Abstract
Ceramides (Cers) are important in embryogenesis, but no comprehensive analysis of gene expression for Cer metabolism nor the Cer amounts and subspecies has been conducted with an often used model: mouse embryonic stem cells (mESCs) versus embroid bodies (EBs). Measuring the mRNA levels by quantitative RT-PCR and the amounts of the respective metabolites by LC-ESI/MS/MS, notable differences between R1 mESCs and EBs were: EBs have higher mRNAs for CerS1 and CerS3, which synthesize C18- and C≥24-carbons dihydroceramides (DH)Cer, respectively; EBs have higher CerS2 (for C24:0- and C24:1-); and EBs have lower CerS5 + CerS6 (for C16-). In agreement with these findings, EBs have (DH)Cer with higher proportions of C18-, C24- and C26- and less C16-fatty acids, and longer (DH)Cer are also seen in monohexosylCers and sphingomyelins. EBs had higher mRNAs for fatty acyl-CoA elongases that produce C18-, C24-, and C26-fatty acyl-CoAs (Elovl3 and Elovl6), and higher amounts of these cosubstrates for CerS. Thus, these studies have found generally good agreement between genomic and metabolomic data in defining that conversion of mESCs to EBs is accompanied by a large number of changes in gene expression and subspecies distributions for both sphingolipids and fatty acyl-CoAs.
Keywords: embryonic stem cell, embryoid body, sphingolipid, differentiation, ceramide synthase, fatty acyl-CoA elongase
Complex sphingolipids (SLs), including glycosphingolipids (GSLs), are a highly diverse family of compounds (1) that have long been regarded to be important in biological processes such as embryogenesis (2–4). The ceramide (Cer) moieties of SLs and GSLs also change in amount during embryogenesis (2–6), and appear to participate in developmental signaling (4, 7–10).
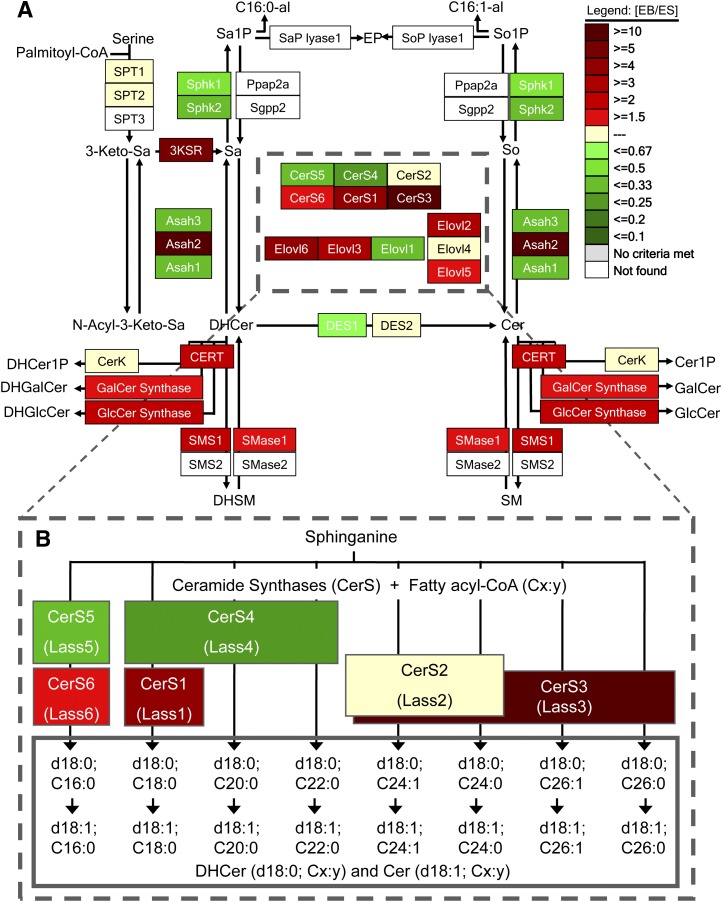
Ceramides are biosynthesized de novo (1, 11) and by recycling (12, 13) pathways that have been well characterized with respect to the intermediates and genes, as depicted in a scheme (Fig. 1) recently described in this journal (14). De novo biosynthesis from serine and palmitoyl-CoA by serine palmitoyltransferase (SPT) (15) first forms 3-ketosphinganine (3-Keto-Sa), which is reduced to sphinganine (Sa, which is also abbreviated d18:0), then acylated to dihydroceramides (DHCer) by a family of ceramide synthases, CerS, that differ in fatty acyl-CoA selectivity as shown in the expanded pathway diagram, Fig. 1B (16–18). DHCer are incorporated into more complex dihydroSL [dihydrosphingomyelins (DHSM), dihydroglucosylceramides (DHGlcCer), etc.] or converted to Cer by addition of a 4-trans-double bond to produce the sphingosine backbone (abbreviated d18:1) (19), then incorporated into more complex SLs.
Fig. 1.
Depiction of the early steps of sphingolipid biosynthesis using a modified KEGG pathway scheme (Sphingo-GenMAPP) (14) to facilitate visualization of differences in gene expression in R1 mESCs and EBs. A: Beginning in the upper left of this scheme, serine and palmitoyl–CoA are condensed by serine palmitoyltransferase (represented by 3 SPT genes) to form 3-ketosphinganine (3-Keto-Sa), which is reduced to sphinganine (Sa) by 3-keto-Sa reductase (3KSR, the FVT1 gene) and either N-acylated by ceramide synthases (CerS) to dihydroceramides (DHCer) (shown by arrows leading to the right) or phosphorylated by sphingosine kinase (SphK1 or SphK2) to Sa 1-phosphate (Sa1P), which can be hydrolyzed back to Sa (by phosphatases Ppap2a or Sgpp2) or cleaved to hexadecanal (C16:0-al) and ethanolamine phosphate (EP) by the shown lyase (sphingosine 1-phosphate lyase). DHCer can be hydrolyzed to Sa by ceramidases (Asah1, 2 or 3); incorporated into more complex sphingolipids (DHCer 1-phosphate, DHCer1P, by Cer kinase, CERK), glycosylated to dihydrogalactosylCer (DHGalCer), dihydroglucosylCer (DHGlcCer), converted to dihydrosphingomyelin (DHSM by SM synthases SMS1 or SMS2); or converted to Cer by DHCer desaturase (DES1 or DES2) (or hydroxylated to phytoceramide by DES2, not shown) and either hydrolyzed by ceramidases or converted to downstream metabolites, etc. Also shown are the fatty acyl-CoA elongases (Elovl) because some of these are involved in production of the cosubstrates for CerS. B: Expansion of the pathway to depict the individual molecular species made by different CerS isoforms (formerly named Lass), with abbreviation of the sphingoid base as d18:0 for sphinganine (citing the number of hydroxyl groups, d for dihydroxyl), d18:1 for sphingosine, and the N-acyl chain length and number of double bonds as Cx:y, respectively. As described in the text, the “heat” scale depicts the relative amounts of transcripts for each of these genes for R1 EBs versus mESCs as measured by qRT-PCR (see Fig. 2).
From this diagram, one would predict that shifts in the expression of the various CerS, and possibly of the availability of the corresponding fatty acyl-CoAs, can affect the N-acyl chain length subspecies of DHCer and Cer (and downstream more complex SLs and GSLs) in cells. To test this hypothesis, we chose mouse embryonic stem cells (mESCs) and embryoid bodies (EBs) because this is a widely studied developmental model (20). EBs are aggregates of cells derived from ESCs that, to some extent, recapitulate early stages of mammalian embryonic development, including cell types derived from endoderm, mesoderm, and ectodermal lineages (20). Described in this report are the expression of the CerS genes (16, 21) and the amounts and types of Cer subspecies as part of a broader investigation of all of the known genes for glycoprotein and (glyco)sphingolipid biosynthesis (22). Also analyzed were the cosubstrate fatty acyl-CoAs and expression of transcripts for fatty acid elongases and desaturases as these may also influence the types of Cer subspecies. These studies found that there were shifts in the expression of many of the genes of this pathway as well as in the associated metabolites in EBs versus ESCs.
MATERIALS AND METHODS
Growth and characterization of mESCs and EBs
The R1 and D3 mESCs (23, 24) (ATCC, Manassas, VA) were cultured in the absence of feeder cells on tissue-culture-grade plastic ware precoated with 0.1% gelatin-PBS, as described previously (25). The mESC culture medium consisted of DMEM (Gibco BRL) supplemented with 10% fetal calf serum (GIBCO BRL), 1 mM L-glutamine, 0.1 mM 2-mercaptoethanol, 100 U/ml penicillin, 100 U/ml streptomycin, and 1000 U/ml recombinant human LIF (leukemia inhibitory factor; ESGRO, Chemicon International) at 37°C under 10% CO2.
Differentiation of mESCs into EBs was carried out as described previously (26). mESCs were harvested by trypsinization to convert suspensions of single ESCs into aggregates that were seeded into 10-cm bacteriological dishes at a density of 1 × 105 cells/ml, in 10 ml mESC-medium lacking LIF. EBs were harvested daily, the medium was changed every 2 days and cultures were split one into two at day 4. Cell populations of mESCs and EBs were characterized by flow cytometry using the lineage specific marker CD9 and by transcript analysis using qRT-PCR. Changes in morphology and lineage-specific gene expression for mESC and EB cell populations produced by the Dalton laboratory were previously described (27).
Measurement of gene expression levels using qRT-PCR
Primer design and validation.
Primer pairs for SL synthesis genes and control genes were designed within a single exon using conditions described in Nairn et al. (22). Primer sequences and accession numbers for genes in this study are presented in Nairn et al. (27). Individual quantitative (q)RT-PCR reactions were also checked for efficiency 100 ± 5% using reaction raw fluorescence data in the LinReg PCR program (28).
Total RNA isolation and cDNA synthesis.
mESCs and EBs (day 6) cell pellets were harvested and flash-frozen in liquid nitrogen and stored at −80°C until use. Cell pellets were homogenized followed by RNA isolation and cDNA synthesis, which were performed as previously described (27).
qRT-PCR reactions and calculation of relative gene expression levels.
Triplicate reactions (20 μl each) containing 5 μl of diluted cDNA or genomic DNA template (primer validation reactions), 5 μl of primer pair mix (125 μM final concentration), and 10 μl of iQ™ SYBR Green Supermix (BioRad, Hercules, CA) were assembled in 96-well microtitre plates. Amplification reactions and the calculation of relative gene expression levels were carried out as previously described (27). The normalization gene was selected by determining the expression level for several housekeeping genes across all samples to determine the most stable gene as described in (22). The control gene, Ribosomal Protein L4 (RPL4, NM_024212) was included on each plate to control for run variation and to normalize individual gene expression.
Integration of gene expression data with Sphingo-GenMAPP pathway
GenMAPP (Gene MicroArray Pathway Profiler) v2.1 (29) is a tool to visualize global gene-expression profiles in the context of cell signaling pathways or KEGG metabolic pathways (31). In the context of the present study, the transcript expression data for R1 mESCs and EBs from the qRT-PCR analysis was imported into a GenMAPP pathway for SL metabolism that had been modified (14) based on the current literature (32) (Fig. 1).
LC-ESI MS/MS
SLs.
The SLs were extracted from cells using published methods (33–35). Briefly, ∼1 to 10 × 106 cells were scraped from culture dishes in a small volume of PBS, then centrifuged to remove excess PBS for a final volume of 0.05–0.1 ml. The cells were suspended in 0.5 ml of CH3OH and 0.25 ml of CHCl3, spiked with a single chain-length internal standard for each category of SL being monitored (which we have found to allow quantitative analysis of all of the compounds in this study, as reported in reference 35)(see details below), disrupted using a bath-type sonicator at room temperature, and incubated overnight in a heating block at 48°C. After cooling the tubes to room temperature, 75 μl of 1 M KOH in CH3OH was added and the mixture was sonicated for 30 s, then incubated for 2 h at 37°C. The solution was neutralized with several μl of glacial acetic acid, 1 ml of CHCl3 was added followed by 2 ml of water, and sonicated. After centrifugation, the upper phase was carefully removed and discarded. The lower phase was dried using a SpeedVac and redissolved in 0.3 ml of 98% of 97:2:1 CH3CN/CH3OH/CH3COOH (v/v/v) (mobile-phase A) and 2% of 64:15:20 CH3OH/H2O/CH3(CH2)3OH/CH3COOH (v/v/v/v) (mobile-phase B); both contained 5 mM ammonium acetate.
The methods used for LC-ESI MS/MS analysis of SL metabolites have been reported in detail (34, 35). SL data was collected using a Perkin Elmer Series 200 MicroPump system (Perkin- Elmer, Norwalk, CT) coupled to a PE Sciex API 3000 triple quadrupole mass spectrometer equipped with a turbo ion-spray source (Applied Biosystems, Foster City, CA). Complex SLs such as Cers, CMHs (ceramide monohexoses, the sum of glucosylceramide and galactosylceramide), and SMs were separated by normal-phase LC using 2.1 mm × 5 cm Supelco NH2 column (Supelco, Bellefonte, PA) at a flow rate of 1.5 ml/min. The elution protocol was: 0.5 min of preequilibration of the column with 98:2 A/B (v/v), sample injection, elution with 98:2 A/B (v/v) for 1.1 min followed by a linear gradient to 82:18 A/B (v/v) in 0.2 min, elution at this composition for 0.4 min, then a linear gradient to 100% B over 0.8 min. Afterwards, the column was reequilibrated by a 0.5 min linear gradient to 98:2 A/B (v/v) and a wash of the column with 98:2 A/B (v/v) for 0.3 min.
Quantitation was performed by comparison of each unknown with the spiked internal standard (IS) for that SL class (the IS for each class was an uncommon chain length homolog that has been validated for analysis of other chain lengths) (35): C12-ceramide, C12-sphingomyelin, and C12-glucosylceramide (for CMH), all of which were obtained from Avanti Polar Lipids (Alabaster, AL). The ISs were added as 10 µl of a 50 µM of stock for a total of 500 pmol of each IS molecular species per sample (see above). Student's t-test was used for statistical analyses.
Fatty Acyl-CoAs.
The fatty acyl-CoAs were extracted from cells using a published method (36). The medium from cells cultured in 100 mm dishes was aspirated and cells were gently washed twice with 5 ml of 4°C PBS, ∼1 ml of ice-cold PBS was added, and the cells were scraped from the dishes using a cell scraper and transferred to chilled 13 × 100 mm screw-cap borosilicate tubes with Teflon-lined caps (Kimble Chase, Vineland, NJ). An additional ∼1 ml of PBS was used to recover the remaining cells by this same procedure and these were added to the first. After centrifugation (1500 rpm, 3 min), the PBS was removed by aspiration and 0.5 ml of CH3OH containing 1 mM EDTA was added together with 10 μl of an internal standard mixture that contained 100 pmol each of C17:0- and C25:0-CoA, which were prepared in CH3OH/CHCl3 (2:1 v/v) containing 30 mM triethylammonium acetate. After sonication (three times at ∼0.5 min each), 0.25 ml of CHCl3 was added followed by another brief sonication.
The single-phase extraction mixture was incubated in a 50°C heating block for 30 min. After cooling to room temperature, CHCl3 and water (0.25 ml each) were added with mixing by vortexing after each addition. After brief centrifugation using a table-top centrifuge, the fatty acyl-CoAs were in the upper phase and interface and most of the other lipids (which would interfere with the subsequent reverse-phase LC) were in the lower phase. The upper phase was removed with a Pasteur pipette and transferred into a clean screw-cap glass tube. The upper layer was adjusted with 5% CH3OH (v/v), 5% butanol (v/v), and 5% CHCl3 (v/v). These organic additives improved the stability of the very long-chain acyl-CoAs for at least 24 h at room temperature in the autosampler.
HPLC separations used a Shimadzu SCL-10A VP system controller, two LC-10AD VP pumps, a DGU-14A degassing unit, a Perkin-Elmer series 200 autosampler, a Phenomenex (Torrance, CA) Gemini C18 column (2 mm ID × 150 mm with 3 μm particles, 110 Angstrom pores) and a 2 × 4 mm guard column with the same packing material. A Metatherm column oven (Torrance, CA) was used to maintain the column temperature at 40°C.
The flow rate was 0.2 ml/min in binary gradient mode with the following elution program: the column was equilibrated with mobile phase C [water/ CH3CN (85:15, v/v), containing 0.05% triethylamine], the sample was injected, and mobile phase C was continued for 5 min, followed by a 13 min gradient to 48% mobile phase C and 52% mobile phase D (CH3CN containing 0.05% triethylamine) during which the long-chain- and very long-chain-fatty acyl-CoAs eluted. Afterward, the column was washed by a 1 min gradient to 100% D and a 5 min hold at 100% D, followed by reequilibration of the column by a 1 min gradient to 100% C and a 5 min hold at 100% C before injection of the next sample.
MS/MS of fatty acyl-CoAs in positive ion mode was performed on a 4000 QTrap triple quadrupole linear ion trap mass spectrometer (Applied Biosystems) as recently described (36).
RESULTS
Characterization of R1 mESCs and EBs
The R1 mESCs and EBs were examined for gross morphological changes and transcript abundances as markers for these cells. The mESCs expressed Zinc finger protein 42 (zfp42/Rex1), Gastrulation brain homeobox (GBX2), and Nanog homeobox (Nanog) as pluripotent markers, and the qRT-PCR transcript profiles for Goosecoid (Gsc), Zinc finger protein of the celebellum 1 (Zic1), SRY-box containing gene 17 (Sox17), Alpha fetoprotein (Afp), and GATA binding protein 6 (Gata6) were typical for the transition from mESCs to EBs (22).
Differences in the expression of SL metabolism genes in R1 mESCs and EBs using Sphingo-GenMAPP
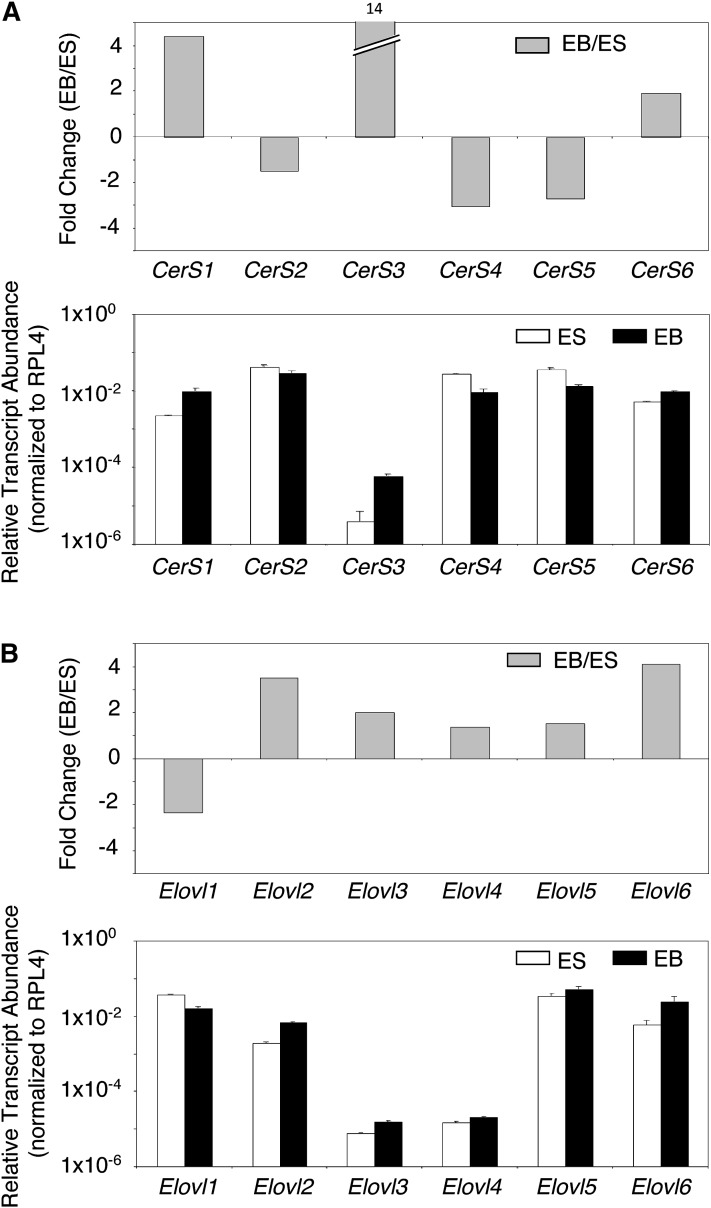
The relative transcript expression levels of R1 EBs versus mESCs are depicted in the pathway diagram (Fig. 1) and with the fold differences shown by the “heat” scale defined on the right side of the figure. There were no differences in the transcript levels for serine palmitoyltransferases (SPT1 and SPT2), the first enzymes of the pathway, but EBs had higher expression of transcripts for 3-ketosphinganine reductase and some of the CerS isoenzymes (CerS1, 3, and 6), as is also shown by bar graphs in Fig. 2 (upper panel). The transcripts were also higher in EBs for enzymes that use Cer for biosynthesis of more complex SLs, i.e., both of the SM synthases and the glycosyltransferases that produce GlcCer and GalCer (Fig. 1). EBs had lower transcript abundances for the major desaturase that synthesizes Cer from DHCer (dihydroCer desaturase, DES1) and transcripts for the other desaturase (DES2, which makes phytoceramides) (37) were barely detectable in both mESCs and EBs.
Fig. 2.
Relative transcript abundance of CerS (Lass) genes (A) and fatty acyl-CoA elongase (Elovl) genes (B) by qRT-PCR for R1 mouse embryonic stem cells and day 6 EBs. A: Upper panel, fold change in relative transcript abundance of CerS genes for EBs/mESCs. A value of zero represents no change in transcript abundance. Values greater than zero represent an increase in transcript abundance in EBs relative to mESCs and values less than zero represent a decrease in EBs relative to mESCs. Values that are off-scale are indicated by a hatched bar and the fold-change value is given above the bar. Lower panel, bars represent average Ct values from triplicate samples normalized to the housekeeping gene RPL4 (as described in the Materials and Methods). Relative transcript abundance for mESCs (white bars) and EBs (black bars) are plotted on a log scale for each gene assayed. Error bars represent 1 SD from the mean. Transcript values less than 1 × 106 are below the threshold of detection. B: Relative transcript abundance of Elovl genes in R1 mESCs versus EBs determined by qRT-PCR. Upper panel, fold change in relative transcript abundance of fatty acyl-CoA elongase gene family members (Elovl1-6) for EBs/mESCs as described for A above. Lower panel, relative transcript abundance for mESCs (white bars) and EBs (black bars) are plotted as described for A above.
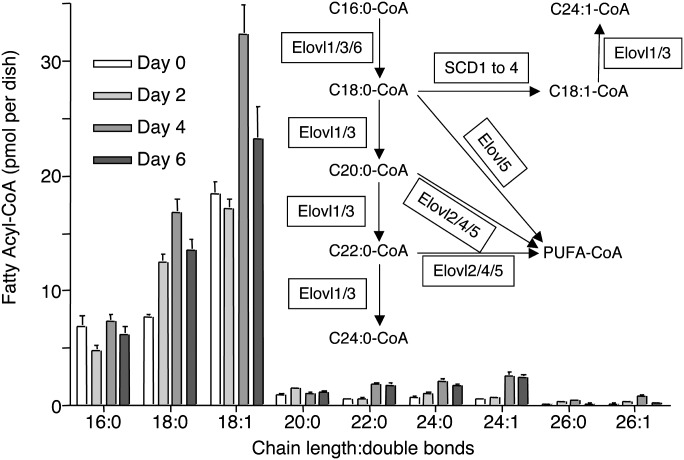
Because the CerS enzymes use very long-chain fatty acyl-CoAs that are made by elongation of palmitoyl- and stearoyl-CoAs, the transcripts for the pertinent fatty acyl-CoA elongases were also analyzed by qRT-PCR. As shown in Fig. 1 and 2B, the transcript level for Elovl6, a fatty acyl-CoA elongase that catalyzes the elongation of saturated and monosaturated acyl-CoAs with 12-16 carbons (38, 39), was 4-fold higher for EBs than mESCs. A fatty acyl-CoA elongase that extends C16:0-CoA to C24:0-CoA, Elovl3 (38, 40, 41), showed 3-fold higher transcript levels for EBs than mESCs, although lower transcript levels were observed for Elovl1, which is also responsible for elongating very long-chain saturated fatty acyl-CoAs (38, 41). EBs also had ∼3.5-fold higher Elovl2 (37, 42) transcript levels, no evident change in Elovl4 (38, 44) transcripts, and slightly higher Elovl5 (38, 43) transcript abundance (which are all elongases for polyunsaturated fatty acids).
These differences in gene expression imply that the SL backbones of EBs might differ from mESCs by having: i) higher proportions of subspecies with longer acyl chains (i.e., >C16) because EBs have a higher transcript abundance of CerS1, which is relatively specific for stearoyl-CoA (C18:0) (45), and CerS3, which is selective for very-long-chain fatty acyl-CoAs (>C20) (46); ii) higher, lower, or unchanged C16-Cer because an elevation in CerS6 (17), one of the CerS responsible for biosynthesis of C16-Cer, may be offset by a decrease in the transcripts encoding a CerS that uses C16-fatty acyl-CoA, CerS5 (47); iii) increases in DHSL as DES1 is lower and DES2 unchanged; and iv) possibly lower amounts of (DH)Cer because the transcripts encoding most of the enzymes that use (DH)Cer were elevated (including the transcripts for one of the ceramidases, Asah2). The gene expression data also suggest that the EBs have the capacity to elongate fatty acyl-CoAs to produce the cosubstrates that are used to make these Cer subspecies. To test these hypotheses, the Cer subspecies (and fatty acyl-CoAs) were analyzed by LC-ESI MS/MS.
Characterization of the Cer backbone subspecies of R1 mESCs and EBs
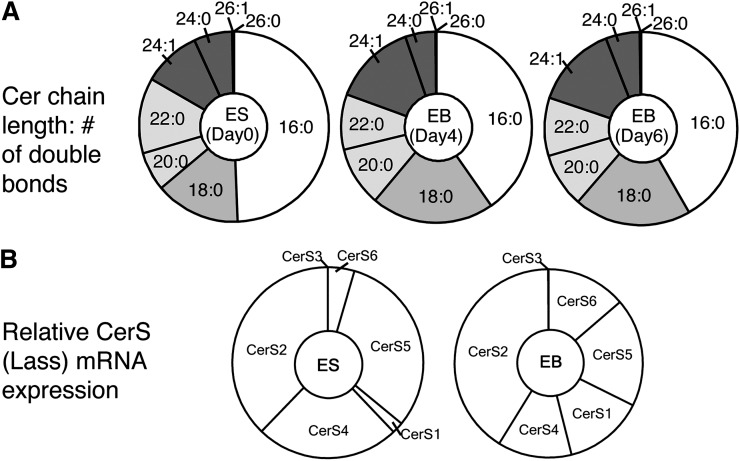
Fig. 3A shows the relative proportions of the Cer subspecies in R1 mESCs (day 0) and EBs on days 4 and 6 as “donut” charts.2 The rationale for the latter depiction is that CerSs have approximately the same Km for the sphingoid base substrate (48); therefore, as a first approximation, one would predict that the relative amounts of the different subspecies might correlate with the relative abundances of the respective CerS transcripts, which have been regraphed in Fig. 3B to show the differences between mESCs and EBs. The order of the CerS genes has been arranged so they correspond to the respective types of Cer that are made, i.e., C16-Cer (CerS6 and CerS5), C18-Cer (CerS1), C20 ± 2 Cer (CerS4), C24-Cer (CerS2), and C26-Cer (CerS3) (c.f. Fig. 1).
Fig. 3.
Ceramide composition of R1 mouse embryonic stem cells at different timepoints during EB formation. A: Cer subspecies distribution with the chain length and number of double bonds depicted by x:y, respectively, for mESCs and EBs after days 4 and 6. The Cer subspecies were analyzed by LC ESI-MS/MS as described in references 34–36. B: Representation of relative changes in CerS transcript abundance of mESCs versus day 6 EBs arranged with the approximate order of the fatty acyl-CoA selectivities of the CerS to match the acyl-chain lengths of the Cer in A.
Comparison of Fig. 3A and B reveals a close correspondence between the differences in transcript abundances for CerS in mESC versus EBs and the Cer subspecies distribution: a lower CerS6 + CerS5 for EBs versus mESC corresponding to a decrease in C16-Cer, higher CerS1 in EBs corresponding to an increase in C18-Cer, and higher CerS2 corresponding to a small increase in C24-Cer. Lower CerS4 transcripts in EBs did not translate into a decrease in the sum of C18 + C20 + C22-Cer; however, this might be due to the large increase in CerS1 transcripts.
Characterization of the DHCer and Cer backbone subspecies of (DH)SL in R1 mESCs and EBs
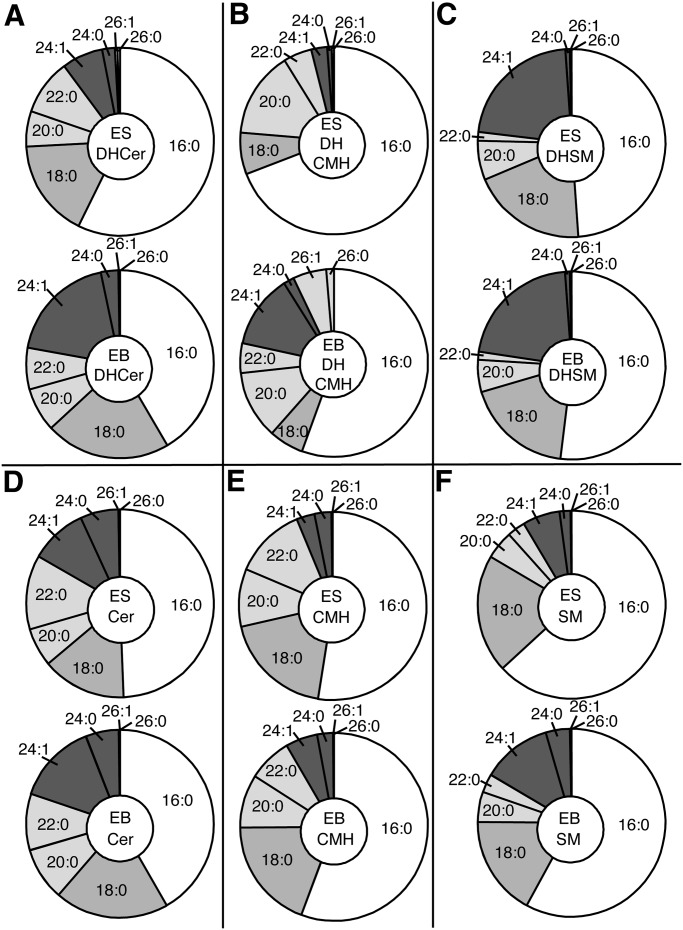
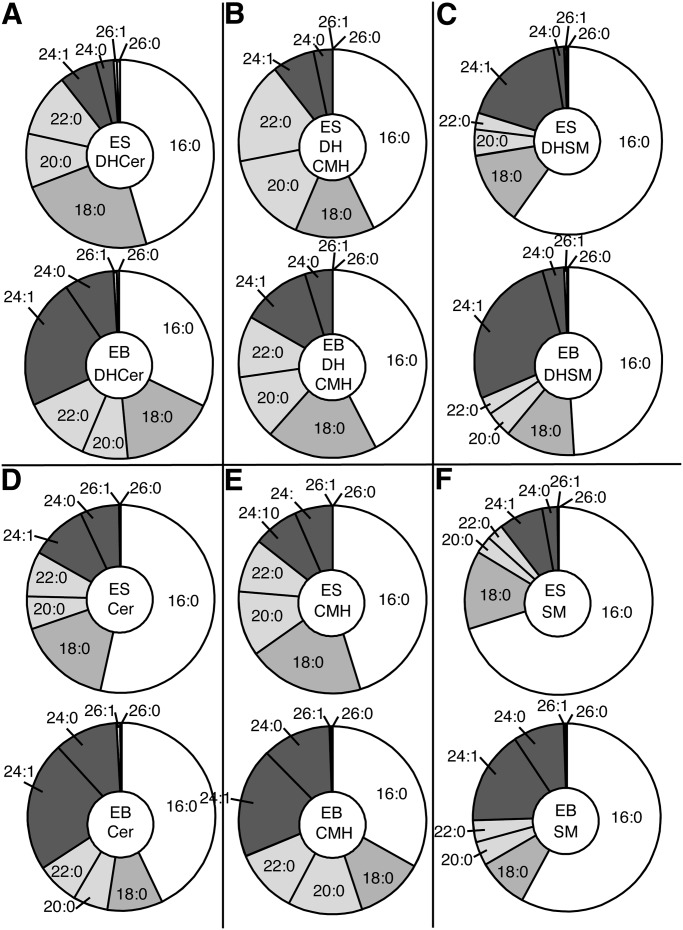
The DHCer of mESCs and day 6 EBs also exhibited shifts in fatty acyl-chain length (Fig. 4A) that correlated with the differences in CerS expression that were similar to the changes in Cer (c.f., Fig. 4A and Fig. 4D), namely, lower proportions of C16-DHCer and more C18-and C24/24:1 DHCer with the greatest increase in C24:1-Cer. Because DHCer is the first N-acylated intermediate of de novo sphingolipid biosynthesis (Fig. 1) versus Cer, which is formed next and during the turnover of more complex sphingolipids, perhaps this accounts for the closer resemblance between the chain length subspecies of DHCer and the transcript abundance of the CerS isoforms (c.f., Fig. 3A and 4).
Fig. 4.
Subspecies distributions of (dihydro)ceramides, (dihydro)ceramide monohexoses and (dihydro)sphingomyelins of R1 mouse embryonic stem cells and day 6 embryoid bodies. The sphingolipids were analyzed by LC-ESI MS/MS as described in references 34–36 and are shown with the ESCs as the upper member of each pair and EBs as the lower dataset. A: Dihydroceramides (DHCer); B: dihydroceramide monohexoses (DHCMH); C: dihydrosphingomyelins (DHSM); D: ceramides (Cer); E: ceramide monohexoses (CMH); and F: sphingomyelins (SM).
The (DH)CMH (Fig. 4B and E) and SM (Fig. 4F) of EBs displayed higher proportions of the C24/C24:1 backbones, however, most of the other subspecies differences that were seen in Cer and DHCer were not evident in these downstream metabolites.
There were also differences in the proportion of DHCer versus Cer backbones in EBs versus mESCs (which are not evident in because the data are shown only as proportions within each category). When summed across all chain lengths and categories, the total amount of these sphingolipids did not differ between mESCs and day 6 EBs cells (i.e., 780 vs. 740 pmol/mg protein, respectively), but the DHSL percentage increased from ∼8% for mESCs to 19% for EBs. This finding was consistent with the lower DES1 expression in EBs versus mESCs (Fig. 1).
Analysis of the fatty acyl-CoA species in R1 mESCs and EBs
The amounts of the fatty acyl-CoAs in R1 mESCs and EBs are shown in Fig. 5. During the time course for conversion of mESCs to EBs, C18:0- and C18:1-fatty acyl-CoAs increased by approximately 2-fold, as did many of the very long-chain (C22:0-, C24:0- and C24:1-) fatty acyl-CoAs. In contrast, palmitoyl-CoA derivatives (C16:0-) were approximately the same in mESCs and EBs. Thus, these results are consistent with the observed changes in the higher amounts of C18:1-, C22:0-, C24:0-, and C24:1-fatty acyl-CoAs in EBs versus mESCs. These results additionally indicate that the CerS that would use these fatty acyl-CoAs to produce the pertinent Cer subspecies are not hampered by a lack of cosubstrate availability during EB development.
Fig. 5.
Fatty acyl-CoA composition of R1 mouse embryonic stem cells at different timepoints during EB formation. Fatty acyl-CoA subspecies of mESCs and EBs for days 2, 4, and 6. Each bar represents the mean ± SD of 6 dishes and analyzed by LC-ESI MS/MS as described in reference 37. The inset pathway shows the metabolic reactions catalyzed by the respective fatty acyl-CoA elongases (Elovl1-6). PUFA-CoA stands for polyunsaturated fatty acyl-CoAs.
Characterization of the DHCer and Cer backbone subspecies of (DH)SL in D3 mESCs and EBs
To determine if these compositional changes were unique to the R1 cells, the lipids of another frequently studied mESC cell line, D3 cells (25), were analyzed by LC/MS/MS. The two similarities observed were decreases in the proportion of C16-(DH)Cer and increases in C24/C24:1 (DH)Cer in D3 EBs versus mESCs, (Fig. 6A, D). Furthermore, these differences were also more prevalent in the downstream metabolites (Fig. 6B, C, E, F). Based on these similarities, it is possible that the shifts in (DH)Cer subspecies are common in this developmental transition.
Fig. 6.
Subspecies distributions of (dihydro)ceramides, (dihydro)ceramide monohexoses and (dihydro)sphingomyelins of D3 mouse embryonic stem cells and day 6 embryoid bodies. The sphingolipids were analyzed by LC ESI-MS/MS as described in reference 34–36 and are shown with the ESCs as the upper member of each pair and EBs as the lower dataset. A: Dihydroceramides (DHCer); B: dihydroceramide monohexoses (DHCMH); C: dihydrosphingomyelins (DHSM); D: ceramides (Cer); E: ceramide monohexoses (CMH); and F: sphingomyelins (SM).
DISCUSSION
The goal of these studies was to explore whether mESCs undergo changes in the mRNA levels for the early steps of de novo SL biosynthesis upon conversion to EBs, then to compare these with the Cer subspecies that are present. There were increases in the mRNAs for many of the early enzymes of SL biosynthesis (Fig. 1), including some of the CerS (1, 3, and 6) and downstream synthases for SMs and GSLs, and decreases in other CerS (4 and 5, with 2 unchanged), the sphingosine kinases, and two of three ceramidases. Although all of these are worthy of further investigation, this study focused on the lipid backbone subspecies because they can be readily analyzed by LC-ESI MS/MS and serve as a relatively easy way to determine how closely changes in gene expression match changes in these important compounds.
For all of the mESC and EB SLs investigated in this study, the prevalent N-acyl-linked fatty acid was C16:0 (palmitate), which is added by CerS5 and CerS6. Consistent with this, the transcript abundance of CerS5 plus CerS6 is higher than that for the other CerSs (1, 3, and 4) except CerS2 (Fig. 2). Considering the relative abundance of CerS2 mRNA, it is somewhat surprising that the C24-chain-length backbones are not more prevalent (supplementary ). We do not know if this is because CerS2 protein amount or enzymatic activity are not equivalently high, but it is worth noting that another factor might be a limited availability of the cosubstrate C24- and C24:1-fatty acyl-CoAs as these are <10% of the levels of the C16- and C18-fatty acyl-CoAs (Fig. 5). Consistent with this possibility, the higher proportions of C24-chain length Cer in EBs versus mESC occur without an increase in CerS2 mRNA whereas the amounts of the very long-chain fatty acyl-CoAs doubled.
The findings for the fatty acyl-CoAs are, by themselves, interesting because there have been no previous studies of whether mESC and EB differ in the mRNA levels for this pathway. As shown in Fig. 1, the mRNA for Elovl6 was higher for EBs than mESCs, which matched the ∼2-fold higher stearoyl- and oleoyl-CoAs (Fig. 5). Furthermore, the higher C24-chain-length fatty acyl-CoAs correlated with only one of the two elongases (Elovl3, which increased, vs. Elovl1, which decreased), therefore, it is likely that Elovl3 is responsible for the observed increases in very long-chain fatty acyl-CoAs.
Decreases in the proportion of C16- and increases in very long-chain DHCer and Cer were two of the most consistent shifts in backbone composition not only for these compounds but also the downstream (DH)SM and (DH)CMH (Figs. 3 and 4) and were even more evident in the D3 cell line (Fig. 6). The functional significance of these subspecies shifts for embryogenesis, if any, is not known; however, synthesis of C16-Cer is elevated during the apoptosis (49–51), and differences in the time course of changes in C16- versus C24-Cer have been associated with different phases of apoptosis l (52, 53). Changes in C18-Cer might also be important because this subspecies is associated with cell growth control (53).
EBs have been widely used as a model system for studies of the formation of the inner cell mass of the mouse embryo shortly before implantation, which has been proposed to involve apoptosis (54) and, thus, might involve Cer. Another important function for Cer-induced apoptosis might be to eliminate residual, pluripotent EBs in later stages of differentiation (9).
It is also interesting that the ratio of DHCer to Cer was higher in most of the SLs of EBs than mESCs (c.f. left vs. right columns of supplementary ), which correlated with the lower DHCer desaturase (DES1) mRNA in EBs versus mESCs (Fig. 1). Dihydroceramides are generally thought not to play an important role in apoptosis, but can participate in other biological processes such as autophagy (55), which has also been proposed to be involved in the formation of the inner cell mass of the mouse embryo (56).
In addition to shifts in the backbone subspecies, the total amounts of (DH)Cers were higher in R1 and D3 EBs than mESCs (supplementary ), which might participate in the regulation of primitive ectoderm cell polarity and/or Cer-induced apoptosis in embryonic development (8, 57). In contrast, the CMH of R1 and D3 EBs were much lower than for mESCs, despite increases in the transcripts for the synthases for GlcCer and GalCer (Fig. 1), which may suggest that these simple GSLs are being used for the biosynthesis of more complex GSLs that are important during development (3, 58, 59). This would be an interesting direction for future studies.
Since the development of tools for analysis of mRNAs using microarrays and high-throughput qRT-PCR for transcript profiling, there have been many studies of changes in gene expression during development (60–62); however, prior to this study, the CerS gene family has not been included because it was only recently characterized (16). There have also been no previous studies of this pathway that have combined analysis of the mRNAs for these genes with the quantitation of the related metabolites. Thus, it is encouraging that, for the most part, there was reasonable agreement between the predictions from the genomic analysis and what was found by this focused lipidomic survey. This type of approach might uncover new ways in which SLs are involved in stem cell biology and embryonic development, including where disruption of SL metabolism results in birth defects (63, 64).
Supplementary Material
Acknowledgments
The authors thank Dr. Michael Pierce at the University of Georgia for useful comments throughout the course of this work, and Dr. Akemi Suzuki (Tokai University) for helpful discussions concerning annotation of sphingolipid metabolic genes.
Footnotes
Abbreviations:
- Cer
- ceramide
- CMH
- ceramide monohexose
- DHCer
- dihydroceramide
- DHCMH
- dihydroceramide monohexoses
- DHGalCer
- dihydrogalactosylceramide
- DHGlcCer
- dihydroglucosylceramide
- (DH)SL
- dihydrosphingolipid
- DHSM
- dihydrosphingomyelin
- EB
- embroid bodies
- GalCer
- galactosylceramide
- GlcCer
- glucosylceramide
- GSL
- glycosphingolipid
- IS
- internal standard
- 3-Keto-SA
- 3-ketosphinganine
- LIF
- leukemia inhibitory factor
- mESC
- mouse embryonic stem cell
- qRT-PCR
- quantitative RT-PCR
- Sa
- sphinganine
- Sa1P
- sphinganine-1-phosphate
- SL
- sphingolipid
- So
- sphingosine
- So1P
- sphingosine-1-phosphate
Bar graphs with the amounts of each subspecies of (DH)Cer, (DH)CMH, and (DH)SM for R1 and D3 mESCs and EBs are available as in the supplementary data.
This work was funded by the National Institutes of Health grant RR018502 for the Integrated Technology Resource for Biomedical Glycomics at the University of Georgia and GM069338 (Lipid Maps). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of two figures.
REFERENCES
- 1.Merrill A. H., Jr, Wang M. D., Park M., Sullards M. C. 2007. (Glyco)sphingolipidology: an amazing challenge and opportunity for systems biology. Trends Biochem. Sci. 32: 457–468. [DOI] [PubMed] [Google Scholar]
- 2.Fenderson B. A., Eddy E. M., Hakomori S. 1990. Glycoconjugate expression during embryogenesis and its biological significance. Bioessays. 12: 173–179. [DOI] [PubMed] [Google Scholar]
- 3.Muramatsu T. 2000. Essential roles of carbohydrate signals in development, immune response and tissue functions, as revealed by gene targeting. J. Biochem. 127: 171–176. [DOI] [PubMed] [Google Scholar]
- 4.Bieberich E. 2004. Integration of glycosphingolipid metabolism and cell-fate decisions in cancer and stem cells: review and hypothesis. Glycoconj. J. 21: 315–327. [DOI] [PubMed] [Google Scholar]
- 5.Regina Todeschini A., Hakomori S. I. 2008. Functional role of glycosphingolipids and gangliosides in control of cell adhesion, motility, and growth, through glycosynaptic microdomains. Biochim. Biophys. Acta. 1780: 421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ngamukote S., Yanagisawa M., Ariga T., Ando S., Yu R. K. 2007. Developmental changes of glycosphingolipids and expression of glycogenes in mouse brains. J. Neurochem. 103: 2327–2341. [DOI] [PubMed] [Google Scholar]
- 7.Bieberich E., MacKinnon S., Silva J., Noggle S., Condie B. G. 2003. Regulation of cell death in mitotic neural progenitor cells by asymmetric distribution of prostate apoptosis response 4 (PAR-4) and simultaneous elevation of endogenous ceramide. J. Cell Biol. 162: 469–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krishnamurthy K., Wang G., Silva J., Condie B., Bieberich E. 2007. Ceramide regulates atypical PKCzeta/lambda-mediated cell polarity in primitive ectoderm cells. A novel function of sphingolipids in morphogenesis. J. Biol. Chem. 282: 3379–3390. [DOI] [PubMed] [Google Scholar]
- 9.Bieberich E., Silva J., Wang G., Krishnamurthy K., Condie B. G. 2004. Selective apoptosis of pluripotent mouse and human stem cells by novel ceramide analogues prevents teratoma formation and enriches for neural precursors in ES cell-derived neural transplants. J. Cell Biol. 167: 723–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bieberich E., MacKinnon S., Silva J., Yu R. K. 2001. Regulation of apoptosis during neuronal differentiation by ceramide and b-series complex gangliosides. J. Biol. Chem. 276: 44396–44404. [DOI] [PubMed] [Google Scholar]
- 11.Lahiri S., Futerman A. H. 2007. The metabolism and function of sphingolipids and glycosphingolipids. Cell. Mol. Life Sci. 64: 2270–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tettamanti G. 2004. Ganglioside/glycosphingolipid turnover: new concepts. Glycoconj. J. 20: 301–317. [DOI] [PubMed] [Google Scholar]
- 13.Kitatani K., Idkowiak-Baldys J., Hannun Y. A. 2008. The sphingolipid salvage pathway in ceramide metabolism and signaling. Cell. Signal. 20: 1010–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Merrill A. H., Jr., Stokes T. H., Momin A., Park H., Portz B. J., Kelly S., Wang E., Sullards M. C., Wang M. D. 2009. Sphingolipidomics: a valuable tool for understanding the roles of sphingolipids in biology and disease. J. Lipid Res. 50(Suppl): S97–S102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei J., Tokumbo Y., Liepelt M., Momin A., Wang E., Hanada K., Merrill A. 2006. Serine Palmitoyltransferase. In Sphingolipid Biology Hirabayashi Y., Igarashi Y., Merrill A., Springer, Tokyo: 25–47. [Google Scholar]
- 16.Pewzner-Jung Y., Ben-Dor S., Futerman A. H. 2006. When do Lasses (longevity assurance genes) become CerS (ceramide synthases)? Insights into the regulation of ceramide synthesis. J. Biol. Chem. 281: 25001–25005. [DOI] [PubMed] [Google Scholar]
- 17.Mizutani Y., Kihara A., Igarashi Y. 2005. Mammalian Lass6 and its related family members regulate synthesis of specific ceramides. Biochem. J. 390: 263–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laviad E. L., Albee L., Pankova-Kholmyansky I., Epstein S., Park H., Merrill A. H., Jr., Futerman A. H. 2008. Characterization of Ceramide Synthase 2: tissue distribution, substrate specificity, and inhibition by sphingosine 1-phosphate. J. Biol. Chem. 283: 5677–5684. [DOI] [PubMed] [Google Scholar]
- 19.Ternes P., Franke S., Zahringer U., Sperling P., Heinz E. 2002. Identification and characterization of a sphingolipid delta 4-desaturase family. J. Biol. Chem. 277: 25512–25518. [DOI] [PubMed] [Google Scholar]
- 20.Desbaillets I., Ziegler U., Groscurth P., Gassmann M. 2000. Embryoid bodies: an in vitro model of mouse embryogenesis. Exp. Physiol. 85: 645–651. [PubMed] [Google Scholar]
- 21.Futerman A. H., Riezman H. 2005. The ins and outs of sphingolipid synthesis. Trends Cell Biol. 15: 312–318. [DOI] [PubMed] [Google Scholar]
- 22.Nairn A. V., York W. S., Harris K., Hall E. M., Pierce J. M., Moremen K. W. 2008. Regulation of glycan structures in animal tissues: transcript profiling of glycan-related genes. J. Biol. Chem. 283: 17298–17313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doetschman T. C., Eistetter H., Katz M., Schmidt W., Kemler R. 1985. The in vitro development of blastocyst-derived embryonic stem cell lines: formation of visceral yolk sac, blood islands and myocardium. J. Embryol. Exp. Morphol. 87: 27–45. [PubMed] [Google Scholar]
- 24.Hadjantonakis A. K., Gertsenstein M., Ikawa M., Okabe M., Nagy A. 1998. Generating green fluorescent mice by germline transmission of green fluorescent ES cells. Mech. Dev. 76: 79–90. [DOI] [PubMed] [Google Scholar]
- 25.Rathjen J., Lake J., Bettess M., Washington J., Chapman G., Rathjen P. 1999. Formation of a primitive ectoderm like cell population, EPL cells, from ES cells in response to biologically derived factors. J. Cell Sci. 112: 601–612. [DOI] [PubMed] [Google Scholar]
- 26.Lake J., Rathjen J., Remiszewski J., Rathjen P. 2000. Reversible programming of pluripotent cell differentiation. J. Cell Sci. 113: 555–566. [DOI] [PubMed] [Google Scholar]
- 27.Nairn A. V., Kinoshita-Toyoda A., Toyoda H., Xie J., Harris K., Dalton S., Kulik M., Pierce J. M., Toida T., Moremen K. W., et al. 2007. Glycomics of proteoglycan biosynthesis in murine embryonic stem cell differentiation. J. Proteome Res. 6: 4374–4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramakers C., Ruijter J. M., Deprez R. H., Moorman A. F. 2003. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339: 62–66. [DOI] [PubMed] [Google Scholar]
- 29.Gene Map Annotator and Pathway Profiler. Accessed at http://www.genmapp.org/.
- 30.Salomonis N., Hanspers K., Zambon A. C., Vranizan K., Lawlor S. C., Dahlquist K. D., Doniger S. W., Stuart J., Conklin B. R., Pico A. R. 2007. GenMAPP 2: new features and resources for pathway analysis. BMC Bioinformatics. 8: 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kyoto Encyclopedia of Genes and Genomes. Accessed at http://www.genome.jp/kegg/.
- 32.Taniguchi N., Honke K., Fukuda M., 2002. Handbook of Glycosyltransferases and Related Genes. Springer-Verlag, Tokyo. [Google Scholar]
- 33.Sullards M. C., Merrill A. H., Jr. 2001. Analysis of sphingosine 1-phosphate, ceramides, and other bioactive sphingolipids by high-performance liquid chromatography-tandem mass spectrometry. Sci. STKE. 2001: PL1. [DOI] [PubMed] [Google Scholar]
- 34.Merrill A. H., Jr., Sullards M. C., Allegood J. C., Kelly S., Wang E. 2005. Sphingolipidomics: high-throughput, structure-specific, and quantitative analysis of sphingolipids by liquid chromatography tandem mass spectrometry. Methods. 36: 207–224. [DOI] [PubMed] [Google Scholar]
- 35.Shaner R. L., Allegood J. C., Park H., Wang E., Kelly S., Haynes C. A., Sullards M. C., Merrill A. H., Jr. 2009. Quantitative analysis of sphingolipids for lipidomics using triple quadrupole and quadrupole linear ion trap mass spectrometers. J. Lipid Res. 50: 1692–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haynes C. A., Allegood J. C., Sims K., Wang E. W., Sullards M. C., Merrill A. H., Jr. 2008. Quantitation of fatty acyl-coenzyme As in mammalian cells by liquid chromatography-electrospray ionization tandem mass spectrometry. J. Lipid Res. 49: 1113–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Omae F., Miyazaki M., Enomoto A., Suzuki M., Suzuki Y., Suzuki A. 2004. DES2 protein is responsible for phytoceramide biosynthesis in the mouse small intestine. Biochem. J. 379: 687–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miyazaki M., Ntambi J. M. 2008. Fatty acid desaturation and chain elongation in mammals. In Biochemistry of Lipids, Lipoproteins and Membranes Vance D. E., Vance J. E., Elsevier, Amsterdam: 191–212. [Google Scholar]
- 39.Matsuzaka T., Shimano H., Yahagi N., Yoshikawa T., Amemiya-Kudo M., Hasty A. H., Okazaki H., Tamura Y., Iizuka Y., Ohashi K., et al. 2002. Cloning and characterization of a mammalian fatty acyl-CoA elongase as a lipogenic enzyme regulated by SREBPs. J. Lipid Res. 43: 911–920. [PubMed] [Google Scholar]
- 40.Westerberg R., Mansson J. E., Golozoubova V., Shabalina I. G., Backlund E. C., Tvrdik P., Retterstol K., Capecchi M. R., Jacobsson A. 2006. ELOVL3 is an important component for early onset of lipid recruitment in brown adipose tissue. J. Biol. Chem. 281: 4958–4968. [DOI] [PubMed] [Google Scholar]
- 41.Westerberg R., Tvrdik P., Unden A. B., Mansson J. E., Norlen L., Jakobsson A., Holleran W. H., Elias P. M., Asadi A., Flodby P., et al. 2004. Role for ELOVL3 and fatty acid chain length in development of hair and skin function. J. Biol. Chem. 279: 5621–5629. [DOI] [PubMed] [Google Scholar]
- 42.Tvrdik P., Westerberg R., Silve S., Asadi A., Jakobsson A., Cannon B., Loison G., Jacobsson A. 2000. Role of a new mammalian gene family in the biosynthesis of very long chain fatty acids and sphingolipids. J. Cell Biol. 149: 707–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leonard A. E., Kelder B., Bobik E. G., Chuang L. T., Lewis C. J., Kopchick J. J., Mukerji P., Huang Y. S. 2002. Identification and expression of mammalian long-chain PUFA elongation enzymes. Lipids. 37: 733–740. [DOI] [PubMed] [Google Scholar]
- 44.Zhang K., Kniazeva M., Han M., Li W., Yu Z., Yang Z., Li Y., Metzker M. L., Allikmets R., Zack D. J., et al. 2001. A 5-bp deletion in ELOVL4 is associated with two related forms of autosomal dominant macular dystrophy. Nat. Genet. 27: 89–93. [DOI] [PubMed] [Google Scholar]
- 45.Venkataraman K., Riebeling C., Bodennec J., Riezman H., Allegood J. C., Sullards M. C., Merrill A. H., Jr., Futerman A. H. 2002. Upstream of growth and differentiation factor 1 (uog1), a mammalian homolog of the yeast longevity assurance gene 1 (LAG1), regulates N-stearoyl-sphinganine (C18-(dihydro)ceramide) synthesis in a fumonisin B1-independent manner in mammalian cells. J. Biol. Chem. 277: 35642–35649. [DOI] [PubMed] [Google Scholar]
- 46.Mizutani Y., Kihara A., Igarashi Y. 2006. LASS3 (longevity assurance homologue 3) is a mainly testis-specific (dihydro)ceramide synthase with relatively broad substrate specificity. Biochem. J. 398: 531–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Riebeling C., Allegood J. C., Wang E., Merrill A. H., Jr., Futerman A. H. 2003. Two mammalian longevity assurance gene (LAG1) family members, trh1 and trh4, regulate dihydroceramide synthesis using different fatty acyl-CoA donors. J. Biol. Chem. 278: 43452–43459. [DOI] [PubMed] [Google Scholar]
- 48.Lahiri S., Lee H., Mesicek J., Fuks Z., Haimovitz-Friedman A., Kolesnick R. N., Futerman A. H. 2007. Kinetic characterization of mammalian ceramide synthases: determination of K(m) values towards sphinganine. FEBS Lett. 581: 5289–5294. [DOI] [PubMed] [Google Scholar]
- 49.Thomas R. L., Jr., Matsko C. M., Lotze M. T., Amoscato A. A. 1999. Mass spectrometric identification of increased C16 ceramide levels during apoptosis. J. Biol. Chem. 274: 30580–30588. [DOI] [PubMed] [Google Scholar]
- 50.Eto M., Bennouna J., Hunter O. C., Hershberger P. A., Kanto T., Johnson C. S., Lotze M. T., Amoscato A. A. 2003. C16 ceramide accumulates following androgen ablation in LNCaP prostate cancer cells. Prostate. 57: 66–79. [DOI] [PubMed] [Google Scholar]
- 51.Eto M., Bennouna J., Hunter O., Lotze M., Amoscato A. 2006. Importance of C16 ceramide accumulation during apoptosis in prostate cancer cells. Int. J. Urol. 13: 148–156. [DOI] [PubMed] [Google Scholar]
- 52.Kroesen B. J., Jacobs S., Pettus B. J., Sietsma H., Kok J. W., Hannun Y. A., de Leij L. F. 2003. BcR-induced apoptosis involves differential regulation of C16 and C24-ceramide formation and sphingolipid-dependent activation of the proteasome. J. Biol. Chem. 278: 14723–14731. [DOI] [PubMed] [Google Scholar]
- 53.Koybasi S., Senkal C. E., Sundararaj K., Spassieva S., Bielawski J., Osta W., Day T. A., Jiang J. C., Jazwinski S. M., Hannun Y. A., et al. 2004. Defects in cell growth regulation by C18:0-ceramide and longevity assurance gene 1 in human head and neck squamous cell carcinomas. J. Biol. Chem. 279: 44311–44319. [DOI] [PubMed] [Google Scholar]
- 54.Murray P., Edgar D. 2000. Regulation of programmed cell death by basement membranes in embryonic development. J. Cell Biol. 150: 1215–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zheng W., Kollmeyer J., Symolon H., Momin A., Munter E., Wang E., Kelly S., Allegood J. C., Liu Y., Peng Q., et al. 2006. Ceramides and other bioactive sphingolipid backbones in health and disease: lipidomic analysis, metabolism and roles in membrane structure, dynamics, signaling and autophagy. Biochim. Biophys. Acta. 1758: 1864–1884. [DOI] [PubMed] [Google Scholar]
- 56.Qu X., Zou Z., Sun Q., Luby-Phelps K., Cheng P., Hogan R. N., Gilpin C., Levine B. 2007. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell. 128: 931–946. [DOI] [PubMed] [Google Scholar]
- 57.Hoekstra D., Maier O., van der Wouden J. M., Slimane T. A., van IJzendoorn S. C. 2003. Membrane dynamics and cell polarity: the role of sphingolipids. J. Lipid Res. 44: 869–877. [DOI] [PubMed] [Google Scholar]
- 58.Muramatsu T., Muramatsu H. 2004. Carbohydrate antigens expressed on stem cells and early embryonic cells. Glycoconj. J. 21: 41–45. [DOI] [PubMed] [Google Scholar]
- 59.Yamashita T., Wada R., Sasaki T., Deng C., Bierfreund U., Sandhoff K., Proia R. L. 1999. A vital role for glycosphingolipid synthesis during development and differentiation. Proc. Natl. Acad. Sci. USA. 96: 9142–9147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Palmqvist L., Glover C. H., Hsu L., Lu M., Bossen B., Piret J. M., Humphries R. K., Helgason C. D. 2005. Correlation of murine embryonic stem cell gene expression profiles with functional measures of pluripotency. Stem Cells. 23: 663–680. [DOI] [PubMed] [Google Scholar]
- 61.zur Nieden N. I., Price F. D., Davis L. A., Everitt R. E., Rancourt D. E. 2007. Gene profiling on mixed embryonic stem cell populations reveals a biphasic role for beta-catenin in osteogenic differentiation. Mol. Endocrinol. 21: 674–685. [DOI] [PubMed] [Google Scholar]
- 62.Hailesellasse Sene K., Porter C. J., Palidwor G., Perez-Iratxeta C., Muro E. M., Campbell P. A., Rudnicki M. A., Andrade-Navarro M. A. 2007. Gene function in early mouse embryonic stem cell differentiation. BMC Genomics. 8: 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marasas W. F., Riley R. T., Hendricks K. A., Stevens V. L., Sadler T. W., Gelineau-van Waes J., Missmer S. A., Cabrera J., Torres O., Gelderblom W. C., et al. 2004. Fumonisins disrupt sphingolipid metabolism, folate transport, and neural tube development in embryo culture and in vivo: a potential risk factor for human neural tube defects among populations consuming fumonisin-contaminated maize. J. Nutr. 134: 711–716. [DOI] [PubMed] [Google Scholar]
- 64.Gelineau-van Waes J., Starr L., Maddox J., Aleman F., Voss K. A., Wilberding J., Riley R. T. 2005. Maternal fumonisin exposure and risk for neural tube defects: mechanisms in an in vivo mouse model. Birth Defects Res. A Clin. Mol. Teratol. 73: 487–497. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.