Abstract
The partitioning of apolipoprotein A-I (apoA-I) molecules in plasma between HDL-bound and -unbound states is an integral part of HDL metabolism. We used the surface plasmon resonance (SPR) technique to monitor in real time the reversible binding of apoA-I to HDL. Biotinylated human HDL2 and HDL3 were immobilized on a streptavidin-coated SPR sensor chip, and apoA-I solutions at different concentrations were flowed across the surface. The wild-type (WT) human and mouse apoA-I/HDL interaction involves a two-step process; apoA-I initially binds to HDL with fast association and dissociation rates, followed by a step exhibiting slower kinetics. The isolated N-terminal helix bundle domains of human and mouse apoA-I also exhibit a two-step binding process, consistent with the second slower step involving opening of the helix bundle domain. The results of fluorescence experiments with pyrene-labeled apoA-I are consistent with the N-terminal helix bundle domain interacting with proteins resident on the HDL particle surface. Dissociation constants (Kd) measured for WT human apoA-I interactions with HDL2 and HDL3 are about 10 µM, indicating that the binding is low affinity. This Kd value does not apply to all of the apoA-I molecules on the HDL particle but only to a relatively small, labile pool.
Understanding the structure and function of HDL is significant because of the beneficial cardioprotective properties of this lipoprotein (1). The anti-atherogenic effects of HDL arise, in part, from its participation in the reverse cholesterol transport pathway where the principal HDL protein, apolipoprotein A-I (apoA-I), plays a central role (2). As a result, the structure-function relationships of apoA-I have been studied extensively (for reviews, see Refs. 3–5). Perhaps the most important characteristic of the apoA-I molecule is its ability to bind lipids; this interaction is mediated by the amphipathic α-helices present in the protein molecule (6). ApoA-I binds well to phospholipid (PL)-water interfaces and, under appropriate conditions, can solubilize the PL to create discoidal HDL particles (7, 8). The binding of apoA-I to a PL surface involves a two-step mechanism. First, α-helices in the C-terminal domain of the protein interact with the surface, and, second, the N-terminal helix bundle domain opens to allow more helix-lipid interactions to occur (5, 9). Although the binding of apoA-I to model PL particles has been studied extensively, the binding of apoA-I to HDL particles has not been investigated much because of the difficulty of separating free and bound apoA-I in this system. This lack of information about apoA-I/HDL interactions is significant because the cycling of apoA-I molecules on and off HDL particles occurs during the metabolism of HDL particles (10, 11), in particular to release apoA-I molecules into the preβ-HDL pool (10, 12). This recycling is consistent with the well-established ability of apolipoproteins, such as apoA-I, to exchange spontaneously between different populations of lipoprotein particles (13–16) and PL vesicles (17, 18). As a rule, any remodeling event that depletes HDL particles of PL induces particle fusion and dissociation of that fraction of the apoA-I molecules that is in a labile pool (19). At this stage, quantitative understanding of the kinetics of apoA-I interactions with HDL particles is unavailable.
Here, we exploit surface plasmon resonance (SPR) to monitor in real time the association and dissociation reactions in the apoA-I/HDL system. SPR has been used to derive quantitative information about the binding of both lipoproteins (20) and apoE (21–23) to proteoglycans. As far as the application of SPR to the HDL system is concerned, the binding of several plasma remodeling factors to HDL immobilized on a sensor chip has been investigated successfully (24–26). Also, the conformation of apoA-I in HDL was explored by comparing the binding of HDL particles to anti-apoA-I monoclonal antibodies immobilized on an SPR chip (27). We have extended these approaches to study the binding of apoA-I to HDL particles. The results show that apoA-I can bind reversibly and with low affinity to HDL particles by a two-step mechanism.
EXPERIMENTAL PROCEDURES
Materials
HDL2 and HDL3 were purified by sequential density ultracentrifugation (28, 29) from a pool of fresh human plasma obtained by combining several single units. Human apoA-I was isolated from the total HDL fraction (1.063 < d < 1.21g/ml) of human plasma as described previously (30). Human apoA-I and engineered variants were expressed in Escherichia coli and isolated as reported earlier (9, 31). Murine apoA-I and engineered variants were prepared similarly as described before (32). Human apoE3 was obtained as formerly reported (31).
Biotinylation of HDL particles
HDL2 and HDL3 were dialyzed into PBS (pH 7.4) prior to biotinylation. The EZ-link sulfo-NHS-LC-biotinylation kit from Pierce Chemical (Rockford, IL) was used for attaching biotin molecules through a 2.24 nm spacer arm to lysine residues on the surface of HDL particles. HDL2 and HDL3, each at 1.0 mg protein/ml, were mixed with a freshly made 10 mM sulfo-NHS-LC-biotin solution at a 10-fold molar exess of biotin. The lipoproteins were incubated under nitrogen at 4°C overnight before dialysis against TBS (pH 7.4) to remove unreacted sulfo-NHS-LC-biotin. The degree of biotinylation of the particles was determined using conditions recommended by Pierce. Briefly, solutions containing biotinylated lipoproteins were added to a mixture of 2-(4′-hydroxyphenyl) azobenzoic acid (HABA) reagent and immunopure avidin (Pierce Chemical). Because of its higher affinity for avidin, biotin, from the biotinylated lipoproteins, displaced avidin-bound HABA. Therefore, the absorbance at 500 nm of the HABA-avidin complex was reduced. The change in absorbance was used to calculate the level of biotin incorporated into the lipoprotein particles. This procedure ordinarily yielded one to two biotin molecules per HDL particle. Such a modification did not change the binding properties of the HDL particle surface as assessed by 14C-apoA-I binding in an HDL2/HDL3 partition assay using control and biotinylated HDL particles (S. Lund-Katz and M. Nickel, unpublished observations).
SPR
Studies of the binding of apolipoproteins (association and dissociation) to HDL were performed with a Biacore 3000 SPR instrument (Uppsala, Sweden) using SA sensor chips (Biacore, Uppsala, Sweden), as described before for the binding of apoE (33). This chip is designed to bind biotinylated ligands through a high-affinity capture process. Prior to immobilization of HDL on the sensor chip, the streptavidin surface was conditioned with three consecutive 1-min injections of 1 M NaCl in 50 mM NaOH (50 µl/min). The biotinylated HDL2 or HDL3 was then immobilized onto the surface through the quasicovalent biotin-streptavidin interaction by exposing the surface to the biotinylated lipoprotein solutions in running buffer (50 mM TBS, pH 7.4) until 2,500–3,000 response units (RUs) of biotinylated HDL were bound to the surface. This was achieved by a 10 µl injection of biotinylated HDL2 or HDL3 (1.0 mg protein/ml) at a flow rate of 2 µl/min at room temperature. After 5 min, the chip was washed with degassed TBS to remove unattached HDL. A 50 µg/ml human apoE3 solution was flowed over the chip at 20 µl/min for 2 min to block any remaining hydrophobic surface areas and reduce apoA-I binding to nonHDL sites; the chip was then washed with TBS until the SPR signal reached a steady background value. The surface of the immobilized HDL was then exposed to a 4 min injection of apoA-I dissolved in degassed TBS at a flow rate of 20 µl/min to monitor association, and then TBS alone was passed over the sensor surface to monitor apoA-I dissociation from the immobilized HDL particles. For these experiments, two flow cells were monitored simultaneously with flow cells 1 and 2 containing immobilized biotinylated HDL2 and HDL3, respectively. A sensor chip lacking immobilized HDL could not be used as a reference cell because apoA-I bound more to this surface than to an HDL-coated chip. The apolipoproteins were dialyzed from 6 M guanidine HCl into TBS, filtered (Ultrafree, MC centrifugal filter devices, 0.1 µm filter unit; Millipore, Bedford, MA) and degassed before serial dilutions (20 to 400 µg/ml) were made just prior to injection. The sensor chip was washed two times with 20 µl TBS between each injection of apolipoprotein. The chips were used for 2 days in repetitive experiments. Regeneration of the sensor chip surface was not possible since the lipoproteins were directly immobilized via biotin-streptavidin interaction. The apoA-I sensorgrams were independent of flow rate in the range 10–40 μl/min, indicating that the apoA-I binding at 20 μl/min was not limited by mass transport (diffusion) effects. Steady state binding isotherms and Kd values of the binding to HDL were obtained by generating sensorgrams at different apoA-I concentrations. The sensorgrams were analyzed with the BIA evaluation software version 4.1 (Biacore). The response curves of various apolipoprotein (analyte) concentrations were fitted to the two-state binding model described by the following equation (34, 35).
| (Eq. 1) |
The equilibrium constants of each binding steps are K1 = ka1/kd1 and K2 = ka2/kd2, and the overall equilibrium binding constant is calculated as Ka = K1 (1 + K2) and Kd = 1/Ka. In this model, the analyte (A) binds to the ligand (HDL) (B) to form an initial complex (AB) and then undergoes subsequent binding or conformational change to form a more stable complex (ABx). A further check of the two-state binding mechanism was obtained by variation of the contact time for association between the apoA-I and the HDL. For a two-state reaction, an increase in the contact time between the analyte and the ligand decreases the dissociation rate since more of the stable ABx complex is formed. For the apolipoproteins, binding responses in the steady-state region of the sensorgrams (Req) were also plotted against apolipoprotein concentration (C) to determine the overall equilibrium binding affinity. The data were subjected to nonlinear regression fitting (Prism 4; GraphPad) according to the following equation:
| (Eq. 2) |
Rmax is the maximum binding response, and Kd is the dissociation constant. We have validated this SPR approach for measuring Kd by monitoring the binding of apoE4 to VLDL by SPR as well as ultracentrifugation (36) and obtained the same Kd (data not shown).
Gel filtration chromatography
The distribution between the free and bound states of trace-labeled 14C-apoA-I incubated with HDL2 was determined using gel filtration chromatography to separate the two pools of apoA-I. 14C- apoA-I (specific activity 2–3 µCi/mg) was prepared by reductive methylation using 14C-formaldehyde as described before (37); the surface activity of apoA-I is unaffected by this conversion of a few lysine residues to monomethylysine (37). The 14C-apoA-I (10 µg) in TBS (pH 7.4) was mixed with unlabeled apoA-I to give the desired total apoA-I concentration and added to HDL2 to give a final volume of 1.0 ml that contained 1 mg of HDL2 protein. The mixture was incubated for 1 h at room temperature to allow the apoA-I to bind to the HDL2 particles and then subjected to gel filtration chromatography on a calibrated Superdex 200 column (60 × 1.6 cm) (38). The Kav values on this column for HDL2 and apoA-I were 0.21 and 0.49, respectively. The fractions corresponding to the HDL2 and free apoA-I peaks were pooled, and the distribution of 14C-apoA-I determined by liquid scintillation counting. The amount of bound apoA-I per mg HDL2 was plotted as a function of free apoA-I concentration and fitted to Equation 2 to derive a Kd value.
Fluorescence measurement of the binding of pyrene-apoA-I to lipid emulsions and HDL3
Site-specific pyrene labeling of cysteine-containing apoA-I variants (V53C and F229C) was performed as described (39). Small triolein/egg PC emulsion (SEM) particles (average diameter of 35 ± 5 nm) were prepared by sonication and purified by ultracentrifugation as described (40). Fluorescence measurements were carried out with a Hitachi F-7000 fluorescence spectrophotometer at 25°C. In SEM or HDL3 binding experiments, pyrene emission fluorescence of apoA-I (25 μg/ml in TBS, pH 7.4) was recorded from 360–500 nm using a 342 nm excitation wavelength at increasing concentrations of SEM or HDL3 (ratios of PL to protein were 0−80 w/w). To reduce the effect of light scattering caused by lipid particles, the sample was excited with vertically polarized light and measured with a horizontal emission polarizer using a 4 × 4 mm cuvette (41). Kinetic data for the increase in fluorescence of pyrene-labeled apoA-I variants upon binding to SEM or HDL3 were obtained by monitoring the emission intensity at 385 nm. Parameters of apoA-I binding to SEM and HDL3 particles were derived from the fluorescence results as follows. The fraction of bound apoA-I, θ was calculated according to where Pb and PT are bound and total apoA-I concentrations, respectively, F and F0 are integrated fluorescence intensities for pyrene-apoA-I in the presence and absence of lipid particles, respectively, and Fmax represents the fluorescence intensity when pyrene-apoA-I completely binds to particles. Binding of apoA-I to lipid particles is expressed by a one-site binding model assuming that apoA-I binds to discrete, equivalent, and noninteracting binding sites on the particle surface (42–45): where Pf is the unbound apoA-I concentration, [PL] is the PL concentration of particles, and Kd and Bmax are the dissociation constant and the maximal binding capacity, respectively (39).
RESULTS
HDL Immobilization on an SPR chip
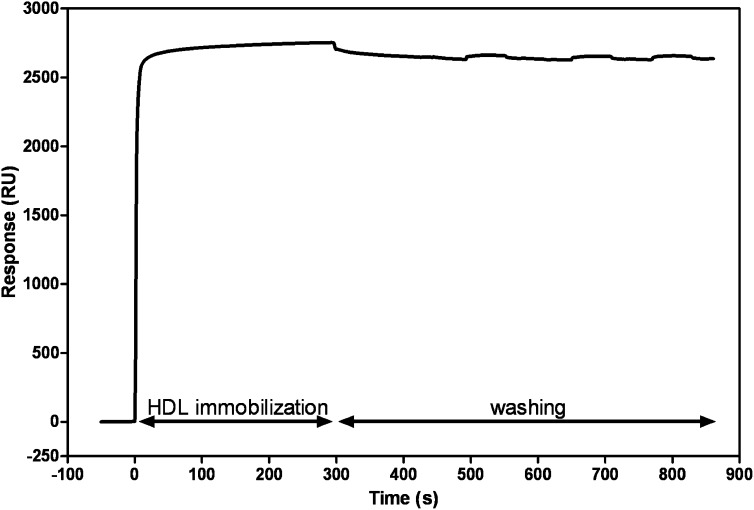
Figure 1 shows the immobilization of biotinylated HDL on a streptavidin-coated sensor chip when an HDL solution is flowed across the surface. It is apparent that there is rapid binding to give a steady state level. Flowing buffer solution alone across the chip washes away a small amount of weakly bound HDL, but the remainder is essentially irreversibly bound by the biotin-streptavidin interaction. In multiple experiments, the amounts of HDL2 and HDL3 immobilized by this procedure were 2909 ± 729 and 2761 ± 449 RU (mean ± SD), respectively. The immobilization of HDL in this fashion permits evaluation of the binding of apoA-I, which is difficult to monitor in solution because of problems in readily separating HDL and unbound apoA-I. Another advantage of the SPR method is that labeling the apoA-I molecules is not required. A potential problem in the SPR experiment is steric restriction of apoA-I binding by the proximity of the sensor chip surface, but, because HDL particles are attached to the surface by a 2.24 nm spacer arm, protein molecules can access the HDL surface. For example, LCAT (24) and cholesteryl ester transfer protein (26) have been shown to bind to HDL in similar SPR experiments.
Fig. 1.
Immobilization of HDL on an SPR sensor chip. Biotinylated HDL2 was immobilized on a pretreated streptavidin-coated sensor chip by flowing a 1 mg/ml solution across the chip for 5 min at 2 µl/min and washing with TBS at 100 µl/min.
Kinetics of human ApoA-I binding to HDL
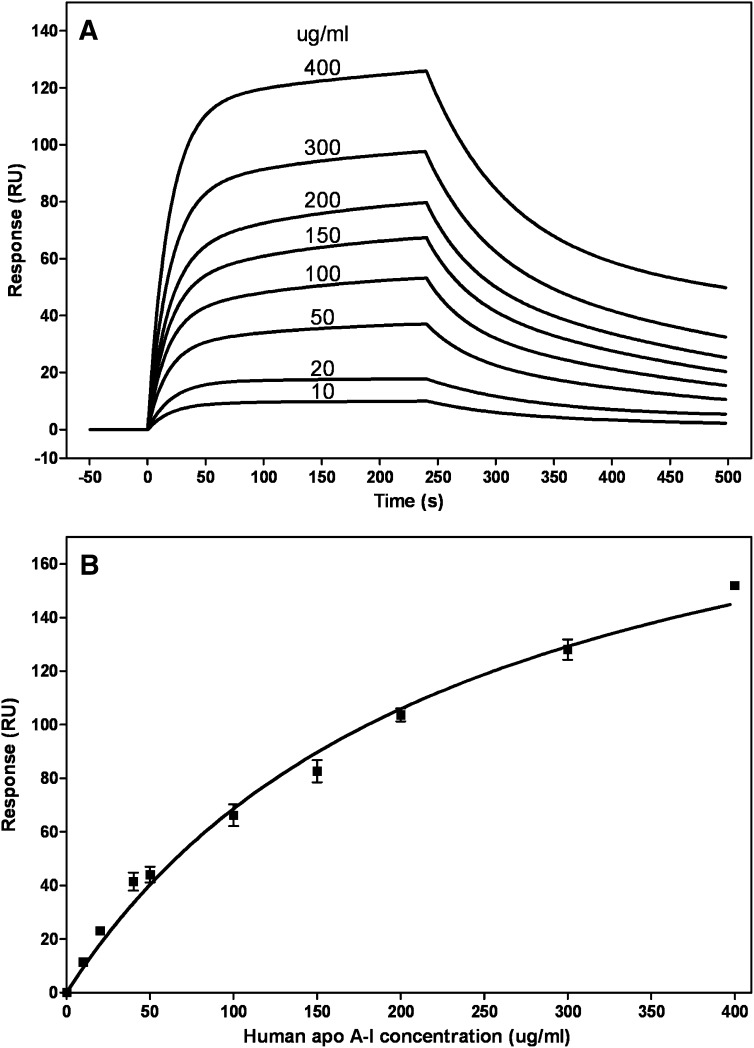
Figure 2A shows a typical series of sensorgrams for binding of wild-type (WT) apoA-I at different concentrations to HDL3. Similar results were obtained with human plasma apoA-I and when HDL2 was immobilized on the sensor chip (data not shown). The data are corrected for bulk refractive index effects and fitted using a two-state binding model (see Experimental Procedures). The kinetic data were not fitted well by a 1:1 Langmuir binding model, as reflected by the large value of the goodness of fit parameter (χ2 > 20 compared with <10 for a global fit using the two-state model). It follows that the binding of apoA-I to HDL particles involves either a sequential two-step process or some conformational change (22, 34, 35). Fitting of the sensorgrams in Fig. 2A gives an association constant Ka∼4 × 105 M−1, indicating weak binding of apoA-I to HDL3 particles. Because the fitting of the sensorgrams obtained at apoA-I concentrations >100 µg/ml is relatively poor, there is some uncertainty associated with this Ka value. Some of the reasons for the difficulty in modeling the apoA-I/HDL interaction include (1) the interaction is low affinity, (2) the HDL particles are heterogeneous and do not contain a discrete number of binding sites, and (3) the tendency of apoA-I to self-associate at higher concentrations. Consequently, we employed the steady-state method to compare the binding affinities of various apoA-I molecules to HDL (see below).
Fig. 2.
SPR analysis of binding of human apoA-I to immobilized HDL3. A: Sensorgrams obtained when apoA-I solutions at the indicated concentrations were flowed across the sensor chip and the experimental data were fitted with the two-state binding model, where A + B ↔ AB ↔ ABx (see Experimental Procedures). B: Binding isotherm. The maximal response (RUmax) was derived by fitting sensorgrams obtained over a range of apoA-I concentrations (cf. Fig. 2A) to the two-state binding model. These RUmax values are plotted (mean ± SD, n = 3) as a function of apoA-I concentration and fitted to a one-site binding model.
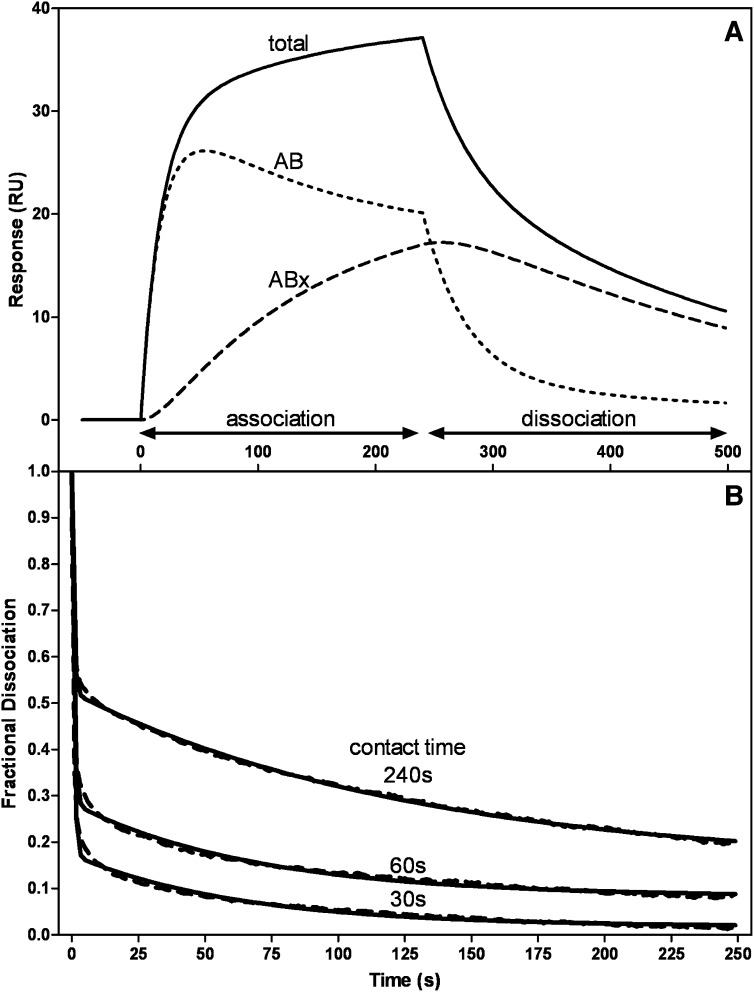
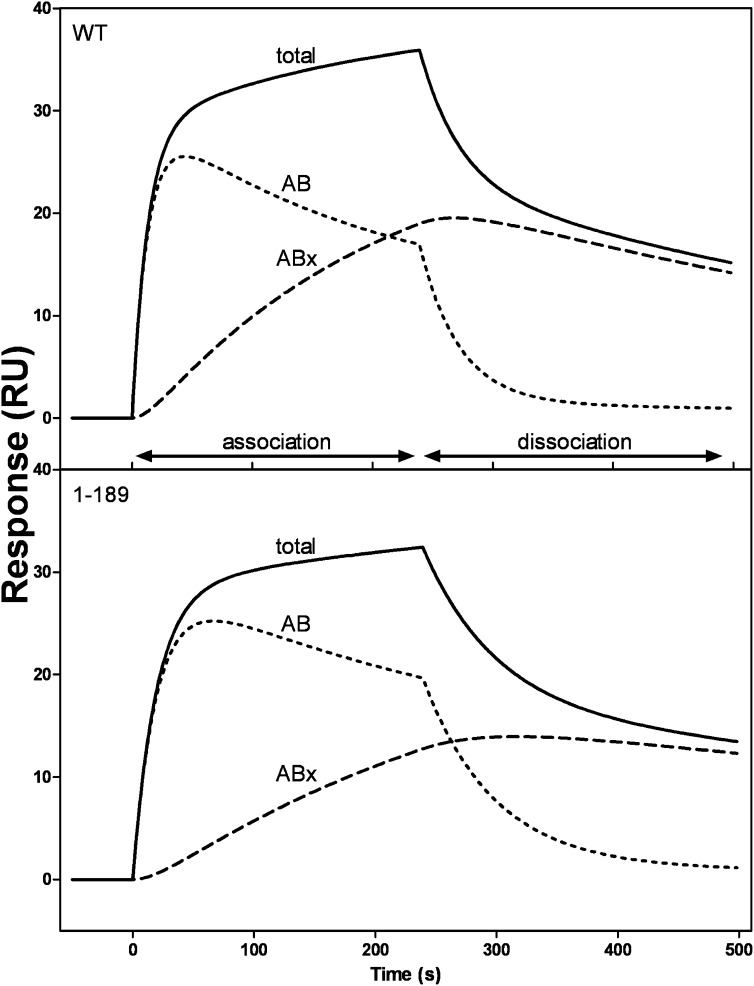
The response curve in Fig. 2A for 50 µg/ml apoA-I flowed across the sensor chip with immobilized HDL3 is analyzed in more detail in Fig. 3A. Here the deconvoluted curves show the initial binding (AB) and subsequent binding or conformational changes (ABx). Because of the uncertainties mentioned above about the valences of the HDL particles, it is difficult to interpret the kinetics of association in detail. However, inspection of the deconvoluted curves in Fig. 3A reveals that at the end of the 4 min association phase of the experiment, the sizes of the AB and ABx pools of bound apoA-I are approximately equal in size. It is also apparent that in the dissociation phase (after 4 min), apoA-I molecules in the AB pool dissociate more readily than those in the ABx pool. Analyzing many sensorgrams, the value for the dissociation rate constant kd1 for the AB pool is in the range of 2–6 × 10−2s−1, while the value of kd2 for the ABx pool is an order of magnitude smaller. These kd values correspond to halftimes of dissociation of 17 s and ∼3 min for the AB and ABx pools, respectively. The rate of dissociation of apoA-I from the HDL surface is dependent upon the length of time the apoA-I is in contact with the particle. As shown in Fig. 3B, progressively shortening the injection (contact) time from 240 s to 60 and 30 s enhances apoA-I dissociation. The fraction of apoA-I in the AB pool increases from ∼0.5 to 0.8 as the contact time is decreased to 30 s, and essentially all the apoA-I is removed at the end of the 4 min dissociation phase. The value of kd1 for the AB pool is not affected by the change in contact time, whereas kd2 approximately doubles as the contact time is decreased to 30 s. These results are consistent with the second step in the two-step mechanism of apoA-I binding to HDL involving a conformational change in the bound apoA-I molecules.
Fig. 3.
Two-state binding of apoA-I to HDL as detected by SPR measurement of the kinetics of association and dissociation. A: The sensorgram (solid line) is for 50 µg/ml apoA-I flowed at 20 µl/min (cf. Fig. 2A). The experimental data were fitted to the two-state binding model where A + B ↔ AB ↔ ABx. The deconvoluted curves are the additive components of the fitted curve and show the initial binding (AB) and the subsequent binding or conformational change (ABx). B: The dissociation curves show the effect of increased injection or contact time on the stability of the apoA-I/HDL3 complex. The experimental dissociation data (dashed line) were fitted to a biexponential decay equation (solid line).
Fitting of the family of sensorgrams depicted in Fig. 2A to the two-state binding model yields a maximal binding value (Rmax) for each concentration of apoA-I. Figure 2B shows a steady-state binding isotherm plotted using such Rmax values and fitted to a one-site binding model (Scatchard equation). The data in Fig. 2B yield a Kd value of 236 ± 27µg apoA-I/ml (8 ± 1 µM, mean ± SEM) for the binding of human apoA-I to HDL3. This Kd value indicates that the apoA-I/HDL3 interaction is of low affinity and is in a similar range to the Kd value of 2–3 µM reported recently by Carnemolla et al. (46). Electrostatic interactions apparently do not play a significant role in the binding process because increasing the sodium chloride concentration in the apoA-I-containing running buffer from 0.15–0.5 M did not alter the sensorgram (data not shown). Furthermore, the sensorgrams (Fig. 3A) and binding isotherm (Fig. 2B) are not affected significantly by any self-association of the apoA-I because preparing 50 µg/ml running solutions by dilution of 0.08 and 0.8 mg/ml stock solutions [self-association only occurs in the latter case (47)] gave rise to similar sensorgrams (data not shown).
Contributions of tertiary structure domains of human apoA-I to HDL binding
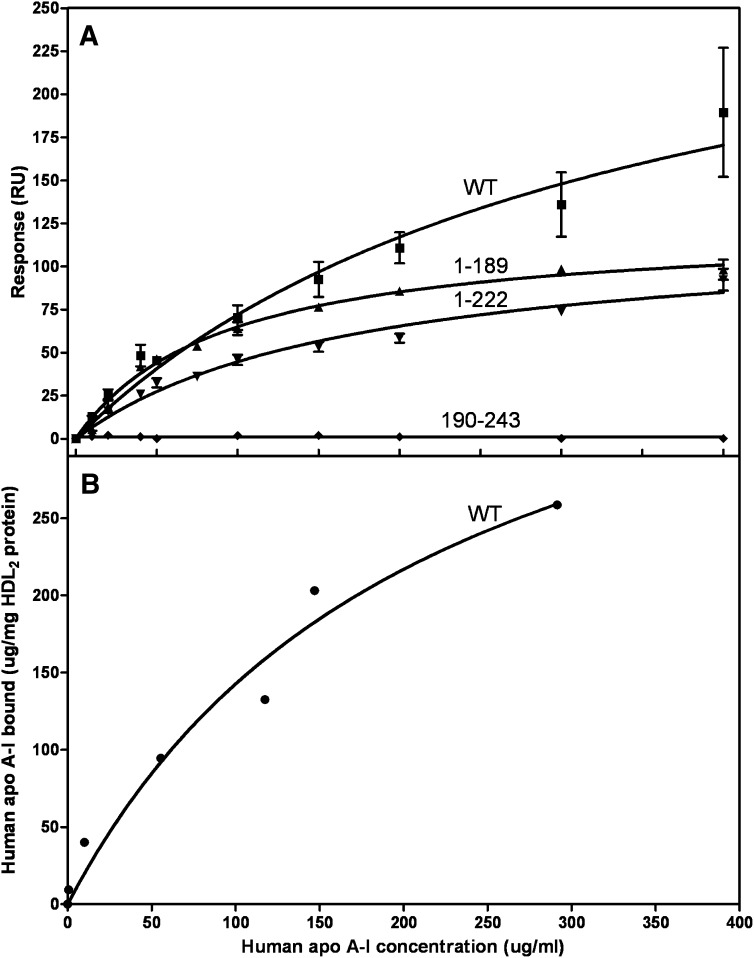
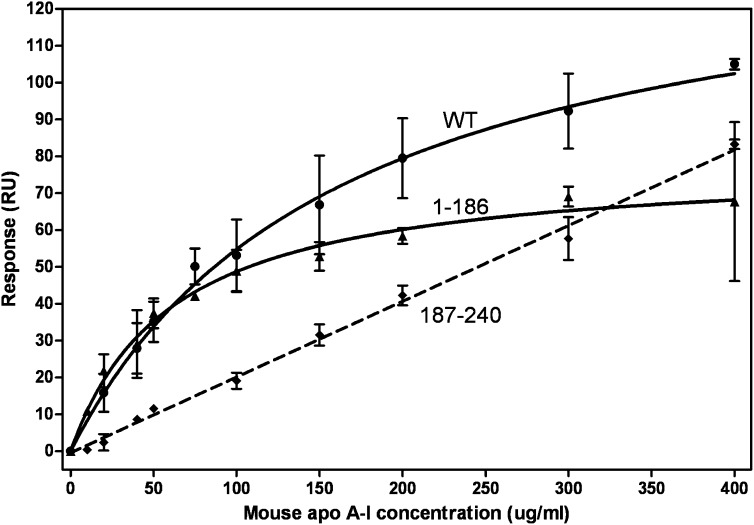
Figure 4A compares the isotherms obtained by SPR for WT human apoA-I and its separate N- (residues 1–189) and C-terminal (residues 190–243) domains binding to HDL2. The equivalent binding isotherms to HDL3 are similar (data not shown). The binding of WT apoA-I to HDL2 was monitored by both SPR and gel filtration chromatography to confirm that the SPR results are reliable. The binding isotherms for WT apoA-I in Fig. 4A and B are consistent in that the Kd values are 12 ± 4 and 8 ± 3 μM, respectively. The Kd and Bmax values derived from the isotherms obtained by SPR are listed in Table 1. It is apparent that removal of the C-terminal domain reduces the amount of apoA-I that binds (lower Bmax) but increases the binding affinity (lower Kd). Removal of the C-terminal α-helix [to give apoA-I (1–222)] also reduces Bmax but does not significantly reduce Kd. The isolated apoA-I C-terminal domain does not bind to HDL (Fig. 4). The precision in the determinations of the kinetic parameters for the association of WT apoA-I and its N-terminal domain with HDL was insufficient to detect significant differences in the ka and kd values for the two proteins.
Fig. 4.
Comparison of the binding of human apoA-I and its tertiary structure domains to HDL2. A: The binding isotherms were obtained and analyzed as described in the legend to Fig. 2B. Squares, WT apoA-I; upwards triangles, N-terminal domain (residues 1–189); diamonds, C-terminal domain (residues 190–243); downwards triangles, apoA-I variant with C-terminal α-helix deleted. B: The binding of 14C-apoA-I to HDL2 was monitored by gel filtration chromatography, as described in Experimental Procedures.
TABLE 1.
Binding constants for apoA-I interaction with HDL2
|
Kda |
Bmaxa |
|||
|---|---|---|---|---|
| ApoA-I | µg/ml | µM | RU | Relative |
| Human | ||||
| WT | 335 ± 111 | 12 ± 4 | 313 ± 61 | 1 |
| 1-189 | 90 ± 8 | 4 ± 0.4 | 124 ± 4 | 0.4 |
| 1-222 | 171 ± 27b | 7 ± 0.3 | 121 ± 9 | 0.4 |
| Mouse | ||||
| WT | 163 ± 26b | 6 ± 1 | 144 ± 11 | 0.5 |
| 1-186 | 61 ± 12 | 3 ± 0.6 | 79 ± 5 | 0.25 |
The Kd and Bmax values are listed as mean ± SEM and are derived from at least two independent binding isotherms of the type shown in Fig. 4.
These values are not significantly different (P > 0.05) to the Kd for WT human apoA-I.
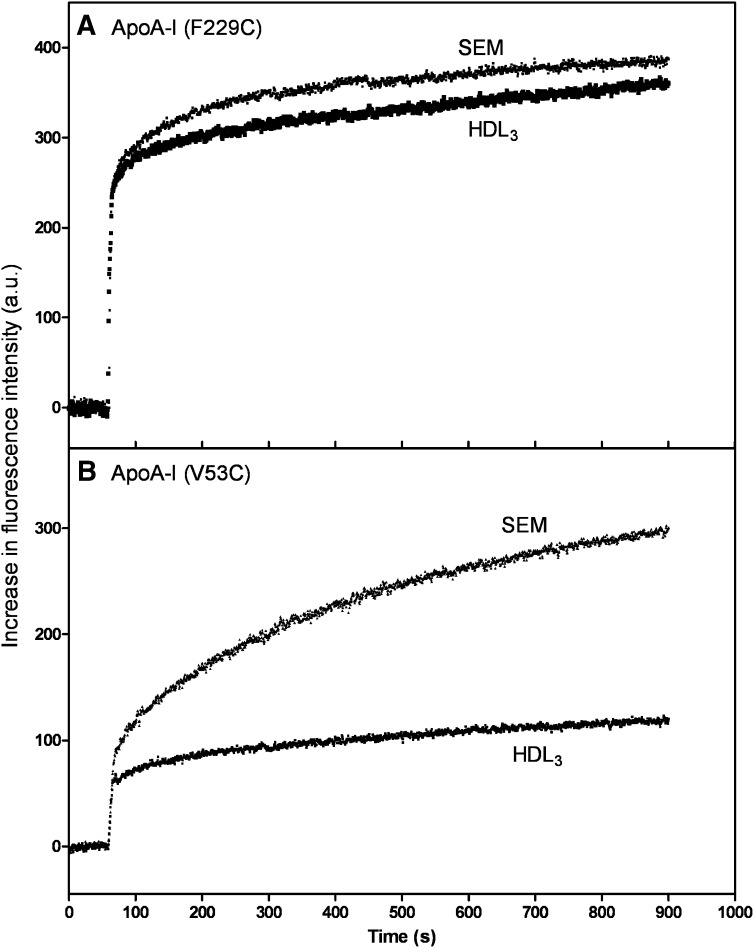
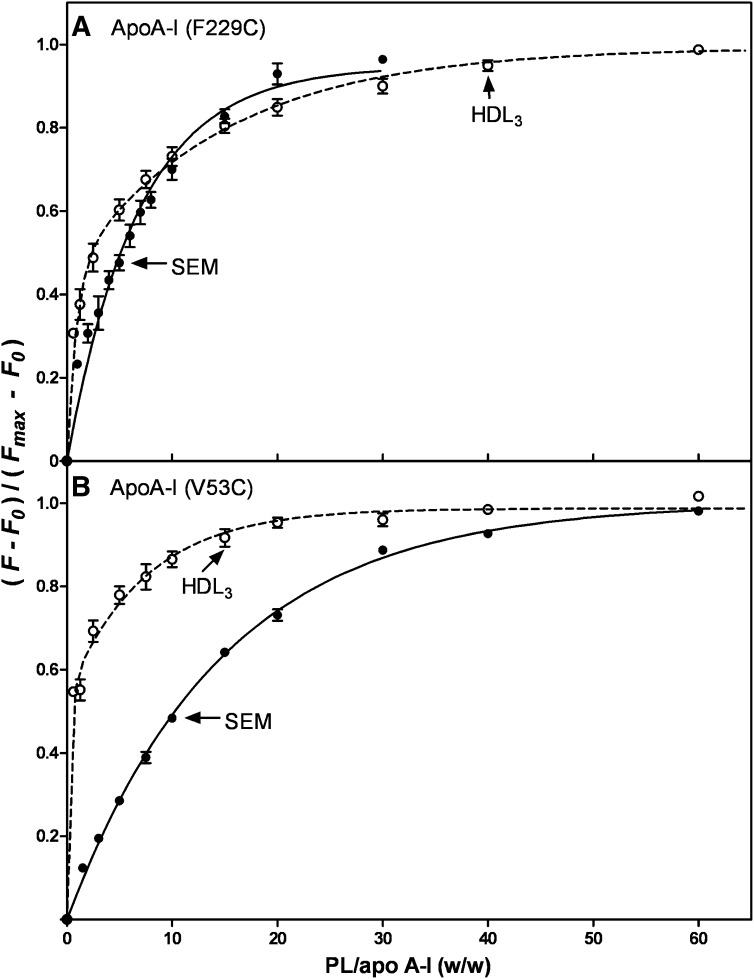
To further explore the binding of the N- and C-terminal domains of apoA-I to HDL3, we monitored the binding of human apoA-I V53C-pyrene and apoA-I F229C-pyrene by fluorescence spectroscopy, as we have done before in a small unilamellar vesicle (SUV) system (39). When these labeled apoA-I molecules bind to a lipid particle, there is an increase in fluorescence intensity due to movement of the pyrene moiety into a hydrophobic environment (Fig. 5). It is apparent from the results in Fig. 5A that the kinetics of binding of the apoA-I C-terminal domain (F229C-pyrene) to SEM and HDL3 are similar, consistent with the binding being lipid-mediated in both cases. In contrast, the kinetics of interaction of the apoA-I N-terminal helix bundle domain (V53C-pyrene) with the SEM and HDL3 particles are markedly different (Fig. 5B). The binding titration curves depicted in Fig. 6 reveal the same trends. Thus, the C-terminal domain of apoA-I binds similarly to the SEM and HDL3 and reaches saturation at about the same PL/apoA-I ratio (Fig. 6A). This result indicates that interactions between the hydrophobic apoA-I C-terminal α-helix and lipid dominate the binding to both the SEM and HDL3 particles. However, the binding behavior of the N-terminal domain is quite different in that it exhibits enhanced binding to HDL3 compared with the SEM (Fig. 6B). This effect is most likely due to protein-protein interactions between the apoA-I N-terminal helix bundle domain and apolipoproteins resident in the HDL3 particle surface. Such protein-protein interactions can occur readily because it can be estimated from the particle lipid/protein composition and size (48) that the resident apolipoproteins occupy about 80% of the HDL3 particle surface area.
Fig. 5.
Time courses of increases in fluorescence intensity upon addition of either emulsion particles (SEM) or HDL3 to apoA-I F229C-pyrene (A) and apoA-I V53C-pyrene (B). SEM or HDL3 particles were added to the apoA-I variants at final concentrations of 10 μg apoA-I variant/ml and 0.4–0.6 mg/ml PL. Pyrene fluorescence was monitored at 385 nm with excitation at 342 nm.
Fig. 6.
Fraction of apoA-I F229C-pyrene (A) and apoA-I V53C-pyrene (B) bound to emulsions (SEM) (closed circles) and HDL3 (open circles) as a function of the weight ratio of PL to pyrene-apoA-I. The protein concentration was 25 μg/ml.
Influence of tertiary structure domain properties on apoA-I binding to HDL
We have taken advantage of the differences in the N- and C-terminal domain properties of human and mouse apoA-I (32) to explore the influence of these properties on the binding of apoA-I to HDL. Figure 7 shows the binding isotherms of WT mouse apoA-I to HDL2, and the corresponding binding constants are listed in Table 1. It is apparent that the Bmax value for WT mouse apoA-I is lower than that of WT human apoA-I, but the Kd values for the two proteins are not significantly different; the Kd value for WT mouse apoA-I binding to HDL2 is in excellent agreement with a prior report (49). As is observed with human apoA-I, deletion of the C-terminal domain of mouse apoA-I to give mouse apoA-I (1–186) increases the binding affinity (reduces Kd) and also reduces the amount of binding (lower Bmax). In contrast to the situation with the isolated C-terminal domain of human apoA-I, which does not bind to HDL2 (Fig. 4), the isolated mouse C-terminal domain binds to HDL2 in a low affinity, nonsaturable fashion giving rise to a linear binding isotherm in the concentration range studied (Fig. 7). The SPR experiments did not reveal significant differences in the binding constants for WT mouse apoA-I interacting with HDL2 and HDL3 (data not shown), although Reschly et al. (49) using different methods showed that this protein binds relatively weakly to HDL3.
Fig. 7.
Comparison of the binding of mouse apoA-I and its tertiary structure domains to HDL2. The binding isotherms were obtained and analyzed as described in the legend to Fig. 2B. Circles, WT apoA-I; triangles, N-terminal domain (residues 1–186); diamonds, C-terminal domain (residues 187–240).
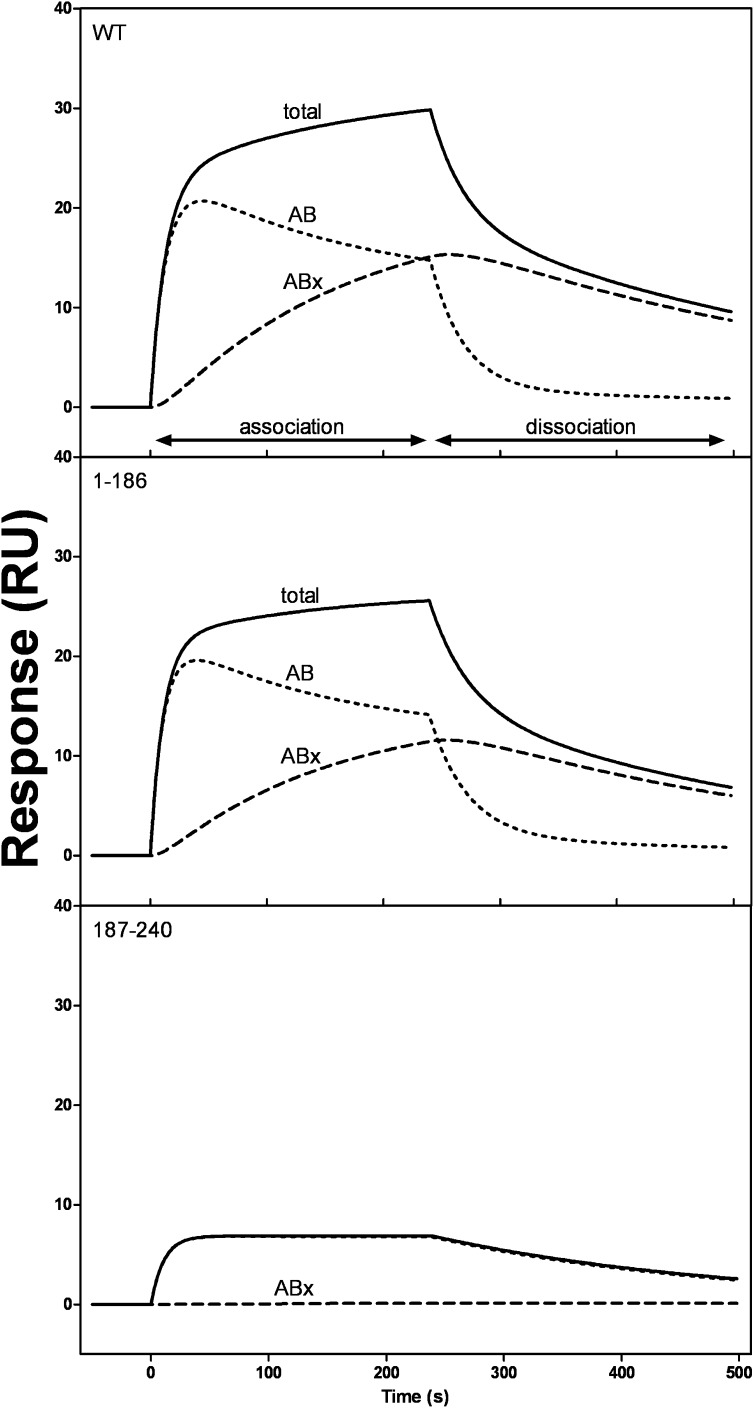
Figures 8 and 9 compare the initial binding (AB) and subsequent binding or conformational change (ABx) components for WT human and mouse apoA-I and their isolated N- and C-terminal domains binding to HDL3. In the case of human apoA-I (Fig. 8), the deconvolution analysis shows that the AB and ABx components are similar for the intact protein and for the isolated N-terminal helix-bundle domain. The level of binding of the isolated C-terminal domain of human apoA-I was too low for the deconvolution analysis to be possible. However, in the case of mouse apoA-I, it was possible to compare the behavior of the isolated N- and C-terminal domains. The results in Fig. 9 demonstrate that the N-terminal domain exhibits two-state binding, whereas the C-terminal domain displays essentially one-state binding with only a very minor ABx component being apparent.
Fig. 8.
SPR sensorgrams of binding of WT human apoA-I and its N-terminal tertiary structure domain (residues 1–189) to immobilized HDL3. The sensorgram for total binding (solid line) is for 50 μg/ml apoA-I flowed at 20 μl/min, and the initial binding (AB) and subsequent binding (ABx) contributions were obtained by deconvolution, as described in the legend to Fig. 3.
Fig. 9.
SPR sensorgrams of binding of WT mouse apoA-I and its N- (residues 1–186) and C-terminal (residues 187–240) tertiary structure domains to immobilized HDL3. The data were analyzed as described for human apoA-I in the legend to Fig. 8.
DISCUSSION
Relatively little detailed understanding is available about the interaction of apoA-I with preformed spherical HDL particles, despite the importance of this phenomenon in the cycling of apoA-I between lipid-bound and lipid-free states in the plasma compartment (10). This SPR study of the real-time apoA-I association with and dissociation from HDL particles provides novel insights into the mechanism of interaction and information about the affinity of binding.
Kinetics of apoA-I/HDL interaction
The fact that the sensorgrams depicted in Fig. 2A are best fitted to a two-state model indicates that some sequential binding events occur when apoA-I molecules bind to HDL particles. The time courses of these two processes (AB and ABx) are shown in Fig. 3A, from which it is apparent that the dissociation of the initial bound form of apoA-I (AB) is more rapid than that of the subsequent bound form (ABx). The fact that the length of time apoA-I molecules are in contact with the HDL surface affects the rate of dissociation (Fig. 3B) provides strong evidence for the second (ABx) kinetic phase involving a conformational change in the apoA-I molecule. The question arises as to what is the nature of these two bound forms of apoA-I.
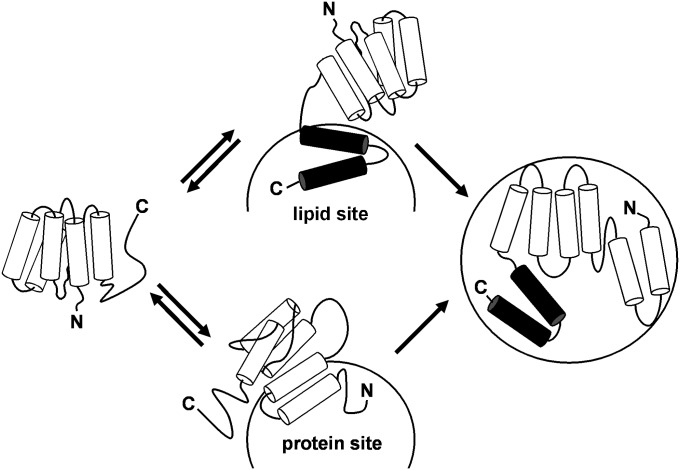
Because apoA-I adopts a two-domain tertiary structure (5, 9), an obvious possibility is that these two domains bind in a sequential fashion to give the observed kinetics. However, the fact that the WT human apoA-I molecule and its isolated N-terminal domain (residues 1–189) exhibit the same reaction kinetics (Fig. 8) indicates that this is not the case. Apparently, either the N-terminal helix bundle domain or the C-terminal domain can initiate binding to the HDL particle surface. In the SUV situation where the particle surface contains only PL molecules, the rapid initial binding step occurs through the C-terminal domain and the second step involves the subsequent slow opening of the N-terminal helix bundle domain (5, 9). As depicted in Fig. 10, this series of events may occur when apoA-I binds to an HDL particle, but the situation is more complex in that an apoA-I molecule can also first bind via its helix bundle domain. Regardless of the mode of initial interaction, the second slower step is proposed to involve helix bundle unfolding (Fig. 10). In support of this, binding of the isolated mouse N-terminal domain (residues 1–186) exhibits a clear second, slow ABx phase, whereas binding of the isolated mouse C-terminal domain (residues 187–240) does not (Fig. 9).
Fig. 10.
Model of the two-step binding mechanism of apoA-I to a spherical HDL particle. Initial binding (Step 1) is rapid and readily reversible and can occur through amphipathic α-helices (cylinders) in either the C-terminal domain (upper pathway) or the N-terminal helix bundle domain (lower pathway) of the apoA-I molecule. In the upper pathway, the C-terminal domain of apoA-I (solid cylinders) first interacts with a lipid site on the HDL particle surface, and Step 2 involves the subsequent opening of the helix bundle whereby hydrophobic helix-helix interactions are converted to helix-lipid interactions. Step 2 is relatively slow and less readily reversible. In the lower pathway, the apoA-I N-terminal helix bundle domain interacts with a protein site on the HDL particle surface.
Affinity of apoA-I/HDL interaction
The isotherms depicted in Figs. 4 and 7 together with the binding constants listed in Table 1 give insight into how the properties of the apoA-I tertiary structure domains influence the apoA-I/HDL interaction. In marked contrast to what is seen with binding to SUV, where high affinity binding is eliminated (44, 50), removal of either the C-terminal domain or C-terminal α-helix does not disrupt the high affinity binding of human apoA-I to HDL (Fig. 4). Furthermore, the isolated C-terminal domain of human apoA-I (residues 190–243) cannot bind to HDL (Fig. 4), but it can bind well to pure lipid particles (32, 51). Removal of the C-terminal domain from WT mouse apoA-I also does not eliminate high affinity binding to HDL (Fig. 7). However, this result is consistent with what occurs with binding to SUV and is a consequence of the C-terminal domain of mouse apoA-I (residues 187–240) being relatively polar and having weak lipid binding ability (32). Unlike its human counterpart, the isolated mouse C-terminal domain can bind to HDL, albeit in a low affinity fashion (Fig. 7). The above differences in behavior point to interactions besides direct apoA-I/lipid interactions being involved in binding of apoA-I to HDL. Indeed, the pyrene-apoA-I fluorescence data in Figs. 5 and 6 demonstrate that the apoA-I helix bundle domain interacts with apolipoproteins resident in the HDL particle surface. Such protein-protein interactions are responsible for the lower pathway of apoA-I binding depicted in Fig. 10.
Physiological implications
A question arises as to the implications of the Kd value of ∼10 µM for WT human apoA-I/HDL interaction (Table 1) for the distribution of apoA-I between lipid-bound and lipid-free states in plasma. Assuming that a typical concentration of HDL particles in human plasma is about 16 µM, then it follows that ∼40% of any apoA-I molecules interacting with a Kd of 10 µM are in the lipid-free state. Only 5–10% of total plasma apoA-I is in the lipid-free state (10), so it follows that most apoA-I molecules in HDL particles are more strongly bound and have a lower Kd value. The Kd of ∼10 µM must apply to a smaller pool of weakly associated apoA-I molecules that are perhaps being held in a “reservoir” state and are not essential structural components of the HDL particle. In support of this idea, several approaches have demonstrated that HDL contains two populations of apoA-I. Thus, there is a labile pool that is readily released by chemical denaturants and susceptible to mild trypsin digestion and another more strongly bound pool that is not released by chemical denaturants and is resistant to trypsin digestion (19, 52). Thermal denaturation of spherical HDL also demonstrates the existence of two pools of apoA-I; dissociation of one apoA-I subpopulation causes HDL particle fusion, while dissociation of the second is associated with particle rupture (53). Studies of disruption of plasma HDL particles by Streptococcal serum opacity factor indicate that most HDL particles contain at least one labile apoA-I molecule that can dissociate readily (54). Finally, the existence of a labile apoA-I pool is consistent with the observation that certain discoidal HDL particles can spontaneously rearrange with the release of some apoA-I molecules (55). The strongly bound apoA-I molecules in spherical HDL particles are aligned in a “trefoil” arrangement (56), while at least some of the weakly bound molecules are probably attached to lipid sites only through the C-terminal domain (39).
Footnotes
Abbreviations:
- apo
- apolipoprotein
- Bmax
- saturating amount bound
- HABA
- 2-(4′-hydroxyphenyl) azobenzoic acid
- PL
- phospholipid
- RU
- resonance unit
- SEM
- small emulsion
- SPR
- surface plasmon resonance
- SUV
- small unilamellar vesicle
- WT
- wild type
This research was supported by National Institutes of Health Grant HL-22633, by the Takeda Science Foundation, and by the Suzuken Memorial Foundation. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1.Rader D. J. 2006. Molecular regulation of HDL metabolism and function: implications for novel therapies. J. Clin. Invest. 116: 3090–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cuchel M., Rader D. J. 2006. Macrophage reverse cholesterol transport. Key to the regression of atherosclerosis? Circulation. 113: 2548–2555. [DOI] [PubMed] [Google Scholar]
- 3.Brouillette C. G., Anantharamaiah G. M., Engler J. A., Borhani D. W. 2001. Structural models of human apolipoprotein A-I: a critical analysis and review. Biochim. Biophys. Acta. 1531: 4–46. [DOI] [PubMed] [Google Scholar]
- 4.Marcel Y. L., Kiss R. S. 2003. Structure-function relationships of apolipoprotein A-I: a flexible protein with dynamic lipid associations. Curr. Opin. Lipidol. 14: 151–157. [DOI] [PubMed] [Google Scholar]
- 5.Saito H., Lund-Katz S., Phillips M. C. 2004. Contributions of domain structure and lipid interaction to the functionality of exchangeable human apolipoproteins. Prog. Lipid Res. 43: 350–380. [DOI] [PubMed] [Google Scholar]
- 6.Segrest J. P., Jones M. K., De Loof H., Brouillette C. G., Venkatachalapathi Y. V., Anantharamaiah G. M. 1992. The amphipathic helix in the exchangeable apolipoproteins: a review of secondary structure and function. J. Lipid Res. 33: 141–166. [PubMed] [Google Scholar]
- 7.Pownall H. J., Massey J. B., Kusserow S. K., Gotto A. M. 1978. Kinetics of lipid-protein interactions: interaction of apolipoprotein A-I from human plasma high density lipoproteins with phosphatidylcholine. Biochemistry 17: 1183–1188. [DOI] [PubMed] [Google Scholar]
- 8.Vedhachalam C., Duong P. T., Nickel M., Nguyen D., Dhanasekaran P., Saito H., Rothblat G. H., Lund-Katz S., Phillips M. C. 2007. Mechanism of ATP-binding cassette transporter A1-mediated cellular lipid efflux to apolipoprotein A-I and formation of high density lipoprotein particles. J. Biol. Chem. 282: 25123–25130. [DOI] [PubMed] [Google Scholar]
- 9.Saito H., Dhanasekaran P., Nguyen D., Holvoet P., Lund-Katz S., Phillips M. C. 2003. Domain structure and lipid interaction in human apolipoproteins A-I and E: a general model. J. Biol. Chem. 278: 23227–23232. [DOI] [PubMed] [Google Scholar]
- 10.Rye K. A., Barter P. J. 2004. Formation and metabolism of prebeta-migrating, lipid-poor apolipoprotein A-I. Arterioscler. Thromb. Vasc. Biol. 24: 421–428. [DOI] [PubMed] [Google Scholar]
- 11.Pownall H. J., Ehnholm C. 2006. The unique role apolipoprotein A-I in HDL remodeling and metabolism. Curr. Opin. Lipidol. 17: 209–213. [DOI] [PubMed] [Google Scholar]
- 12.Duong P. T., Weibel G. L., Lund-Katz S., Rothblat G. H., Phillips M. C. 2008. Characterization and properties of pre beta-HDL particles formed by ABCA1-mediated cellular lipid efflux to apoA-I. J. Lipid Res. 49: 1006–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shepherd J., Patsch J. R., Packard C. J., Gotto A. M., Jr., Taunton O. D. 1978. Dynamic properties of human high density lipoprotein apoproteins. J. Lipid Res. 19: 383–389. [PubMed] [Google Scholar]
- 14.Grow T. E., Fried M. 1978. Interchange of apoprotein components between the human plasma high density lipoprotein subclasses HDL2 and HDL3 in vitro. J. Biol. Chem. 25: 8034–8041. [PubMed] [Google Scholar]
- 15.Cheung M. C., Wolf A. C., Knopp R. H., Foster D. M. 1992. Protein transfer between A-I-containing lipoprotein subpopulations: Evidence of non-transferable A-I in particles with A-II. Biochim. Biophys. Acta. 1165: 68–77. [DOI] [PubMed] [Google Scholar]
- 16.Boyle K. E., Phillips M. C., Lund-Katz S. 1999. Kinetics and mechanism of exchange of apolipoprotein C–III molecules from very low density lipoprotein particles. Biochim. Biophys. Acta. 1430: 302–312. [DOI] [PubMed] [Google Scholar]
- 17.Ibdah J. A., Lund-Katz S., Phillips M. C. 1990. Kinetics and mechanism of transfer of reduced and carboxymethylated apolipoprotein A-II between phospholipid vesicles. Biochemistry 29: 3472–3479. [DOI] [PubMed] [Google Scholar]
- 18.Ibdah J. A., Smith C., Lund-Katz S., Phillips M. C. 1991. Effects of apolipoprotein structure on the kinetics of apolipoprotein transfer between phospholipid vesicles. Biochim. Biophys. Acta. 1081: 220–228. [DOI] [PubMed] [Google Scholar]
- 19.Pownall H. J., Hosken B. D., Gillard B. K., Higgins C. L., Lin H. Y., Massey J. B. 2007. Speciation of human plasma high-density lipoprotein (HDL): HDL stablity and apolipoprotein A-I partitioning. Biochemistry 46: 7449–7459. [DOI] [PubMed] [Google Scholar]
- 20.Lookene A., Savonen R., Olivecrona G. 1997. Interaction of lipoproteins with heparan sulfate proteoglycans and with lipoprotein lipase. Studies by surface plasmon resonance technique. Biochemistry 36: 5267–5275. [DOI] [PubMed] [Google Scholar]
- 21.Libeu C. P., Lund-Katz S., Phillips M. C., Wehrli S., Hernaiz M. J., Capila I., Linhardt R. J., Raffai R. L., Newhouse Y. M., Zhou F., et al. 2001. New insights into the heparin sulfate proteoglycan-binding activity in apolipoprotein E. J. Biol. Chem. 276: 39138–39144. [DOI] [PubMed] [Google Scholar]
- 22.Futamura M., Dhanasekaran P., Handa T., Phillips M. C., Lund-Katz S., Saito H. 2005. Two-step mechanism of binding of apolipoprotein E to heparin. J. Biol. Chem. 280: 5414–5422. [DOI] [PubMed] [Google Scholar]
- 23.Yamauchi Y., Deguchi N., Takagi C., Tanaka M., Dhanasekaran P., Nakano M., Handa T., Phillips M. C., Lund-Katz S., Saito H. 2008. Role of the N- and C-terminal domains in binding of apolipoprotein E isoforms to heparan sulfate and dermatan sulfate: a surface plasmon resonance study. Biochemistry 47: 6702–6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin L., Shieh J. J., Grabbe E., Adimoolam S., Durbin D., Jonas A. 1999. Surface plasmon resonance biosensor studies of human wild-type and mutant lecithin cholesterol acyltransferase interactions with lipoproteins. Biochemistry 38: 15659–15665. [DOI] [PubMed] [Google Scholar]
- 25.Gaidukov L., Tawfik D. S. 2005. High affinity, stability, and lactonase activity of serum paraxonase PON1 anchored on HDL with apoA-I. Biochemistry 44: 11843–11854. [DOI] [PubMed] [Google Scholar]
- 26.Clark R. W., Ruggeri R. B., Cunningham D., Bamberger M. J. 2006. Description of the torcetrapib series of cholesteryl ester transfer protein inhibitors, including mechanism of action. J. Lipid Res. 47: 537–552. [DOI] [PubMed] [Google Scholar]
- 27.Curtiss L. K., Bonnett D. J., Rye K. A. 2000. The conformation of apolipoprotein A-I in high-density lipoproteins is influenced by core lipid composition and particle size: A surface plasmon resonance study. Biochemistry 39: 5712–5721. [DOI] [PubMed] [Google Scholar]
- 28.Havel R. J., Eder H. A., Bragdon J. H. 1955. The distribution and chemical composition of untracentrifugally separated lipoproteins in human serum. J. Clin. Invest. 34: 1345–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hatch F. T., Lees R. S. 1968. Practical methods for plasma lipoprotein analysis. Adv. Lipid Res. 6: 1–68. [PubMed] [Google Scholar]
- 30.Reijngoud D. J., Lund-Katz S., Hauser H., Phillips M. C. 1982. Lipid-protein interactions. Effect of apolipoprotein A-I on phosphatidylcholine polar group conformation as studied by proton nuclear magnetic resonance. Biochemistry 21: 2977–2983. [DOI] [PubMed] [Google Scholar]
- 31.Morrow J. A., Arnold K. S., Weisgraber K. H. 1999. Functional characterization of apolipoprotein E isoforms overexpressed in escherichia coli. Protein Expr. Purif. 16: 224–230. [DOI] [PubMed] [Google Scholar]
- 32.Tanaka M., Koyama M., Dhanasekaran P., Nguyen D., Nickel M., Lund-Katz S., Saito H., Phillips M. C. 2008. Influence of tertiary structure domain properties on the functionality of apolipoprotein A-I. Biochemistry 47: 2172–2180. [DOI] [PubMed] [Google Scholar]
- 33.Nguyen D., Dhanasekaran P., Phillips M. C., Lund-Katz S. 2009. Molecular mechanism of apolipoprotein E binding to lipoprotein particles. Biochemistry 48: 3025–3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karlsson R., Falt A. 1997. Experimental design for kinetic analysis of protein-protein interactions with surface plasmon resonance biosensors. J. Immunol. Methods. 200: 121–133. [DOI] [PubMed] [Google Scholar]
- 35.Lipschultz C. A., Li Y., Smith-Gill S. 2000. Experimental design for analysis of complex kinetics using surface plasmon resonance. Methods. 20: 310–318. [DOI] [PubMed] [Google Scholar]
- 36.Saito H., Dhanasekaran P., Baldwin F., Weisgraber K. H., Phillips M. C., Lund-Katz S. 2003. Effects of polymorphism on the lipid interaction of human apolipoprotein E. J. Biol. Chem. 278: 40723–40729. [DOI] [PubMed] [Google Scholar]
- 37.Phillips M. C., Krebs K. E. 1986. Studies of apolipoproteins at the air-water interface. Methods Enzymol. 128: 387–403. [DOI] [PubMed] [Google Scholar]
- 38.Liu L., Bortnick A. E., Nickel M., Dhanasekaran P., Subbaiah P. V., Lund-Katz S., Rothblat G. H., Phillips M. C. 2003. Effects of apolipoprotein A-I on ATP-binding cassette transporter A1-mediated efflux of macrophage phospholipid and cholesterol. J. Biol. Chem. 278: 42976–42984. [DOI] [PubMed] [Google Scholar]
- 39.Kono M., Okumura Y., Tanaka M., Nguyen D., Dhanasekaran P., Lund-Katz S., Phillips M. C., Saito H. 2008. Conformational flexibility of the N-terminal domain of apolipoprotein A-I bound to spherical lipid particles. Biochemistry 47: 11340–11347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saito H., Dhanasekaran P., Baldwin F., Weisgraber K., Lund-Katz S., Phillips M. C. 2001. Lipid binding-induced conformational change in human apolipoprotein E. J. Biol. Chem. 276: 40949–40954. [DOI] [PubMed] [Google Scholar]
- 41.Ladokhin A. S., Jayasinghe S., White S. H. 2000. How to measure and analyze tryptophan fluorescence in membranes properly, and why bother? Anal. Biochem. 285: 235–245. [DOI] [PubMed] [Google Scholar]
- 42.Derksen A., Gantz D., Small D. M. 1996. Calorimetry of apolipoprotein-AI binding to phosphatidylcholine-triolein-cholesterol emulsions. Biophys. J. 70: 330–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saito H., Miyako Y., Handa T., Miyajima K. 1997. Effect of cholesterol on apolipoprotein A-I binding to lipid bilayers and emulsions. J. Lipid Res. 38: 287–294. [PubMed] [Google Scholar]
- 44.Saito H., Dhanasekaran P., Nguyen D., Deridder E., Holvoet P., Lund-Katz S., Phillips M. C. 2004. Alpha-helix formation is required for high affinity binding of human apolipoprotein A-I to lipids. J. Biol. Chem. 279: 20974–20981. [DOI] [PubMed] [Google Scholar]
- 45.Tajima S., Yokoyama S., Yamamoto A. 1983. Effect of lipid particle size on association of apolipoproteins with lipid. J. Biol. Chem. 258: 10073–10082. [PubMed] [Google Scholar]
- 46.Carnemolla R., Ren X., Biswas T. K., Meredith S. C., Reardon C. A., Wang J., Getz G. S. 2008. The specific amino acid sequence between helices 7 and 8 influences the binding specificity of human apolipoprotein A-I for high density lipoprotein (HDL) subclasses: a potential for HDL preferential generation. J. Biol. Chem. 283: 15779–15788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vitello L. B., Scanu A. M. 1976. Studies on human serum high density lipoproteins. Self-association of apolipoprotein A-I in aqueous solutions. J. Biol. Chem. 251: 1131–1136. [PubMed] [Google Scholar]
- 48.Shen B. W., Scanu A. M., Kezdy F. J. 1977. Structure of human serum lipoproteins inferred from compositional analysis. Proc. Natl. Acad. Sci. USA. 74: 837–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reschly E. J., Sorci-Thomas M., Davidson W. S., Meredith S. C., Reardon C., Getz G. S. 2002. Apolipoprotein A-I alpha helices 7 and 8 modulate high density lipoprotein subclass distribution. J. Biol. Chem. 277: 9645–9654. [DOI] [PubMed] [Google Scholar]
- 50.Tanaka M., Dhanasekaran P., Nguyen D., Ohta S., Lund-Katz S., Phillips M. C., Saito H. 2006. Contributions of the N- and C-terminal helical segments to the lipid-free structure and lipid interaction of apolipoprotein A-I. Biochemistry 45: 10351–10358. [DOI] [PubMed] [Google Scholar]
- 51.Zhu H. L., Atkinson D. 2007. Conformation and lipid binding of a C-terminal (198–243) peptide of human apolipoprotein A-I. Biochemistry 46: 1624–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mendez A. J., Oram J. F. 1997. Limited proteolysis of high density lipoprotein abolishes its interaction with cell-surface binding sites that promote cholesterol efflux. Biochim. Biophys. Acta. 1346: 285–299. [DOI] [PubMed] [Google Scholar]
- 53.Gao X., Yuan S., Jayaraman S., Gursky O. 2009. Differential stability of high-density lipoprotein subclasses: effects of particle size and protein composition. J. Mol. Biol. 387: 628–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Han M., Gillard B. K., Courtney H. S., Ward K., Rosales C., Khant H., Ludtke S. J., Pownall H. J. 2009. Disruption of human plasma high-density lipoproteins by streptococcal serum opacity factor requires labile apolipoprotein A-I. Biochemistry 48: 1481–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cavigiolio G., Shao B., Geier E. G., Ren G., Heinecke J. W., Oda M. N. 2008. The interplay between size, morphology, stability, and functionality of high-density lipoprotein subclasses. Biochemistry 47: 4770–4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Silva R. A. G., Huang R., Morris J., Fang J., Gracheva E. O., Ren G., Kontush A., Jerome W. G., Rye K. A., Davidson W. S. 2008. Structure of apolipoprotein A-I in spherical high density lipoproteins of different sizes. Proc. Natl. Acad. Sci. USA. 105: 12176–12181. [DOI] [PMC free article] [PubMed] [Google Scholar]