Abstract
BCL6 is a transcriptional repressor required for mature B-cell germinal center (GC) formation and implicated in lymphomagenesis. BCL6's physiologic function is only partially known because the complete set of its targets in GC B cells has not been identified. To address this issue, we used an integrated biochemical-computational-functional approach to identify BCL6 direct targets in normal GC B cells. This approach includes (1) identification of BCL6-bound promoters by genome-wide chromatin immunoprecipitation, (2) inference of transcriptional relationships by the use of a regulatory network reverse engineering approach (ARACNe), and (3) validation of physiologic relevance of the candidate targets down-regulated in GC B cells. Our approach demonstrated that a large set of promoters (> 4000) is physically bound by BCL6 but that only a fraction of them is repressed in GC B cells. This set of 1207 targets identifies several cellular functions directly controlled by BCL6 during GC development, including activation, survival, DNA-damage response, cell cycle arrest, cytokine signaling, Toll-like receptor signaling, and differentiation. These results define a broad role of BCL6 in preventing centroblasts from responding to signals leading to exit from the GC before they complete the phase of proliferative expansion and of antibody affinity maturaton.
Introduction
BCL6 has emerged as a critical regulator of germinal centers (GCs), the sites where B cells undergo somatic hypermutation (SHM) and class switch recombination of their immunoglobulin genes (Ig) and are then selected on the basis of the production of antibodies with high affinity for the antigen.1 BCL6 is also a frequently activated oncogene in the pathogenesis of human B-cell lymphomas, most of which derive from the GC B cells. The BCL6 gene encodes a 95-kDA nuclear phosphoprotein belonging to the BTB/POZ zinc-finger (ZF) family of transcription factors.2–4 BCL6 functions as a transcriptional repressor via its C-terminal zinc-finger domain that binds to specific DNA sequences in the promoter region of target genes and 2 transcriptional repression domains5 that interact with distinct corepressor complexes during the GC reaction.6–9 Within the B-cell lineage, the BCL6 protein is expressed at high levels only in mature B cells within GCs.10 GC formation and the development of normal T cell–dependent humoral immune responses require expression of BCL6 because BCL6-null mice do not form GCs and are unable to produce high-affinity antibodies.2,4 BCL6 expression is regulated by several signals that are crucial for GC development. Activation of B-cell receptor (BCR) induces mitogen-activated protein kinase (MAPK)-mediated phosphorylation of the BCL6 protein, which targets BCL6 for rapid degradation by the ubiquitin proteasome pathway.11 Stimulation of the CD40 receptor by CD40 ligands expressed by T cells leads to transcriptional down-regulation of BCL6 via a signaling pathway that involves nuclear factor (NF)-κB–mediated transcriptional activation of interferon regulatory factor 4 (IRF4), which, in turn, directly represses BCL6 transcription.12,13 BCL6 degradation is induced by DNA damage via a pathway that is distinct from the one induced by BCR,14 whereas BCL6 function is also inactivated by acetylation, which triggers its dissociation from corepressor complexes.15 These findings indicate that although BCL6 is required for GC formation, its down-regulation may be critical for B cells to exit the GC and differentiate toward memory and plasma cells.
A variety of structural alterations of the BCL6 gene are associated with its deregulated expression in B-cell lymphomas. Chromosomal translocations juxtaposing heterologous promoters to the BCL6 coding domain are found in approximately 40% of diffuse large B-cell lymphoma (DLBCL) and in a minority (5%-10%) of follicular lymphoma (FL).16–18 The common denominator of these promoters is their constitutive activity in the B-cell lineage and in particular their persistent activity in post-GC cells such as immunoblasts and plasma cells, in contrast with the GC-specific activity of the BCL6 promoter.19 In addition, although alterations of the 5′ noncoding region of BCL6 by SHM is a feature of normal GC B cells,20,21 specific mutations found only in DLBCL lead to the deregulated expression of BCL6 through disruption of the sequences mediating a negative autoregulatory circuit or IRF4-mediated repression.13,22 These genetic alterations suggest that BCL6 deregulation contributes to lymphomagenesis, and this notion has been validated in transgenic mice in which the constitutive expression of BCL6 leads to the development of DLBCL.23
A thorough understanding of the biologic role of BCL6 in normal B-cell development and lymphomagenesis depends upon the identification of the full set of genes that are targets of its transcriptional regulatory function. Toward this end, gene expression profiling (GEP) analysis has identified a large number of genes whose expression responds to variations of BCL6 expression.24 However, this approach cannot distinguish between direct targets and the presumably very large set of secondary targets whose expression is indirectly influenced by BCL6. More recently, a genome-wide chromatin immunoprecipitation (ChIP-on-chip) study has identified a large set of genes whose promoter regions are bound by BCL6 in vivo.25 However, this approach has limitations because physical binding does not necessarily imply functional activity, as shown for other transcription factors.26
Furthermore, a common limitation of the aforementioned 2 approaches is that they have been performed in transformed B cells, leaving open the possibility that BCL6 function may be altered in lymphoma cells. Thus, only a few genes have been fully validated as functionally and physiologically relevant targets in normal cells, including the gene encoding the coactivator molecule CD80,27 genes involved in the sensing and response to DNA damage (TP53, ATR, and CHEK1),28–30 the cell-cycle arrest gene CDKN1A/p21,31 and the plasma cell differentiation master gene BLIMP1.32 A recent publication in which the investigators aimed at the comparison of BCL6 transcriptional program in normal and malignant B cells reported that BCL6 binds to almost 2000 promoters. Approximately 180 genes of the 900 for which GEPs were available also showed down-regulation in GC B cells, representing the first large set of BCL6 physiologic targets.33
Ideally, a comprehensive analysis of the BCL6 physiologic transcriptional program would involve the identification of genes whose promoter is bound by BCL6 and is functionally responsive to BCL6 in its physiologic context, that is, normal GC B cells. Toward this end, we have applied an integrated approach involving 3 complementary analyses (supplemental Figure 1, available on the Blood website; see the Supplemental Materials link at the top of the online article): biochemical (ChIP-on-chip, for physical binding), coupled with a reverse-engineering algorithm that identifies robust transcriptional relationships conserved across a large set of B-cell phenotypes (ARACNe),34,35 and finally filtered by GEP analysis to identify targets that are physiologically repressed in GC B cells. The results identify a large set of novel target genes that provide a comprehensive picture of BCL6 function in normal B cells.
Methods
Primary cells and cell lines
Purification of GC B cells was performed as previously reported36 by the use of magnetic cell sorting of mononucleated cells obtained from human tonsils. Tissue collection from tonsils was approved by the institutional ethical committee of Columbia University with informed consent from patients in accordance with the Declaration of Helsinki. P3HR1 and Ramos (Burkitt lymphoma cell lines) and 293T (fetal kidney cell line) were obtained from ATCC and maintained at the recommended concentration in Iscove modified Dulbecco medium and Dulbecco modified Eagle medium (both Invitrogen), respectively, supplemented with 10% fetal bovine serum and antibiotics.
Plasmids, reporter assays, and lentiviral infections
See the supplemental Methods for details on plasmids and transfection method. Reporter assays were performed as previously described.5,15 Lentiviral vectors for BCL6 siRNA (TRC0000013606 and TRC0000013603) and control siRNA (SHC002) were purchased from Sigma-Aldrich, and lentiviral supernatants were produced and used to infect P3HR1 and Ramos cells as previously reported.13
Immunoblotting
Whole-cell lysates were prepared as previously described.11 Proteins were analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and immunoblotting by use of the following antibodies: anti-BCL6 (Cell Signaling); anti–β-actin (A5441, Sigma-Aldrich); and anti-HA (3F10; Roche).
ChIP, qChIP, and ChIP-on-chip assays
ChIP was performed as previously reported22 starting from 40 × 106 purified GC B cells or 20 × 106 P3HR1 cells and by the use of 4 μg of anti-BCL6 antibody (N3; Santa Cruz Biotechnology) or isotype-matched polyclonal IgG (Sigma-Aldrich). DNA fragments enriched by ChIP were analyzed for the expression of individual targets including BCL6 (positive control) and β-actin (negative control) by the use of primers reported in supplemental Table 1 and SYBR Green PCR Master Mix (Applied Biosystems) as recommended by the manufacturer. ChIP-on-chip was performed independently on GC B cells isolated from 3 patients. BCL6-ChIP DNA and whole-cell extract (input) DNA were purified, amplified by ligation-mediated polymerase chain reaction, and labeled (CGH Labeling kit; Invitrogen) with Cy5 and Cy3 (PerkinElmer), respectively. The labeled ChIP DNA samples were mixed in 1:1 ratio with their respective input DNA and hybridized on the Human Promoter ChIP-on-chip Microarray Set (2 × 244K; Agilent Technologies). Hybridization was performed for 40 hours at 65°C in a rotating oven, and then the slides were washed and scanned (Agilent Scanner; Agilent Technologies) following the manufacturer's indications. The data were analyzed by ChIP-on-chip significance analysis (CSA).37 Details on quantitative ChIP (qChIP) and ChIP-on-chip analyses are reported in the supplemental Methods.
Quantitative reverse-transcription PCR
Total RNA was isolated by the use of the TRIzol Reagent (Invitrogen) following the manufacturer's instructions. One microgram of total RNA was reverse-transcribed by use of the Superscript First Strand Synthesis System for RT-PCR (Invitrogen), and 1/100 of cDNA was used as template for PCR amplification by use of the primers reported in supplemental Table 1 and SYBR Green PCR Master Mix (Applied Biosystems) as recommended by the manufacturer. GAPDH was used as a control. The quantitative reverse-transcription PCRs were performed in triplicate, and each experiment was repeated at least twice.
DNA binding motifs discovery
The details of this analysis are reported in the supplemental Methods. In brief, pattern enrichment was measured by the use of the classification relative error rate.38 De novo motifs were identified with DME,39 and de novo motif discovery was followed by module discovery by the use of motif anchoring.40 Potential BCL6 cofactors were inferred by identifying enriched TRANSFAC motifs.41
GEP, differential expression, and inferred networks
GEPs were generated as previously reported34 by use of the HG-U133Plus2.0 GeneChip and HG-U95A GeneChip platforms (Affymetrix). ARACNe was run with bootstrapping at a P value less than 10−4 threshold before correction for multiple testing. Gene set enrichment was calculated with the use of Gene Set Enrichment Analysis (GSEA)42 with t test–based P values for weighting statistics. See the supplemental Methods for details on the samples and the data analysis. The data discussed in this publication have been deposited in the Gene Expression Omnibus (GEO; National Center for Biotechnology Information) and are accessible through GEO Series accession numbers GSE2350 and GSE12195 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE2350 and http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE12195).
Results
Identification of promoter regions bound by BCL6 in GC B cells
Genome-wide array-based ChIP (ie, ChIP-on-chip) was performed on human tonsil GC B cells purified from 3 donors by the use of a commercially available array (Human Promoter ChIP-on-chip Microarray Set 2 × 244K; Agilent Technologies) with probes mapped to 18 625 promoters. CSA37 was used to assign localization P values for individual probes and to combine probe measurements across promoters and experiments. By the use of a false discovery rate cutoff of 0.0001, the use of CSA identified 4918 BCL6-bound promoters, corresponding to 4616 genes (supplemental Table 2). To confirm that the adopted threshold for significance allowed the inclusion of truly positive genes, 12 promoters were randomly chosen among the 100 lowest-ranking bound genes for validation by quantitative PCR on ChIP DNA obtained from GC B cells of different donors. All tested BCL6-bound regions but none of the control ChIP-on-chip-negative regions showed enrichment greater than 2-fold in at least 2 of the donors (supplemental Figure 2), confirming the low rate of false positives even among the less significantly bound promoters. In addition, qChIP was performed on 10 targets spacing the whole spectrum of CSA ranking. As expected, qChIP showed enrichment for all tested promoters (supplemental Figure 3). These results indicate that BCL6 is bound to more than 4000 (26%) of the tested promoter regions in normal GC B cells.
Characterization of BCL6 DNA binding motifs in vivo
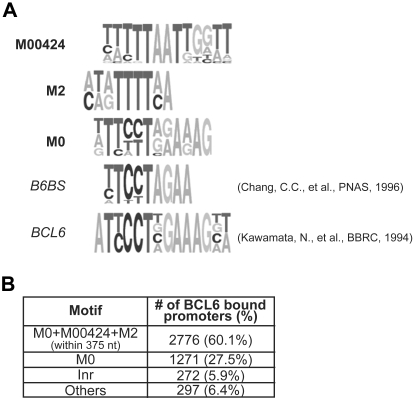
BCL6 DNA binding motifs have been identified on the basis of the binding of recombinant BCL6 to synthetic oligonucleotides in vitro.5,43 The identification of promoters bound by BCL6 in vivo allows the redefinition of its consensus binding motif in normal B cells under physiologic conditions. Toward this end, we analyzed the 4918 BCL6-bound genomic regions associated with 4616 genes for motifs enriched in BCL6-bound regions by using as a negative reference (background set) the 2087 unbound promoters (false discovery rate > 0.5, corresponding to 1937 genes) identified by CSA. Location within a  C
C phosphate
phosphate G
G (CpG) island emerged as the most predictive pattern for regions containing BCL6 binding sites (relative error rate of 0.39). To identify transcription factor binding site patterns that are predictive of BCL6 binding regardless of CpG islands, we searched for enriched motifs and motif combinations (modules) in regions with and without CpG islands. Modules had a stronger than expected predictive value for BCL6 binding compared with single motifs. The most significantly associated module (Figure 1) included (1) the TRANSFAC motif M00424; (2) the novel motif M2, which mimics a M00424 half site; and (3) the M0 motif, which is similar but not entirely compatible with previously identified BCL6 binding motifs in vitro (B6BS and BCL6; Figure 1A). The M00424 motif is a consensus sequence for the NKX-homeobox family of transcription factors, of which only NKX6-3 is expressed in GC B cells, on the basis of the GEP data. The M00424-M2-M0 module had sites in 60% (2776 of 4616) of the bound regions compared with only 30% (579 of 1937) of the unbound regions (P = 0 on the basis of a table contingency test). The statistical significance of this finding was further confirmed by permutation testing. In addition, although enrichment was evaluated only in terms of overrepresentation in the CSA-identified bound relative to the unbound regions, the presence of sites for the module correlated with CSA-based promoter ranking across the entire promoter set (supplemental Figure 4). The inclusion of additional motifs did not improve the classification significance for the module.
(CpG) island emerged as the most predictive pattern for regions containing BCL6 binding sites (relative error rate of 0.39). To identify transcription factor binding site patterns that are predictive of BCL6 binding regardless of CpG islands, we searched for enriched motifs and motif combinations (modules) in regions with and without CpG islands. Modules had a stronger than expected predictive value for BCL6 binding compared with single motifs. The most significantly associated module (Figure 1) included (1) the TRANSFAC motif M00424; (2) the novel motif M2, which mimics a M00424 half site; and (3) the M0 motif, which is similar but not entirely compatible with previously identified BCL6 binding motifs in vitro (B6BS and BCL6; Figure 1A). The M00424 motif is a consensus sequence for the NKX-homeobox family of transcription factors, of which only NKX6-3 is expressed in GC B cells, on the basis of the GEP data. The M00424-M2-M0 module had sites in 60% (2776 of 4616) of the bound regions compared with only 30% (579 of 1937) of the unbound regions (P = 0 on the basis of a table contingency test). The statistical significance of this finding was further confirmed by permutation testing. In addition, although enrichment was evaluated only in terms of overrepresentation in the CSA-identified bound relative to the unbound regions, the presence of sites for the module correlated with CSA-based promoter ranking across the entire promoter set (supplemental Figure 4). The inclusion of additional motifs did not improve the classification significance for the module.
Figure 1.
Motifs associated with BCL6 binding and their distribution across promoters. (A) DNA sequence analysis of BCL6 bound promoters as detected by ChIP-on-chip revealed enrichment for 3 motifs (M00424, M2, and M0), of which 1 (M0) resembles previously reported BCL6 binding sequences (B6BS, BCL6). (B) The module including M0, M00424, and M2 within 375 bp is found in the majority of BCL6-bound promoters, the M0 motif has sites in the promoters of the majority of the remaining genes, and almost half of the M0-free promoters display Inr elements. BCL6 is known to repress some of its targets upon interaction with Miz-1, which binds to Inr elements.
Most of the genes that did not have sites for the module M00424-M2-M0 in their promoters had sites for the M0 motif alone (Figure 1B), whereas the remaining BCL6-bound promoters were characterized either by the presence of Inr elements (∼ 6% of genes), which have been shown to recruit BCL6 via direct binding of the transcription factor Miz-1,31 or by other nonidentifiable motifs (6.4% of genes). Overall, M0 sites were found in approximately 88% of the bound genes. These results provide a refinement of the known BCL6 binding site, confirm the presence of BCL6 in Inr-driven promoters, and indicate the frequent association of BCL6 binding with other motifs.
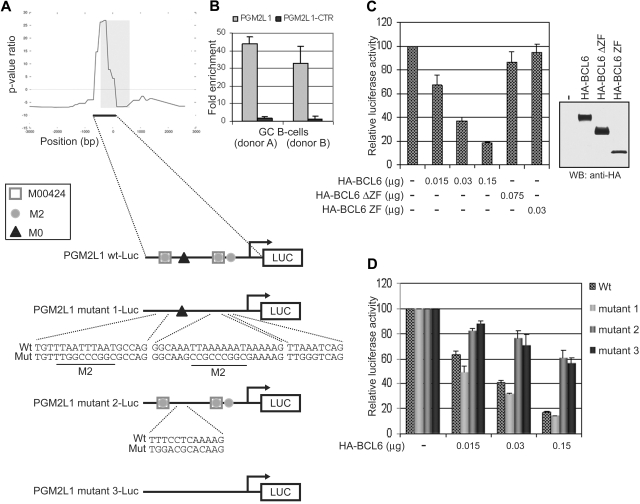
To establish the role of the main module (M00424-M2-M0) identified by ChIP-on-chip as predictive of BCL6 binding, we tested the responsiveness of a luciferase reporter construct driven by the promoter of the PGM2L1 gene (encoding a phosphoglucomutase), which contains the M00424-M2-M0 module. PGM2L1 was selected only because (1) it belonged to the core of BCL6 targets (see next subsection) identified by all 3 approaches (ChIP-on-chip, ARACNe, GEP), (2) it contained the 3 DNA binding motifs that needed to be tested, and (3) its promoter region was in a convenient size range for the experimental validation. The BCL6-bound region identified by ChIP-on-chip in the PGM2L1 promoter extends for approximately 700 bp upstream and 80 bp downstream of the initiation transcription site (Figure 2A). It lacks previously reported BCL6 binding sites but includes 1 M0, 2 M00424, and 3 M2 sites (2 of which partially overlap with M00424; Figure 2A). BCL6 binding in this region but not in a neighboring location (−2351/−2546) was confirmed by qChIP assay (Figure 2B). To examine the effect of BCL6 binding on the PGM2L1 promoter activity, we transiently cotransfected the luciferase reporter gene driven by the PGM2L1 native promoter into 293T cells along with a vector encoding wild-type BCL6 fused to an HA tag (HA-BCL6).
Figure 2.
The M0 site is necessary and sufficient for BCL6 repression on the PGM2L1 promoter. (A) Schematic representation of BCL6 binding to the PGM2L1 promoter as detected by ChIP-on-chip and of luciferase reporter constructs generated to assess the role of M0, M2, and M00424 sites. (B) BCL6 binding to PGM2L1 promoter was confirmed by qChIP assay in GC B cells obtained from 2 donors. A neighboring region (PGM2L1-CTR; −2351/−2546) tested as a control was not enriched. (C) A luciferase reporter construct driven by the PGM2L1 promoter was cotransfected in 293T cells with plasmids expressing HA-BCL6 or its mutants lacking the DNA binding domain (HA-BCL6ΔZF) or the transcriptional repressor domain (HA-BCL6 ZF). The PGM2L1 promoter-driven luciferase construct showed a dose-dependent decreased luciferase activity in the presence of HA-BCL6 but not of its mutants. The relative luciferase activities are displayed as average ± SD of 2 independent transfections. Immunoblotting protein detection of HA-BCL6 wild-type and mutants is displayed on the right. Results are representative of at least 3 independent experiments each performed in duplicate. (D) Disruption of M00424 and M2 sites in PGM2L1 promoter (mutant 1) did not affect repression by BCL6. Conversely, mutations in M0 (mutant 2) were linked to resistance to BCL6 repression, and further mutagenesis of all sites (mutant 3) did not increase the level of luciferase activities. The relative luciferase activities are displayed as average ± SD of 2 independent transfections. Results are representative of at least 3 independent experiments, each performed in duplicate.
To verify that repression by BCL6 is dependent on both its specific trans-repressive activity and DNA binding, HA-BCL6 mutants lacking the transcriptional repressor POZ and PEST domains (HA-BCL6-ZF) or the carboxyl-terminal DNA-binding Zinc-finger domain (HA-BCL6-ΔZF) were tested in parallel. Wild-type BCL6 but not its mutants was able to repress the expression of PGM2L1 reporter gene, indicating that PGM2L1 is a bona fide direct target of BCL6 (Figure 2C). To determine which component of the M00424-M2-M0 module is required for BCL6 function, we constructed PGM2L1 reporter genes with mutations in the M00424 and M2 motifs (PGM2L1 mutant1-luc), in the M0 motif only (PGM2L1 mutant2-luc), or in all 3 putative BCL6 binding correlated motifs (PGM2L1 mutant3-luc; Figure 2A). Transient transfection assays demonstrated that the mutant reporters lacking the M0 motif, including PGM2L1 mutant2-luc, which carries mutations only in M0, are resistant to transcriptional repression by BCL6 (Figure 2D), indicating that the M0 site but not M00424 and M2 is sufficient for BCL6-mediated transcriptional repression. Thus, the commonly associated M00424 and M2 motifs may represent sites for the binding of other transcription factors complementing the biologic activity of BCL6 or facilitating its transcriptional function by appropriately modifying chromatin.
Identification of BCL6-bound and regulated promoters in GC B cells
Because binding may not necessarily reflect BCL6 trans-repressive activity and/or may not involve genes expressed in B cells, the results of ChIP-on-chip analysis (promoter binding) were integrated with those obtained by the use of a reverse engineering algorithm (ARACNe) that identifies genes whose mRNA expression is directly correlated with BCL6 activity in a set of GEPs obtained from both normal B cells and a subset of tumors (FL) in which the BCL6 locus is not altered (supplemental Figure 5). ARACNe complements ChIP-on-chip by providing functional relationships but is limited because it also identifies upstream regulators of BCL6. The ARACNe-predicted BCL6 network includes 657 first neighbor genes, 48% of which were found to be bound by BCL6 in their promoters (supplemental Figures 5-6; supplemental Table 2).
Finally, the results of the ChIP-on-chip and ARACNe analyses were integrated with those obtained by the use of a GEP-based approach identifying genes displaying (1) low expression in GC B cells relative to purified naive and/or memory B cells, as evidence of possible repression by BCL6, or (2) lack of expression in any of the considered normal B-cell subpopulations but expression in a panel of B-NHL primary cases, as evidence of potential expression in B cells. In total we identified 3834 genes whose expression in GC B cells is consistent with BCL6 repression function (supplemental Figure 6; supplemental Table 2). The integration with ChIP-on-chip data identified 1207 genes bound by BCL6 and down-regulated or not expressed in GC B cells. The combined approach identified 120 “core” BCL6 target genes (supplemental Figure 6; supplemental Table 2) bound by BCL6, dynamically correlated with its expression, and physiologically repressed in GC B cells.
BCL6 “core” target genes identify cellular pathways controlled by BCL6
The 120 “core” target genes include only 1 previously reported validated BCL6 target (CCND2),24 a few additional ones (n = 9) for which BCL6 binding on their promoter was detected by ChIP-on-chip performed in a lymphoma cell line,25 and 33 similarly identified in GC B cells (see “Discussion”).33 Analysis of the BCL6 “core” targets revealed the presence of genes pointing to major pathways such as NF-κB activation (NFkBIE), MAPK signaling (MAP3K5), T cell–mediated B-cell activation (CD274), apoptosis (BCL2), response to genotoxic stress (SUB1), cytokine signaling (IL10RB), Toll-like receptor signaling (TLR7), transforming growth factor-β (TGF-β) signaling (TGFBR2), and the WNT pathway (FZD4). Nonetheless, some known targets were not present in the “core” list because they failed to reach the required threshold for differential expression (eg, TP53, CD80) or because they were excluded from the ARACNe-inferred BCL6 network as the result of an insufficient dynamic range in the expression data (eg, STAT3, PRDM1).
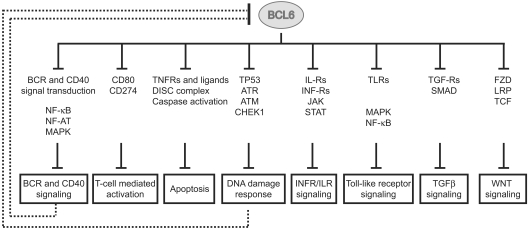
These results are consistent with the reported low false positive but potentially greater false negative rates among ARACNe-inferred targets of transcription factors. This observation, together with the fact that the number of “core” targets was too limited to allow pathway enrichment analysis (by the use of DAVID and databases from KEGG, Biocarta, and Panther),44,45 led us to explore the possibility that some of the genes present in the “core” list may reflect a broader activity of BCL6 on their respective pathways. Hence, we performed pathway enrichment analysis on the larger dataset obtained by crossing the ChIP-on-chip and the GEP data (1207 genes). The results identified an expanded set of targets involved in cellular signaling pathways at multiple levels, ranging from the cell surface (receptors), through signaling molecules, to the nucleus (supplemental Table 3). BCL6 appears to repress mostly positive but also some negative regulators of several signaling pathways, suggesting that it may play a complex role in modulating the activity of these pathways. Overall, the results (shown schematically in Figure 3 and detailed in supplemental Figure 7) can be summarized in the subsections that follow.
Figure 3.
BCL6 modulates multiple pathways in normal GC B cells. BCL6 represses multiple genes at different levels from the membrane surface (receptors), through signal transduction molecules, into the nucleus (transcription factors). Representative genes and gene families, found to be targeted by BCL6, are displayed for each pathway. Details on BCL6 targets and their pathways are reported in supplemental Figure 7. Several pathways (BCR and CD40 signaling; DNA damage response) have been previously reported to have a repressor function on BCL6, leading to its transcriptional down-regulation and protein degradation.
BCR and CD40 signaling.
These pathways were represented by several genes encoding molecules involved in signal transduction (eg, the tyrosine kinase LYN and the tyrosine phosphatase PTP6), MAPK activation (supplemental Figure 7), NF-AT activation (PPP3CA and PPP3R1), and NF-κB activation (NFKB1, NFKB2, and several IκB family members). Because some of these signaling pathways (MAPK, NF-κB) have been previously shown to regulate BCL6,11–13 these findings suggest an automodulatory circuit in which BCL6 regulates its own regulatory signals.
T cell–mediated B-cell activation.
CD274 (alias B7-H1, PDL1) has been shown to bind CD80 and to regulate the balance of activation and inhibition of the T-cell response.46 Because CD80 is also a BCL6 direct target gene (not represented in the dataset because of its poor dynamic range),27 the inclusion of CD274 suggests a broad role for BCL6 in controlling the expression of molecules essential for B-T–cell interaction.
Apoptosis.
BCL2 was prominently represented in the list of “core” targets, consistent with evidence that BCL6 binds to its promoter via Miz-1 and suppresses its expression in the GC.47 In addition, newly identified BCL6 targets included molecules associated with both positive and negative regulation of the DISC complex and caspase activation (Figure 3; supplemental Figure 7), suggesting a fine-tuning role for BCL6 in balancing pro- and antiapoptotic programs in GC B cells.
Response to DNA damage.
The role of BCL6 in modulating the sensing and execution of responses to DNA damage has been previously suggested by the discovery of TP53, CDKN1A, ATR, and CHEK1 as BCL6 direct targets.28–31 The results herein extend this notion by identifying an additional set of genes in the same pathway, including TP53BP1, a conserved checkpoint protein that enhances TP53 transactivation function48; SUB1, an activator of TP53 that is overexpressed upon DNA damage49,50; USP3, a H2A/H2B deubiquitinating enzyme involved in maintenance of genome integrity51; TLK1, a nuclear serine/threonine kinase acting as a molecular chaperone in DNA repair52; CHD2, a chromodomain helicase binding protein affecting DNA damage signaling and processing at the chromatin level53; and several not yet functionally characterized TP53 transcriptional targets (TOR1AIP1, STX6, IFI16, IRF2BP2).54–57
Interferon and cytokine signaling.
Multiple interferon-type (IFNAR1, IFNAR2, IFNGR1) and interleukin receptors (IL-10RA, IL-10RB, IL-12RB1, IL-13RA1, IL-2RG, IL-6R, IL-7R) that lead to activation of Janus kinase/signal transducers and activator of transcription (JAK/STAT) were broadly represented among BCL6 targets. Furthermore, STAT family members (STAT1, STAT3, and STAT5A) also were found to be directly repressed by BCL6. Together with the cited modulatory activity on multiple MAPK components and on components (JUN/FOSB) of the AP1 transcriptional complex, these findings point to a broad function of BCL6 in modulating incoming signals from a variety of soluble signals that may prematurely activate centroblasts in the GC.
Toll-like receptor signaling.
A modulatory activity on this signaling pathway is suggested by the presence among BCL6 target genes of those encoding both Toll-like receptors (TLR1, TLR7, TLR9), and transducers of the Toll-derived signals (MYD88 and TIRAP). Because this pathway has been reported to have a role in T-dependent immune response and the development of memory B cells,58,59 our findings suggest that its silencing by BCL6 may also be necessary to avoid activating stimuli during the proliferative stage of GC reaction.
TGFβ receptor signaling.
Previous studies in TGFβ−/− mice showed the role of TGF-β in promoting the differentiation of IgA-secreting plasma cells and in attenuating B-cell response to low-affinity antigens,60 suggesting a role for TGF-β signaling in regulating post-GC differentiation. Accordingly, the results herein identify genes encoding TGF-β–type receptors (TGFBR2, BMPR2), a ligand (BMP2), and a nuclear effector (SMAD7) as BCL6 direct targets, suggesting a modulatory action of BCL6 on the ability of TGF-β to regulate post-GC differentiation.
WNT signaling.
This pathway appears also to be affected by BCL6 through the control of genes encoding its receptors (FZD4, FZD8, LRP6), signal transducers (AXIN1), and downstream transcription factors (TCF7L2, LEF1). In conjunction with the report that mice bearing B cell–specific deletion of β-catenin show defective plasma cell formation in vitro,61 these results suggest a role of WNT signaling in the late stage of B-cell differentiation and support the silencing of this pathway by BCL6 in the early stage of the GC reaction.
BCL6 is required for target gene suppression
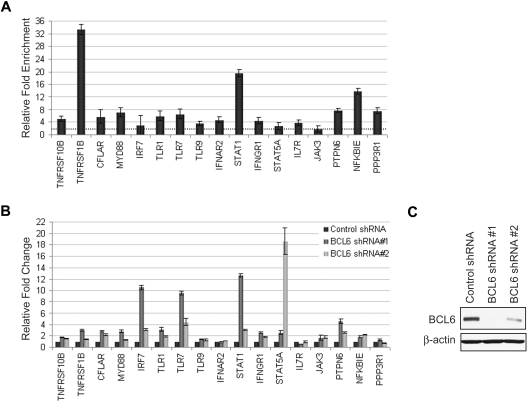
To provide at least a partial functional validation of the results, 17 BCL6 targets, chosen to be representative of the aforementioned pathways, were further experimentally investigated. Because normal GC B cells undergo apoptosis within hours of ex vivo culture and therefore cannot be experimentally manipulated in vitro, we relied on the use of a Burkitt lymphoma cell line (P3HR1) to asses whether BCL6 inactivation leads to target gene expression, as expected for direct targets of transcriptional suppression. First, we verified by qChIP assay that BCL6 binding identified in normal GC B cells (supplemental Figure 8) was retained in this cell line (Figure 4A). Then, different levels of BCL6 silencing were obtained in P3HR1 cells by the use of 2 sh-RNAs targeting BCL6 (Figure 4C). Reverse-transcription PCR-based analysis of target gene expression (Figure 4B) demonstrated that 11 of 17 of the tested genes are up-regulated at least 2-fold upon BCL6 silencing, suggesting that BCL6 is indeed required for their repression. The lack of up-regulation of a minority of these genes is not surprising because the removal of the repressor may not be enough to induce expression of these genes in absence of activators. These results indicate that the subset of BCL6 target genes tested as representative of relevant cellular pathways is in fact under active repression by BCL6.
Figure 4.
BCL6 targets involved in multiple pathways are responsive to BCL6 silencing. (A) Seventeen targets representative of the major pathways affected by BCL6 were analyzed in the P3HR1 cell line. The BCL6-bound promoter region identified by ChIP-on-chip in GC B cells was tested for direct binding in P3HR1 by qChIP. All promoters, except JAK3, showed enrichment of at least 2-fold in the BCL6 immunoprecipitated chromatin fraction. (B) BCL6 silencing was obtained by the use of 2 shRNAs delivered by lentiviral infection in P3HR1. Quantitative reverse-transcription PCR analysis showed that the expression of 11 of 17 targets increased more than 2-fold upon BCL6 silencing by at least one of the shRNA. The target induction appeared consistent with the level of BCL6 silencing in all cases, except STAT5A. (C) BCL6 protein expression as detected by immunoblotting upon infection with control and BCL6 shRNAs.
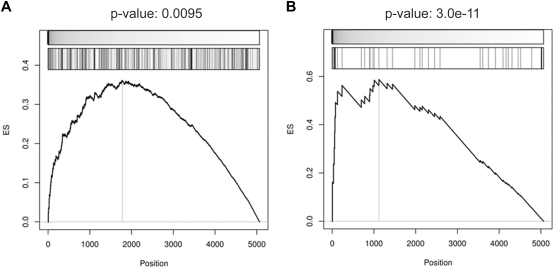
Moreover, provided that a considerable fraction of targets appeared to be responsive to BCL6 removal, we extended the analysis to the complete set of identified BCL6 targets. GEPs were generated from Ramos cells infected with control or BCL6-targeting sh-RNAs and used to investigate the enrichment of the BCL6 targets in the gene signature obtained upon BCL6 silencing. GSEA showed that both the core targets and the targets identified by ChIP-on-chip and GEP were significantly enriched among the genes whose expression increases upon BCL6 silencing (Figure 5). In conclusion, these results suggest that a significant fraction of the reported targets are actively responding to BCL6 repression.
Figure 5.
BCL6-silencing gene signature is significantly enriched in BCL6 target genes. Gene set enrichment according to up-regulation in response to BCL6 silencing was measured for (A) the targets identified as BCL6-anti-coexpressed genes with BCL6-bound promoters according to ChIP-on-chip analysis and (B) the subset of targets that is also inferred by ARACNe. GSEA computed the significance of up-regulation in BCL6-silencing versus control experiments by t test (top meter in each panel) for each target set (second meter). Leading edge analysis was used to predict responsive BCL6 targets (left of vertical line in each panel) and enrichment score (ES) P values were estimated by the use of extrapolation over 100 000 sample-permutation tests.
Discussion
In this study we aimed at elucidating the biologic function of BCL6 by providing a comprehensive analysis of its target genes in the GC. On the assumption, supported by previous studies on other transcription factors, that direct binding on a promoter sequence does not necessarily reflect a functional relationship between a given transcription factor and a target gene,26 we have adopted an integrated biochemical and computational approach that allowed the identification of genes whose promoter is directly bound and repressed by BCL6 in a physiologic GC context. The results identified a set of strictly defined biochemical and functional BCL6 targets that have implications for the understanding of BCL6 activity as transcription factor, of its biologic function, and of its role as an oncogene in lymphomagenesis.
BCL6 binding sites in the genome: binding versus function
The more than 4000 target promoters reported here significantly extend the recent report of less than 2000 BCL6-bound promoters,33 935 of which also were found in our study. This difference is likely the result of the different promoter array used and to the far more sensitive ChIP-on-chip analysis method (CSA) used in our study. The large number of BCL6-bound promoters reported in this study is not an overestimation, as shown by the limited number of false positives detected by qChIP validation even on the promoters identified by CSA at the lowest confidence levels (supplemental Figure 2). Overall, our results suggest that BCL6 binds to a relatively large number of promoters far exceeding the number of genes that are functionally controlled by BCL6 in GC. The observed differences between binding and functional activity is analogous to the one observed for other transcription factors.26 It probably reflects both the requirement of additional cofactors for transcriptional activity as well as the fact that many BCL6-bound promoters may be active in tissues other than GC B cells.
Our findings provide a refinement of the previously in vitro–identified binding sites5,43 and indicate the frequent presence of this site in a complex module (M00424-M2-M0) whose role requires further functional analysis. Furthermore, the data confirmed the possibility for BCL6 to bind to selected promoters characterized by lack of BCL6 DNA binding sites but containing Inr motifs, most likely reflecting the presence of BCL6 bound to Miz-1, as previously shown for the CDKN1A promoter.31 Finally, a minority of BCL6-bound promoters did not display any readily identifiable motif, suggesting that BCL6 may bind to these promoters via alternative, currently unknown, cofactors.
The database of BCL6-bound regions reported here represents a resource for the DNA binding analysis of cofactors associated with BCL6 activity. In fact, several DNA binding motifs were found significantly associated with BCL6 binding sites, including those corresponding to important regulators of GC development (ie, MYB, IRF8)62,63 as well as known competitors of BCL6 functions (IRF4, STATs)13,64–66 (supplemental Table 4). In addition, the ARACNe-predicted hubs for several transcription factors known to compete with BCL6 function (IRF4, STATs, TP53) were significantly enriched in BCL6-bound targets, suggesting that BCL6 has a broad control on a large portion of their transcriptional program (supplemental Table 5). The establishment of the physiologic significance of these associations will require extensive functional validation in vitro as well as in normal tissues.
Implications for the BCL6 biologic program
The authors of previous studies have attempted to identify BCL6 biologic function either by analysis of the transcriptional response to BCL6 induction by GEP24 or through genome-wide analysis of promoters bound by BCL6 in malignant GC B cells.25 A study33 published while this current article was in preparation reported the identification of physiologic BCL6 targets in normal GC B cells with a combined biochemical and GEP analysis. However, down-regulation in GC was shown only for a limited number of genes (n = 187) of the 1970 identified to be bound by BCL6. Thus, the data here reported greatly enlarge the core of BCL6-bound genes validated for their physiologic down-regulation in GC (n = 1207). In addition, GSEA demonstrated that this set of BCL6 targets is significantly more enriched for genes that are derepressed in BCL6-depleted B cells compared with targets reported by Ci et al33 (supplemental Figure 9). The larger number of functional targets reported here was instrumental toward the reconstruction of the multiple pathways affected by BCL6.
Indeed, our results confirmed the activity of BCL6 in modulating B-cell activation and differentiation and significantly extended the role of BCL6 in controlling DNA damage sensing and response via the identification of several additional genes regulated by BCL6 in the same pathway. The latter activity appears to be functionally coordinated with the broad control that BCL6 has on the apoptotic machinery in which it affects multiple genes encoding both pro- and antiapoptotic molecules. More interestingly, these results provide novel insights concerning the function of BCL6 on several biologically relevant cellular pathways by modulating signaling through toll-like receptors, INF-R, a variety of cytokines, TGF-R, and WNT signaling. An intriguing feature of BCL6 function appears to be the broad control of several targets along the same pathway, often involving the simultaneous modulation of the expression of cell surface receptors, signaling molecules, and nuclear effectors.
When examined collectively, these results indicate a complex but remarkably coherent biologic function of BCL6 in GC B cells. In fact, BCL6 appears to modulate a relatively large number of signals that are involved in B-cell activation, survival, and/or differentiation at the end of the GC reaction. This finding is consistent with the role of BCL6 in protecting proliferating centroblasts from signals that may prematurely lead to their GC exit before they have expanded and undergone SHM of their Ig genes. Once these functions have been completed, the high-affinity recognition of the antigen as well as the interaction with T cells (CD40 signaling) down-regulates BCL6 expression,11–13 thus releasing its negative control on the signals that regulate post-GC differentiation. The balance between BCL6-mediated suppression and the activity of multiple signaling pathways seems to be finely tuned by circuits in which BCL6 regulates components of its own regulatory pathways (suppression of CD40 and BCR signaling and the DNA damage response).
Implications for lymphomagenesis
BCL6 is expressed in all GC-derived malignancies, including Burkitt lymphoma, FL, DLBCL, and a subset of Hodgkin lymphoma. In a significant fraction of DLBCL and a small fraction of FL, BCL6 expression is deregulated by chromosomal translocation and somatic mutations.13,16–18,22 The set of physiologic target genes identified in this study will allow comparative analyses aimed at defining whether the BCL6 transcriptional program remains intact in B-cell lymphomas carrying a normal or deregulated BCL6 gene. In addition, the results herein suggest that all lymphoma subtypes expressing BCL6 may be relatively insensitive to a variety of activation and differentiation stimuli. This notion suggests a potential synergic therapeutic activity between molecules aimed at inactivation of BCL6 and biologic agents leading to B-cell activation and differentiation.
Supplementary Material
Acknowledgments
This work was supported by National Cancer Institute grants 5PO1 CA092625-09 and 5R37 CA37295-26 to R.D.-F. and by National Institute of Allergy and Infectious Diseases (R01AI066116) and National Cancer Institute (R01CA109755) grants and by the National Centers for Biomedical Computing National Institutes of Health Roadmap Initiative (U54CA121852) to A.C. C.S. is supported by a Fellowship of the Mildred Scheel Stiftung für Krebsforschung.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: K.B. and M.S. designed and performed the experimental assays and interpreted the results; P.S. performed DNA binding motif discovery and contributed to writing the manuscript; A.A.M. developed the CSA method; K.W. and W.-K. L. generated the ARACNe network; Y.K. and C.S. purified GC B cells; M.J.A. performed GSEA; P.S. and A.A.M. performed ChIP-on-chip and GEP data analysis; K.B., M.S., A.C., and R.D.-F. wrote the manuscript; and A.C. and R.D.-F. contributed to design the experiments, interpreted the results, and edited the final manuscript.
The current address for M.S. is Division of Viral Immunology, Center for AIDS Research, Kumamoto University, Kumamoto, Japan.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Riccardo Dalla-Favera, MD, Columbia University, 1130 St Nicholas Ave, New York, NY 10032; e-mail: rd10@columbia.edu.
References
- 1.Klein U, Dalla-Favera R. Germinal centres: role in B-cell physiology and malignancy. Nat Rev Immunol. 2008;8(1):22–33. doi: 10.1038/nri2217. [DOI] [PubMed] [Google Scholar]
- 2.Dent AL, Shaffer AL, Yu X, Allman D, Staudt LM. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science. 1997;276(5312):589–592. doi: 10.1126/science.276.5312.589. [DOI] [PubMed] [Google Scholar]
- 3.Ye BH, Rao PH, Chaganti RS, Dalla-Favera R. Cloning of bcl-6, the locus involved in chromosome translocations affecting band 3q27 in B-cell lymphoma. Cancer Res. 1993;53(12):2732–2735. [PubMed] [Google Scholar]
- 4.Ye BH, Cattoretti G, Shen Q, et al. The BCL-6 proto-oncogene controls germinal-centre formation and Th2-type inflammation. Nat Genet. 1997;16(2):161–170. doi: 10.1038/ng0697-161. [DOI] [PubMed] [Google Scholar]
- 5.Chang CC, Ye BH, Chaganti RS, Dalla-Favera R. BCL-6, a POZ/zinc-finger protein, is a sequence-specific transcriptional repressor. Proc Natl Acad Sci U S A. 1996;93(14):6947–6952. doi: 10.1073/pnas.93.14.6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dhordain P, Albagli O, Lin RJ, et al. Corepressor SMRT binds the BTB/POZ repressing domain of the LAZ3/BCL6 oncoprotein. Proc Natl Acad Sci U S A. 1997;94(20):10762–10767. doi: 10.1073/pnas.94.20.10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dhordain P, Lin RJ, Quief S, et al. The LAZ3(BCL-6) oncoprotein recruits a SMRT/mSIN3A/histone deacetylase containing complex to mediate transcriptional repression. Nucleic Acids Res. 1998;26(20):4645–4651. doi: 10.1093/nar/26.20.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fujita N, Jaye DL, Geigerman C, et al. MTA3 and the Mi-2/NuRD complex regulate cell fate during B lymphocyte differentiation. Cell. 2004;119(1):75–86. doi: 10.1016/j.cell.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 9.Huynh KD, Bardwell VJ. The BCL-6 POZ domain and other POZ domains interact with the co-repressors N-CoR and SMRT. Oncogene. 1998;17(19):2473–2484. doi: 10.1038/sj.onc.1202197. [DOI] [PubMed] [Google Scholar]
- 10.Cattoretti G, Chang CC, Cechova K, et al. BCL-6 protein is expressed in germinal-center B cells. Blood. 1995;86(1):45–53. [PubMed] [Google Scholar]
- 11.Niu H, Ye BH, Dalla-Favera R. Antigen receptor signaling induces MAP kinase-mediated phosphorylation and degradation of the BCL-6 transcription factor. Genes Dev. 1998;12(13):1953–1961. doi: 10.1101/gad.12.13.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Basso K, Klein U, Niu H, et al. Tracking CD40 signaling during germinal center development. Blood. 2004;104(13):4088–4096. doi: 10.1182/blood-2003-12-4291. [DOI] [PubMed] [Google Scholar]
- 13.Saito M, Gao J, Basso K, et al. A signaling pathway mediating downregulation of BCL6 in germinal center B cells is blocked by BCL6 gene alterations in B-cell lymphoma. Cancer Cell. 2007;12(3):280–292. doi: 10.1016/j.ccr.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 14.Phan RT, Saito M, Kitagawa Y, Means AR, Dalla-Favera R. Genotoxic stress regulates expression of the proto-oncogene Bcl6 in germinal center B cells. Nat Immunol. 2007;8(10):1132–1139. doi: 10.1038/ni1508. [DOI] [PubMed] [Google Scholar]
- 15.Bereshchenko OR, Gu W, Dalla-Favera R. Acetylation inactivates the transcriptional repressor BCL6. Nat Genet. 2002;32(4):606–613. doi: 10.1038/ng1018. [DOI] [PubMed] [Google Scholar]
- 16.Lo Coco F, Ye BH, Lista F, et al. Rearrangements of the BCL6 gene in diffuse large cell non-Hodgkin's lymphoma. Blood. 1994;83(7):1757–1759. [PubMed] [Google Scholar]
- 17.Butler MP, Iida S, Capello D, et al. Alternative translocation breakpoint cluster region 5′ to BCL-6 in B-cell non-Hodgkin's lymphoma. Cancer Res. 2002;62(14):4089–4094. [PubMed] [Google Scholar]
- 18.Ye BH, Chaganti S, Chang CC, et al. Chromosomal translocations cause deregulated BCL6 expression by promoter substitution in B-cell lymphoma. EMBO J. 1995;14(24):6209–6217. doi: 10.1002/j.1460-2075.1995.tb00311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen W, Iida S, Louie DC, Dalla-Favera R, Chaganti RS. Heterologous promoters fused to BCL6 by chromosomal translocations affecting band 3q27 cause its deregulated expression during B-cell differentiation. Blood. 1998;91(2):603–607. [PubMed] [Google Scholar]
- 20.Shen HM, Peters A, Baron B, Zhu X, Storb U. Mutation of BCL-6 gene in normal B cells by the process of somatic hypermutation of Ig genes. Science. 1998;280(5370):1750–1752. doi: 10.1126/science.280.5370.1750. [DOI] [PubMed] [Google Scholar]
- 21.Pasqualucci L, Migliazza A, Fracchiolla N, et al. BCL-6 mutations in normal germinal center B cells: evidence of somatic hypermutation acting outside Ig loci. Proc Natl Acad Sci U S A. 1998;95(20):11816–11821. doi: 10.1073/pnas.95.20.11816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pasqualucci L, Migliazza A, Basso K, Houldsworth J, Chaganti RS, Dalla-Favera R. Mutations of the BCL6 proto-oncogene disrupt its negative autoregulation in diffuse large B-cell lymphoma. Blood. 2003;101(8):2914–2923. doi: 10.1182/blood-2002-11-3387. [DOI] [PubMed] [Google Scholar]
- 23.Cattoretti G, Pasqualucci L, Ballon G, et al. Deregulated BCL6 expression recapitulates the pathogenesis of human diffuse large B-cell lymphomas in mice. Cancer Cell. 2005;7(5):445–455. doi: 10.1016/j.ccr.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 24.Shaffer AL, Yu X, He Y, Boldrick J, Chan EP, Staudt LM. BCL-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity. 2000;13(2):199–212. doi: 10.1016/s1074-7613(00)00020-0. [DOI] [PubMed] [Google Scholar]
- 25.Polo JM, Juszczynski P, Monti S, et al. Transcriptional signature with differential expression of BCL6 target genes accurately identifies BCL6-dependent diffuse large B-cell lymphomas. Proc Natl Acad Sci U S A. 2007;104(9):3207–3212. doi: 10.1073/pnas.0611399104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fernandez PC, Frank SR, Wang L, et al. Genomic targets of the human c-Myc protein. Genes Dev. 2003;17(9):1115–1129. doi: 10.1101/gad.1067003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niu H, Cattoretti G, Dalla-Favera R. BCL6 controls the expression of the B7-1/CD80 costimulatory receptor in germinal center B cells. J Exp Med. 2003;198(2):211–221. doi: 10.1084/jem.20021395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Phan RT, Dalla-Favera R. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature. 2004;432(7017):635–639. doi: 10.1038/nature03147. [DOI] [PubMed] [Google Scholar]
- 29.Ranuncolo SM, Polo JM, Dierov J, et al. Bcl-6 mediates the germinal center B-cell phenotype and lymphomagenesis through transcriptional repression of the DNA-damage sensor ATR. Nat Immunol. 2007;8(7):705–714. doi: 10.1038/ni1478. [DOI] [PubMed] [Google Scholar]
- 30.Ranuncolo SM, Polo JM, Melnick A. BCL6 represses CHEK1 and suppresses DNA damage pathways in normal and malignant B cells. Blood Cells Mol Dis. 2008;41(1):95–99. doi: 10.1016/j.bcmd.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Phan RT, Saito M, Basso K, Niu H, Dalla-Favera R. BCL6 interacts with the transcription factor Miz-1 to suppress the cyclin-dependent kinase inhibitor p21 and cell cycle arrest in germinal center B cells. Nat Immunol. 2005;6(10):1054–1060. doi: 10.1038/ni1245. [DOI] [PubMed] [Google Scholar]
- 32.Tunyaplin C, Shaffer AL, Angelin-Duclos CD, Yu X, Staudt LM, Calame KL. Direct repression of prdm1 by Bcl-6 inhibits plasmacytic differentiation. J Immunol. 2004;173(2):1158–1165. doi: 10.4049/jimmunol.173.2.1158. [DOI] [PubMed] [Google Scholar]
- 33.Ci W, Polo JM, Cerchietti L, et al. The BCL6 transcriptional program features repression of multiple oncogenes in primary B cells and is deregulated in DLBCL. Blood. 2009;113(22):5536–5548. doi: 10.1182/blood-2008-12-193037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Basso K, Margolin AA, Stolovitzky G, Klein U, Dalla-Favera R, Califano A. Reverse engineering of regulatory networks in human B cells. Nat Genet. 2005;37(4):382–390. doi: 10.1038/ng1532. [DOI] [PubMed] [Google Scholar]
- 35.Margolin AA, Nemenman I, Basso K, et al. ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics. 2006;7(suppl 1):S7. doi: 10.1186/1471-2105-7-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klein U, Tu Y, Stolovitzky GA, et al. Transcriptional analysis of the B-cell germinal center reaction. Proc Natl Acad Sci U S A. 2003;100(5):2639–2644. doi: 10.1073/pnas.0437996100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Margolin AA, Palomero T, Sumazin P, Califano A, Ferrando AA, Stolovitzky G. ChIP-on-chip significance analysis reveals large-scale binding and regulation by human transcription factor oncogenes. Proc Natl Acad Sci U S A. 2009;106(1):244–249. doi: 10.1073/pnas.0806445106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith AD, Sumazin P, Zhang MQ. Tissue-specific regulatory elements in mammalian promoters. Mol Syst Biol. 2007;3:73. doi: 10.1038/msb4100114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith AD, Sumazin P, Zhang MQ. Identifying tissue-selective transcription factor binding sites in vertebrate promoters. Proc Natl Acad Sci U S A. 2005;102(5):1560–1565. doi: 10.1073/pnas.0406123102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith AD, Sumazin P, Das D, Zhang MQ. Mining ChIP-chip data for transcription factor and cofactor binding sites. Bioinformatics. 2005;21(suppl 1):i403–412. doi: 10.1093/bioinformatics/bti1043. [DOI] [PubMed] [Google Scholar]
- 41.Matys V, Fricke E, Geffers R, et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31(1):374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kawamata N, Miki T, Ohashi K, et al. Recognition DNA sequence of a novel putative transcription factor, BCL6. Biochem Biophys Res Commun. 1994;204(1):366–374. doi: 10.1006/bbrc.1994.2468. [DOI] [PubMed] [Google Scholar]
- 44.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 45.Dennis G, Jr, Sherman BT, Hosack DA, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4(5):P3. [PubMed] [Google Scholar]
- 46.Butte MJ, Keir ME, Phamduy TB, Sharpe AH, Freeman GJ. Programmed death-1 ligand 1 interacts specifically with the B7-1 costimulatory molecule to inhibit T-cell responses. Immunity. 2007;27(1):111–122. doi: 10.1016/j.immuni.2007.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saito M, Novak U, Piovan E, et al. BCL6 suppression of BCL2 via Miz1 and its disruption in diffuse large B-cell lymphoma. Proc Natl Acad Sci U S A. 2009;106(27):11294–11299. doi: 10.1073/pnas.0903854106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang B, Matsuoka S, Carpenter PB, Elledge SJ. 53BP1, a mediator of the DNA damage checkpoint. Science. 2002;298(5597):1435–1438. doi: 10.1126/science.1076182. [DOI] [PubMed] [Google Scholar]
- 49.Banerjee S, Kumar BR, Kundu TK. General transcriptional coactivator PC4 activates p53 function. Mol Cell Biol. 2004;24(5):2052–2062. doi: 10.1128/MCB.24.5.2052-2062.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kishore AH, Batta K, Das C, Agarwal S, Kundu TK. p53 regulates its own activator: transcriptional co-activator PC4, a new p53-responsive gene. Biochem J. 2007;406(3):437–444. doi: 10.1042/BJ20070390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nicassio F, Corrado N, Vissers JH, et al. Human USP3 is a chromatin modifier required for S phase progression and genome stability. Curr Biol. 2007;17(22):1972–1977. doi: 10.1016/j.cub.2007.10.034. [DOI] [PubMed] [Google Scholar]
- 52.Sunavala-Dossabhoy G, De Benedetti A. Tousled homolog, TLK1, binds and phosphorylates Rad9; TLK1 acts as a molecular chaperone in DNA repair. DNA Repair (Amst) 2009;8(1):87–102. doi: 10.1016/j.dnarep.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 53.Nagarajan P, Onami TM, Rajagopalan S, Kania S, Donnell R, Venkatachalam S. Role of chromodomain helicase DNA-binding protein 2 in DNA damage response signaling and tumorigenesis. Oncogene. 2009;28(8):1053–1062. doi: 10.1038/onc.2008.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koeppel M, van Heeringen SJ, Smeenk L, Navis AC, Janssen-Megens EM, Lohrum M. The novel p53 target gene IRF2BP2 participates in cell survival during the p53 stress response. Nucleic Acids Res. 2009;37(2):322–335. doi: 10.1093/nar/gkn940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krieg AJ, Hammond EM, Giaccia AJ. Functional analysis of p53 binding under differential stresses. Mol Cell Biol. 2006;26(19):7030–7045. doi: 10.1128/MCB.00322-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Song LL, Alimirah F, Panchanathan R, Xin H, Choubey D. Expression of an IFN-inducible cellular senescence gene, IFI16, is up-regulated by p53. Mol Cancer Res. 2008;6(11):1732–1741. doi: 10.1158/1541-7786.MCR-08-0208. [DOI] [PubMed] [Google Scholar]
- 57.Zhang Y, Shu L, Chen X. Syntaxin 6, a regulator of the protein trafficking machinery and a target of the p53 family, is required for cell adhesion and survival. J Biol Chem. 2008;283(45):30689–30698. doi: 10.1074/jbc.M801711200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pasare C, Medzhitov R. Control of B-cell responses by Toll-like receptors. Nature. 2005;438(7066):364–368. doi: 10.1038/nature04267. [DOI] [PubMed] [Google Scholar]
- 59.Meyer-Bahlburg A, Khim S, Rawlings DJ. B cell intrinsic TLR signals amplify but are not required for humoral immunity. J Exp Med. 2007;204(13):3095–3101. doi: 10.1084/jem.20071250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cazac BB, Roes J. TGF-beta receptor controls B-cell responsiveness and induction of IgA in vivo. Immunity. 2000;13(4):443–451. doi: 10.1016/s1074-7613(00)00044-3. [DOI] [PubMed] [Google Scholar]
- 61.Yu Q, Quinn WJ, 3rd, Salay T, Crowley JE, Cancro MP, Sen JM. Role of beta-catenin in B cell development and function. J Immunol. 2008;181(6):3777–3783. doi: 10.4049/jimmunol.181.6.3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thomas MD, Kremer CS, Ravichandran KS, Rajewsky K, Bender TP. c-Myb is critical for B-cell development and maintenance of follicular B cells. Immunity. 2005;23(3):275–286. doi: 10.1016/j.immuni.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 63.Lee CH, Melchers M, Wang H, et al. Regulation of the germinal center gene program by interferon (IFN) regulatory factor 8/IFN consensus sequence-binding protein. J Exp Med. 2006;203(1):63–72. doi: 10.1084/jem.20051450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Klein U, Casola S, Cattoretti G, et al. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat Immunol. 2006;7(7):773–782. doi: 10.1038/ni1357. [DOI] [PubMed] [Google Scholar]
- 65.Walker SR, Nelson EA, Frank DA. STAT5 represses BCL6 expression by binding to a regulatory region frequently mutated in lymphomas. Oncogene. 2007;26(2):224–233. doi: 10.1038/sj.onc.1209775. [DOI] [PubMed] [Google Scholar]
- 66.Diehl SA, Schmidlin H, Nagasawa M, et al. STAT3-mediated up-regulation of BLIMP1 Is coordinated with BCL6 down-regulation to control human plasma cell differentiation. J Immunol. 2008;180(7):4805–4815. doi: 10.4049/jimmunol.180.7.4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.