADS3 were produced using a double-X 28-day protocol (Eurogentec, Angers, France). Briefly, two specific peptides corresponding to the N-terminal sequence (31QIRQHDLPGDKWL) and the carboxyl-terminal sequence (352PKEIGHEKHRDWAS) of rat FADS3 (Fig. 1B) were synthesized and coupled to Keyhole Limpet Hemocyanin; these target peptides display 85% and 100% identity with the human protein sequence respectively. Rabbits were then coimmunized with both peptides for 4 weeks. The two antibodies were then purified from the serum by affinity chromatography and were respectively referred to as anti-NtermFADS3 and anti-CtermFADS3. The specific reactivity of each antibody was checked by blot against the immunizing peptides and by Western blot against recombinant FADS3. A third antibody, referred to as anti-FADS2/3, was also punctually used because of its immunospecificity with rat recombinant FADS2 and FADS3. This third nonpurified polyclonal serum was produced by immunization of rabbits with two peptides as described in Fig. 1B (17).
Protein expression
Protein expression was assessed by Western blot after SDS-PAGE or native PAGE as described elsewhere (21). Rat FADS3 antibodies were incubated at 4°C overnight at 1 µg/ml in tris-buffered saline (20 mM Tris-HCl, 150 mM NaCl; pH7.4) containing 0.05% Tween-20 and 5% BSA. The other antibodies were used according to the manufacturer's instructions. Primary antibodies were then coupled to HRP conjugated anti-IgG and peroxidase activity was determined by chemoluminescent detection using Immobilon reagents. The protein level was estimated using anti-actin and confirmed with anti-GAPDH. Actin was not detected in the rat pancreas, nor was GAPDH detected in the rat intestine, so in this case results exhibit only one antibody to evaluate the tissue protein level. The molecular mass of proteins migrating in the gel was determined using a standard curve constructed with kaleidoscope marker in SDS-PAGE. Experiments were reproduced several times on independent cell cultures or animals as indicated in the legend of figures; results exhibit one case in point from the indicated number of experiments.
Statistics
Data were analyzed using the R software (22). For each Fads transcript level, a linear model was applied on data to estimate the effect of sex and tissue as well as their interaction.
RESULTS
Expression of the Fads genes in rat tissues
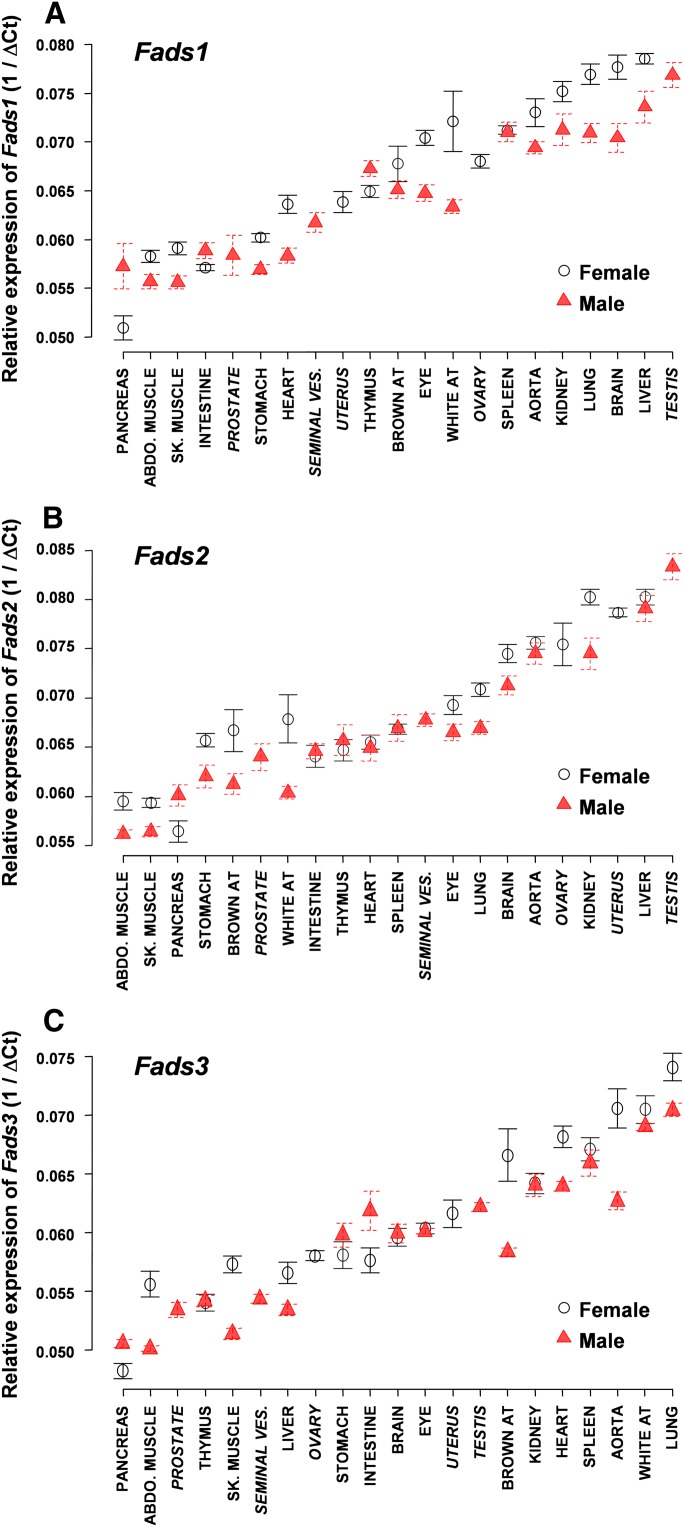
The expression of the Fads3 gene was estimated by determining the tissue distribution of Fads3 transcripts as compared with the other genes of the cluster (Fig. 2). First, this experiment underlined a high significant effect of sex, tissue, and the interaction of sex and tissue on the mRNA level of each Fads gene (P < 10−5). Second, our results showed that in somatic tissues, Fads1 and Fads2 displayed a similar mRNA profile (Fig. 2A, B) with the highest transcript amounts in the liver, kidney, brain, lung, and aorta. Fads3 mRNAs were differently distributed and mainly present in the lung, white AT, aorta, spleen, heart, and kidney (Fig. 2C). The pancreas and skeletal and abdominal muscles shared the lowest transcript level of Fads whereas the aorta, lung, and kidney represented tissues with the highest transcript level commonly found for each Fads gene. Moreover, our data showed a differential Fads mRNA amount according to gender. For instance, Fads3 transcripts were significantly more abundant in females than in males, particularly in the aorta, brown AT, and skeletal and abdominal muscles. In sexual organs, Fads2 mRNA and, to a lesser extent, Fads1 mRNA were highly abundant especially in testis whereas Fads3 mRNA was not particularly represented. These data demonstrated that Fads3 transcripts, as well as Fads1 and Fads2 transcripts, were distributed in a tissue-specific and sex-dependent manner.
Fig. 2.
Transcript level of Fads in rat tissues. Total RNA was extracted from male and female rat tissues, retrotranscripted, and quantified by TaqMan® real-time PCR as described in Methods. Somatic and germinal tissues were then ordered according to the relative expression for each Fads gene. A: Transcript level of Fads1. B: Transcript level of Fads2. C: Transcript level of Fads3. Data are presented as the mean ± SEM of separate experiments on three rats with four measurements per animal. sk. muscle, skeletal muscle; abdo. muscle, abdominal muscle; AT, adipose tissue; seminal ves., seminal vesicle.
Immunospecificity of anti-NtermFADS3 and anti-CtermFADS3 antibodies
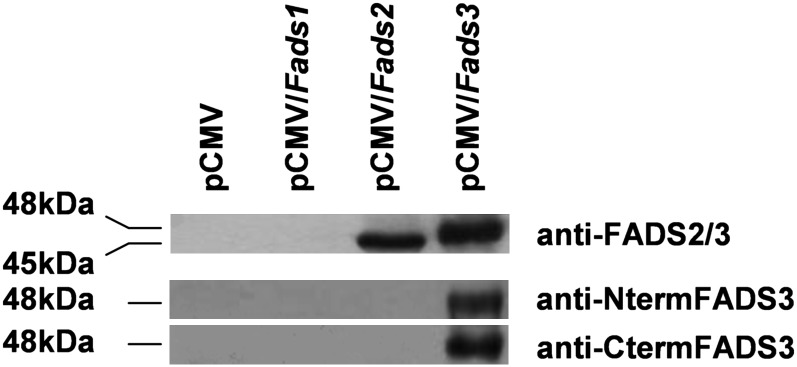
In order to assess the occurrence of FADS3 in rat tissues, we produced antibodies specifically targeted against rat FADS3. Different peptides were thus designed as described in Fig. 1B. The first successful antibody was produced after rabbit immunization with two long peptides. This polyclonal antibody was tested on rat recombinant FADS1, FADS2, and FADS3. Our results showed that this antibody recognized a protein at 48 kDa corresponding to recombinant FADS3, but also a protein corresponding to recombinant FADS2 at 45 kDa (Fig. 3). This antibody was named anti-FADS2/3. Therefore, the nonspecificity toward FADS3 resulted in the production of new antibodies targeted against smaller peptides (Fig. 1B). Of seven antibodies, only two were successfully produced. The specificity of antibodies was tested on recombinant FADS1, FADS2, and FADS3 (Fig. 3). Our results showed that both antibodies detected FADS3 but neither FADS1 nor FADS2. Thus, the antibody directed against the N-terminal end of FADS3 was named anti-NtermFADS3 and the other one targeted toward the carboxyl-terminal end of FADS3 was named anti-CtermFADS3.
Fig. 3.
Rat FADS3 antibody characterization. To characterize the immunospecificity, antibodies were tested on rat recombinant FADS1, FADS2, and FADS3. Cos-7 cells were transfected with pCMV, pCMV/Fads1, pCMV/Fads2, or pCMV/Fads3 plasmids. Western blots were assessed on cell extracts containing recombinant FADS proteins. One polyclonal antibody detected both FADS2 and FADS3 and was referred to as anti-FADS2/3. Two purified polyclonal antibodies detected specifically FADS3 and were referred to as anti-NtermFADS3 and anti-Cterm-FADS3. Experiments were reproduced three times; results exhibit only one case in point.
Identification of the FADS3 protein in rat tissues
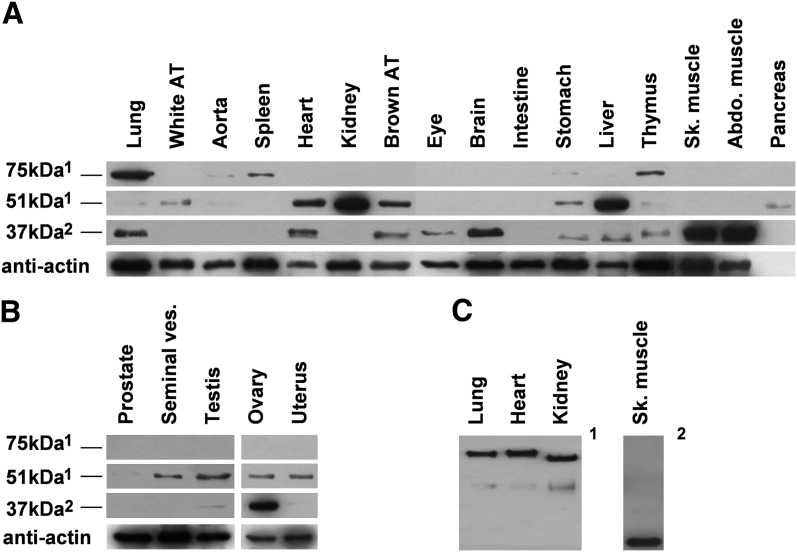
We then investigated the protein level of FADS3 in the same selected tissues as previously described for the mRNA level in rat tissues using the two FADS3 antibodies, anti-NtermFADS3 and anti-CtermFADS3 (Fig. 4) . The predicted molecular mass of rat FADS3 is 51.5 kDa according to the cDNA sequence. Our results showed a band around 51 kDa essentially found in the liver and kidney and to a lesser extent in the heart and brown and white AT (Fig. 4A). This 51 kDa band was also present in a few other tissues but more than 40 µg of protein extracts were needed to visualize it by Western blot (data not shown). A similar result was obtained with the anti-NtermFADS3 antibody but the 51 kDa protein was also identified by the anti-CtermFADS3 antibody. Different bands were also observed at around 75 kDa and 37 kDa depending on the antibody. The anti-NtermFADS3 antibody recognized a 75 kDa protein in several organs, especially in the lung but also in the spleen, thymus, and aorta. The anti-CtermFADS3 antibody revealed a 37 kDa protein, highly abundant in skeletal and abdominal muscles, abundant in the brain and minor in the heart, eye, and thymus. Another band was also found below 37 kDa, for example, in the lung, stomach, and liver. This band may also be suspected in muscles under the extended signal as observed in diluted samples (data not shown). These data were representative for male rats but similar results were also obtained in females (data not shown). In sexual organs, the major protein was detected at 37 kDa in the ovary (Fig. 4B). The 51 kDa protein was equivalently present in all tissues except in the prostate, whereas no protein was observed at 75 kDa.
Fig. 4.
FADS3 distribution in rat tissues. The occurrence of FADS3 was determined in different rat tissues. SDS-PAGE (A and B) and native PAGE (C) were performed on protein lysates from somatic male tissues (A) and from male and female germinal tissues (B). Western blots were then assessed using rat anti-FADS3 antibodies (1anti-NtermFADS3 and 2anti-CtermFADS3). The intensity of band observed in abdominal and skeletal muscles was deliberately reduced by five because of the excessive signal. Experiments were reproduced on three rats; results exhibit only one case in point.
This data showed three potential protein isoforms of FADS3, one of which was expected at 51 kDa. When tissues were presented in the decreasing order of the mRNA level determined in Fig. 2C, we showed that the occurrence of FADS3 was independent of the transcription level. Thus, our results have demonstrated that the Fads3 mRNA level is not correlated with the FADS3 protein level.
We further investigated these three isoforms of FADS3 in native PAGE by focusing on the lung, heart, kidney, and skeletal muscle (Fig. 4C). When the 51 kDa and 75 kDa isoforms from the lung, heart, and kidney were studied in native conditions, two bands were observed. On the contrary, the 37 kDa isoform from skeletal muscle migrated as a single band.
Occurrence of FADS3 in mouse tissues
The expression of FADS3 was also tested on several tissue extracts from mice using our two rat FADS3 antibodies (Fig. 5). Our results showed a similar profile with three protein isoforms. The 75 kDa protein was only observed in the lung. Liver and kidney presented the 51 kDa protein together with the 37 kDa protein, which is particularly abundant as compared with the rat tissues (Fig. 4A). Heart only contained the 51 kDa protein at a very low level and no 37 kDa protein as anticipated. The most unexpected result appeared in skeletal muscle, which displayed no protein, whereas it corresponded to the main tissue expressing the 37 kDa protein in rat. Thus, these results confirmed the presence of three isoforms of FADS3 distributed differently according to the tissues and depending on the animal type.
Fig. 5.
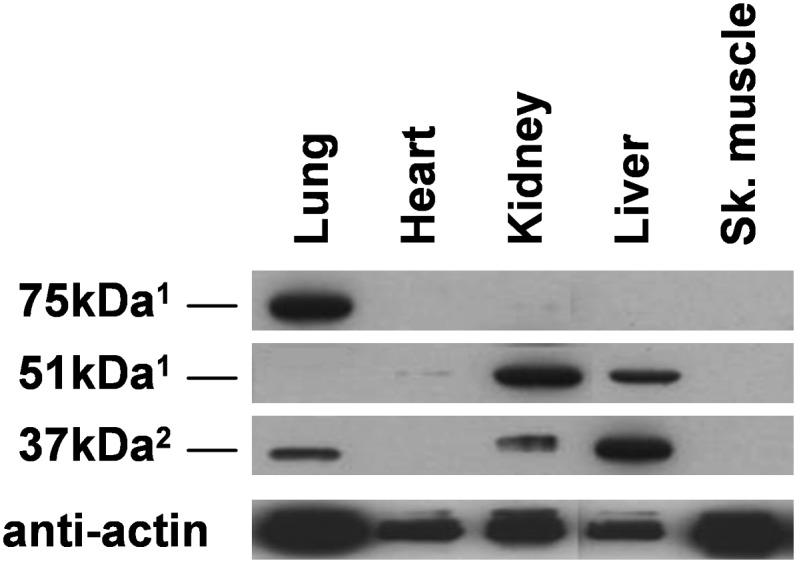
FADS3 occurrence in mouse tissues. The FADS3 profile was determined on mice. SDS-PAGE was performed on protein lysates from female mouse tissues. FADS3 was then detected by Western blot with rat anti-FADS3 antibodies (1anti-NtermFADS3 and 2anti-CtermFADS3). Experiments were reproduced on three mice; results exhibit only one case in point.
Occurrence of FADS3 in human cells and tissues
We then studied the natural FADS3 protein on human cells (SH-SY5Y and HUH7) and tissues using both anti-NtermFADS3 and anti-CtermFADS3 (Fig. 6) . The target peptides designed to produce these antibodies share 85% and 100% homology with the human protein sequence, respectively; therefore, these antibodies conceived to detect the rat FADS3 protein can also interact with the human one.
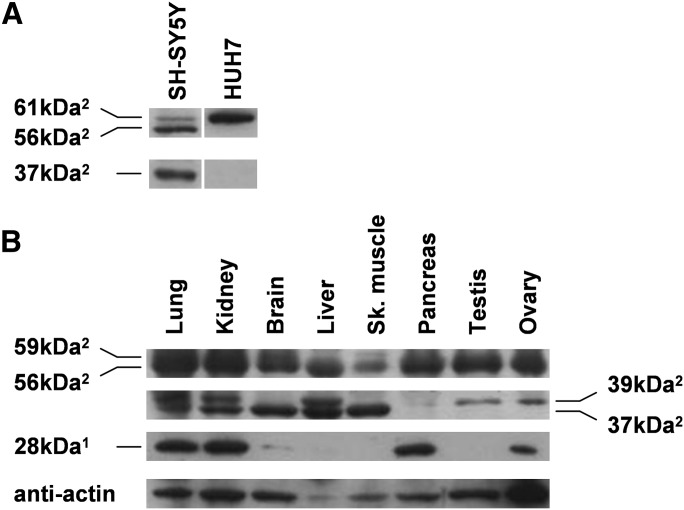
Fig. 6.
FADS3 occurrence in human cells and tissues. The occurrence of natural FADS3 was studied in different human cells and tissues. FADS3 was detected by Western blot with rat anti-FADS3 antibodies (1anti-NtermFADS3 or 2anti-CtermFADS3). A: FADS3 was determined in neuroblastoma SH-SY5Y cells and hepatoma HUH7 cells in the exponential growth phase. Experiments were reproduced on three independent cell cultures; results exhibit only one case in point. B: The tissue distribution of FADS3 was performed on a commercial human tissue blot. Experiments were representative of one sample of one human tissue.
Our results on human cells showed different isoforms of FADS3 detected with the anti-CtermFADS3 antibody (Fig. 6A). In SH-SY5Y cells, we found three proteins with apparent molecular masses of 61 kDa, 56 kDa, and 37 kDa. The 37 kDa isoform was previously found in rat and mouse tissues in comparison with the two other proteins. The 61 kDa isoform was a protein commonly found in SH-SY5Y and HUH7 cells and commonly detected with both anti-NtermFADS3 and anti-CtermFADS3 (data not shown).
In human tissues, first, we found the same 56 kDa isoform located ubiquitously and also another slightly heavier one at 59 kDa, but only in the lung, kidney, brain, and skeletal muscles (Fig. 6B). Second, the 37 kDa isoform and a second one, just above at 39 kDa, were also detected in the same tissues as observed in rat (Fig. 4A). Pancreas did not display any of these isoforms as previously observed in rat, whereas kidney presented both isoforms undetected in rat. Third, the 75 kDa isoform especially present in the rat lung was not observed in the human lung with the anti-NtermFADS3 (data not shown) but this antibody revealed a new isoform of FADS3 at 28 kDa, mainly in the lung, kidney, pancreas, and ovary and to a lesser extent in the brain and liver.
These results together showed that FADS3 is expressed in human cultured cells and tissues under different isoforms commonly found in the murine tissues or specific to humans.
DISCUSSION
This study presented the cloning of Fads3 and, for the first time, the occurrence of the natural FADS3 protein in mammalian tissues or cells, which demonstrated that Fads3 is not a pseudogene.
The analysis of rat tissues confirmed that Fads3 is transcripted in an organ-specific manner. This data was previously exhibited in various human tissues. Marquardt et al. (6) showed, by Northern blot, a consistent level of FADS3 transcripts in the heart, brainstem, and liver as compared with the uterus and lung. We did not obtain the same result in rat using real-time PCR. First, we found the highest transcription level of Fads3 particularly in the lung but also in the heart. Our result was confirmed by Northern blot (data not shown). Second, the most unexpected result lies in the fact that the level of Fads3 mRNA is weak in the liver, which is considered the main organ of FA desaturases such as Fads1 and Fads2 (5, 23, 24). Moreover, the Fads3 mRNA profile in rat tissues was different from those determined for Fads1 and Fads2 suggesting a differential function of Fads3 in lipid metabolism.
Thereafter, taking the translation into consideration, we further investigated the expression of FADS3. We used different specific antibodies directed against the N-terminal and carboxyl-terminal sequences of rat FADS3, corresponding respectively to the cytochrome b5-like domain and the FA desaturase domain (Fig. 1). We found three potential protein isoforms (37 kDa, 51 kDa, and 75 kDa) differently distributed according to the species (rat and mouse). The 51 kDa molecular mass corresponds to the predicted size of murine FADS3 considering the cDNA sequence. When studied in rats, this 51 kDa protein was essentially present in the liver contrary to the transcript level. The data also observed for the other tissues suggest that no correlation exists between the transcription level and the translation rate of Fads3. More than the mRNA amount, the occurrence of FADS3 in liver is consistent with the idea that this protein may be an FA desaturase as invoked by its amino acid sequence.
A 75 kDa protein was also present but merely in a few tissues (present in 24% of all organs as compared with 62% for the 51 kDa protein). This occurred only by using the anti-NtermFADS3 antibody but not with the anti-CtermFADS3 antibody, which suggests that the 75 kDa protein would be devoid of the C-terminal end containing an FA desaturase domain (Fig. 1B). In that case, this suggestion is singular considering a potential desaturase activity of FADS3. Moreover, the unexpected size could result from a cleavage of FADS3 and a subsequent fusion forming a new protein complex. We can hypothesize that this posttranslational modification may confer a new function or a different subcellular targeting to the protein. Such a situation was observed, for instance, on p21-activated protein kinase 2, which is cleaved by a caspase and then myristoylated, redirecting the mature protein from the cytosol to the plasma membrane and thus, promoting cell death (25).
In native PAGE, the 51 kDa and 75 kDa proteins were detected as oligomers or protein complexes, contrary to the 37 kDa protein found as a single protein. That additional information on native proteins argues in favor of a different physiological involvement of FADS3 depending on the isoform. The 37 kDa protein was detected in mammalian tissues with only the anti-CtermFADS3, suggesting it would correspond to the putative FA desaturase domain. This short molecular mass could be explained by protein degradation or cleavage; one is known in the 169VLA•AL motif, which separates the cytochrome b5-like domain from the FA desaturase domain, but that cleavage would generate a 33 kDa protein. We can also hypothesize the possibility of an alternative translation initiation process. In that case, a 37.3 kDa fragment would be produced if the translation is initiated by the second 129Met found in the primary sequence of FADS3, even though the sequence conditions of such a process do not appear optimal here (26). Such a 37 kDa size was also observed for the Δ9-desaturase from rat liver using antibodies against C terminus of rat SCD1 and SCD2 (27). Thus, the 37 kDa protein could present the same structure as the purified Δ9-desaturase naturally devoid of fused cytochrome b5-like domain.
The three potential isoforms of FADS3 identified in rat were also detected in mouse using the same rat FADS3 antibodies. The tissue distribution was, nevertheless, different notably in muscles where no 37 kDa protein was observed in mouse tissues as compared with rat tissues. This unexpected result could be explained by the fact that the experiment was conducted on female mice. Indeed, we previously showed a significant difference between male and female rats on the transcript level of Fads3 especially in skeletal muscles. To confirm this distinction between genders, we looked at the occurrence of the 37 kDa protein in rat tissues and specifically in skeletal muscles but no difference was observed (data not shown). Thus, FADS3 is thought to be located variably depending not only on the tissues but also on the murine species or strains. This may be extended to humans, as demonstrated in our results on human cultured cells where we also identified various isoforms of FADS3 differently distributed according to the cell types (neuroblastoma and hepatoma cells) collected from males (HUH7) or females (SH-SY5Y).
The tissue distribution of FADS3 was also tested on human samples and our results underlined the presence of multiple isoforms of the natural protein. We did not observe the predicted protein at 51 kDa but two heavier ones, which molecular mass probably results from a posttranslational modification. The most unexpected result came from the new isoform at 28 kDa only detected with the anti-NtermFADS3 antibody. This short isoform corresponds to the N-terminal end of FADS3 containing the cytochrome b5-like domain. No explanation can be postulated on the nature of such a size with regard to the known putative cleavage sites, but this may be the product of an alternative transcript with truncated and skipped exons as described by Park et al. (13). This isoform would be, thus, an enzymatic cofactor or a short protein involved in the regulation of the FA metabolism.
Alternately, if different natural isoforms of FADS3 were present in humans, these results obtained on a commercial tissue blot have nevertheless to be considered as the result on one sample of one human being. Further experiments have to be investigated to confirm these results from independent human samples taking into account the heterogeneity of a population.
To conclude, FADS3 does exist. In this study, we reported the occurrence of different potential isoforms of FADS3 in rat, mouse, and human tissues. The demonstration of multiple Fads3 gene products was not unexpected with regard to Park's results on alternative transcripts of FADS3 expressed in baboon tissues, even if we did not find nine proteins (13). If FADS3 is ubiquitously expressed in the organism, its biological function remains unknown. Our preliminary experiments carried out on pCMV-Fads3 transfected Cos-7 and SH-SY5Y cells did not result in a change of the lipid pattern. Our findings on the protein profile exposed in this article have now to be considered for further investigations on the physiological role of FADS3, as an enzyme as predicted by its structure or maybe as a regulation protein in the FA metabolism.
Acknowledgments
We thank the Région Bretagne, the Groupe Lipides et Nutrition, Valorex (Combourtillé, France), and Polaris (Pleuven, France) for their financial support.
Footnotes
Abbreviations:
- AT
- adipose tissue
- FADS
- fatty acid desaturase
- FCS
- fetal calf serum
- LC-PUFA
- long-chain PUFA
- RIPA
- radio immuno precipitation assay
This work was supported by the Région Bretagne, the Groupe Lipides et Nutrition, Valorex (Combourtillé, France), and Polaris (Pleuven, France).
REFERENCES
- 1.Spector A. A. 1999. Essentiality of fatty acids. Lipids. 34 (suppl): S1–S3. [DOI] [PubMed] [Google Scholar]
- 2.Aki T., Shimada Y., Inagaki K., Higashimoto H., Kawamoto S., Shigeta S., Ono K., Suzuki O. 1999. Molecular cloning and functional characterization of rat Δ-6 fatty acid desaturase. Biochem. Biophys. Res. Commun. 255: 575–579. [DOI] [PubMed] [Google Scholar]
- 3.Cho H. P., Nakamura M., Clarke S. D. 1999. Cloning, expression and fatty acid regulation of the human Δ-5 desaturase. J. Biol. Chem. 274: 37335–37339. [DOI] [PubMed] [Google Scholar]
- 4.Cho H. P., Nakamura M., Clarke S. D. 1999. Cloning, expression and nutritional regulation of the mammalian Δ-6 desaturase. J. Biol. Chem. 274: 471–477. [DOI] [PubMed] [Google Scholar]
- 5.Matsuzaka T., Shimano H., Yahagi N., Amemiya-Kudo M., Yoshikawa T., Hasty A. H., Tamura Y., Osuga J., Okazaki H., Iizuka Y., et al. 2002. Dual regulation of mouse Δ5- and Δ6-desaturase gene expression by SREBP-1 and PPARα. J. Lipid Res. 43: 107–114. [PubMed] [Google Scholar]
- 6.Marquardt A., Stöhr H., White K., Weber B. H. F. 2000. cDNA cloning, genomic structure, and chromosomal localization of three members of the human fatty acid desaturase family. Genomics. 66: 175–183. [DOI] [PubMed] [Google Scholar]
- 7.Malerba G., Schaeffer L., Xumerle L., Klopp N., Trabetti E., Biscuola M., Cavallari U., Galavotti R., Martinelli N., Guarini P., et al. 2008. SNPs of the FADS gene cluster are associated with polyunsaturated fatty acids in a cohort of patients with cardiovascular disease. Lipids. 43: 289–299. [DOI] [PubMed] [Google Scholar]
- 8.Tanaka T., Shen J., Abecasis G. R., Kisialiou A., Ordovas J. M., Guralnik J. M., Singleton A., Bandinelli S., Cherubini A., Arnett D., et al. 2009. Genome-wide association study of plasma polyunsaturated fatty acids in the InCHIANTI Study. PLoS Genet. 5: e1000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aulchenko Y. S., Ripatti S., Lindqvist I., Boomsma D., Heid I. M., Pramstaller P. P., Penninx B. W., Janssens A. C., Wilson J. F., Spector T., et al. 2009. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat. Genet. 41: 47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kathiresan S., Willer C. J., Peloso G. M., Demissie S., Musunuru K., Schadt E. E., Kaplan L., Bennett D., Li Y., Tanaka T., et al. 2009. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat. Genet. 41: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma X. H., Hu S. J., Ni H., Zhao Y. C., Tian Z., Liu J. L., Ren G., Liang X. H., Yu H., Wan P., et al. 2006. Serial analysis of gene expression in mouse uterus at the implantation site. J. Biol. Chem. 281: 9351–9360. [DOI] [PubMed] [Google Scholar]
- 12.Tondreau T., Dejeneffe M., Meuleman N., Stamatopoulos B., Delforge A., Martiat P., Bron D., Lagneaux L. 2008. Gene expression pattern of functional neuronal cells derived from human bone marrow mesenchymal stromal cells. BMC Genomics. 9: 166–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park W. J., Kothapalli K. S., Reardon H. T., Kim L. Y., Brenna J. T. 2009. Novel fatty acid desaturase 3 (FADS3) transcripts generated by alternative splicing. Gene. 446: 28–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guillou H., D'Andréa S., Rioux V., Barnouin R., Dalaine S., Pédrono F., Jan S., Legrand P. 2004. Distinct roles of endoplasmic reticulum cytochrome b5 and fused cytochrome b5-like domain for rat Δ6-desaturase activity. J. Lipid Res. 45: 32–40. [DOI] [PubMed] [Google Scholar]
- 15.Stroud C. K., Nara T. Y., Roqueta-Rivera M., Radlowski E. C., Lawrence P., Zhang Y., Cho B. H., Segre M., Hess R. A., Brenna J. T., et al. 2009. Disruption of FADS2 gene in mice impairs male reproduction and causes dermal and intestinal ulceration. J. Lipid Res. 50: 1870–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stoffel W., Holz B., Jenke B., Binczek E., Günter R. H., Kiss C., Karakesisoglou I., Thevis M., Weber A. A., Arnhold S., et al. 2008. Delta6-desaturase (FADS2) deficiency unveils the role of omega3- and omega6-polyunsaturated fatty acids. EMBO J. 27: 2281–2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D'Andrea S., Guillou H., Jan S., Catheline D., Thibault J-N., Bouriel M., Rioux V., Legrand P. 2002. The same rat Δ6-desaturase not only acts on 18- but also on 24-carbon fatty acids in very-long-chain polyunsaturated fatty acid biosynthesis. Biochem. J. 364: 49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guillou H., Rioux V., Catheline D., Thibault J-N., Bouriel M., Jan S., D'Andrea S., Legrand P. 2003. Conversion of hexadecanoic acid to hexadecenoic acid by rat Delta 6-desaturase. J. Lipid Res. 44: 450–454. [DOI] [PubMed] [Google Scholar]
- 19.Park W. J., Kothapalli K. S., Lawrence P., Tyburczy C., Brenna J. T. 2009. An alternate pathway to long chain polyunsaturates: the FADS2 gene product Δ8-desaturates 20:2n-6 and 20:3n-3. J. Lipid Res. 50: 1195–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramakers C., Ruijter J. M., Deprez R. H., Moorman A. F. 2003. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339: 62–66. [DOI] [PubMed] [Google Scholar]
- 21.Iwamura T., Yoneyama M., Yamaguchi K., Suhara W., Mori W., Shiota K., Okabe Y., Namiki H., Fujita T. 2001. Induction of IRF-3/-7 kinase and NF-κB in response to double-stranded RNA and virus infection: common and unique pathways. Genes Cells. 6: 375–388. [DOI] [PubMed] [Google Scholar]
- 22.R Development Core Team. 2008. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: ISBN 3–900051–07–0. Accessed 2009. Available at http://www.R-project.org. [Google Scholar]
- 23.Saether T., Tran T. N., Rootwelt H., Christophersen B. O., Haugen T. B. 2003. Expression and regulation of Δ5-desaturase, Δ6-desaturase, stearoyl-coenzyme A (CoA) desaturase 1, and stearoyl-CoA desaturase 2 in rat testis. Biol. Reprod. 69: 117–124. [DOI] [PubMed] [Google Scholar]
- 24.Zolfaghari R., Cefelli C. J., Banta M. D., Ross C. 2001. Fatty acid Δ5-desaturase mRNA is regulated by dietary vitamin A and exogenous retinoic acid in liver of adult rats. Arch. Biochem. Biophys. 391: 8–15. [DOI] [PubMed] [Google Scholar]
- 25.Vilas G. L., Corvi M. M., Plummer G. J., Seime A. M., Lambkin G. R., Berthiaume L. G. 2006. Posttranslational myristoylation of caspase-activated p21-activated protein kinase 2 (PAK2) potentiates late apoptotic events. Proc. Natl. Acad. Sci. USA. 103: 6542–6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Touriol C., Bornes S., Bonnal S., Audigier S., Prats H., Prats A. C., Vagner S. 2003. Generation of protein isoform diversity by alternative initiation of translation at non-AUG codons. Biol. Cell. 95: 169–178. [DOI] [PubMed] [Google Scholar]
- 27.Heinemann F. S., Ozols J. 2003. Stearoyl-CoA desaturase, a short-lived protein of endoplasmic reticulum with multiple control mechanisms. Prostaglandins Leukot. Essent. Fatty Acids. 68: 123–133. [DOI] [PubMed] [Google Scholar]