Abstract
Background
Inorganic polyphosphate (polyP), a polymer of tens or hundreds of phosphate residues linked by ATP-like bonds, is found in all organisms and performs a wide variety of functions. PolyP is synthesized in bacterial cells by the actions of polyphosphate kinases (PPK1 and PPK2) and degraded by exopolyphosphatase (PPX). Bacterial cells with polyP deficiencies due to knocking out the ppk1 gene are affected in many structural and important cellular functions such as motility, quorum sensing, biofilm formation and virulence among others. The cause of this pleiotropy is not entirely understood.
Results
The overexpression of exopolyphosphatase in bacteria mimicked some pleitropic defects found in ppk1 mutants. By using this approach we found new structural and functional defects in the polyP-accumulating bacteria Pseudomonas sp. B4, which are most likely due to differences in the polyP-removal strategy. Colony morphology phenotype, lipopolysaccharide (LPS) structure changes and cellular division malfunction were observed. Finally, we used comparative proteomics in order to elucidate the cellular adjustments that occurred during polyP deficiency in this bacterium and found some clues that helped to understand the structural and functional defects observed.
Conclusions
The results obtained suggest that during polyP deficiency energy metabolism and particularly nucleoside triphosphate (NTP) formation were affected and that bacterial cells overcame this problem by increasing the flux of energy-generating metabolic pathways such as tricarboxilic acid (TCA) cycle, β-oxidation and oxidative phosphorylation and by reducing energy-consuming ones such as active transporters and amino acid biosynthesis. Furthermore, our results suggest that a general stress response also took place in the cell during polyP deficiency.
Background
Polyphosphate (polyP) is a ubiquitous linear polymer of hundreds of orthophosphate residues (Pi) linked by phosphoanhydride bonds. PolyP has been found in all tree domains of life (Archaea, Bacteria and Eukarya). In bacteria, the main enzymes involved in the metabolism of polyP are the polyphosphate kinases (PPK1 and PPK2) that catalyze the reversible conversion of the terminal phosphate of ATP (or GTP) into polyP and the exopolyphosphatase (PPX) that processively hydrolyzes the terminal residues of polyP to liberate Pi [1,2].
PolyP is a reservoir of phosphate and, as in ATP, of high-energy phosphate bonds. Furthermore, biochemical experiments and studies with ppk1 mutants in many bacteria have indicated additional roles for polyP. These include inhibition of RNA degradation [3], activation of Lon protease during stringent response [4,5], involvement in membrane channel structure [6,7], and contribution to the resistance to stress generated by heat, oxidants, osmotic challenge, antibiotics and UV [8-12]. Particularly, a ppk1 mutant of Pseudomonas aeruginosa PAO1 was impaired in motility, biofilm development, quorum sensing and virulence [13-15].
In addition to PPK1, another widely conserved polyP enzyme is PPK2 [16,17]. In contrast to the ATP-dependent polyP synthetic activity of PPK1, PPK2 preferentially catalyses the polyP-driven synthesis of GTP from GDP. Orthologs to both proteins have been found in many bacterial genomes and curiously there are many bacteria with orthologs of either PPK1 or PPK2, or both, or neither [17].
PolyP in bacteria is localized predominantly in volutin granules, also called polyP granules, or in acidocalcisomes [18]. Many biochemical pathways are connected and a given metabolite such as polyP can be generated and/or consumed by several enzymes or cellular processes. The genetic background, culture conditions and environmental factors can influence polyP levels. Its absence, as mentioned above, causes many structural and functional defects. The link between genotypes and phenotypes observed during polyP deficiency can be the result of complex networks of interaction that can be elucidated by using OMICS technology [19,20].
Recombinant Pseudomonas sp. B4 that overexpressed yeast exopolyphosphatase also showed the functional deficiencies in motility and biofilm development reported for ppk1 mutants from P. aeruginosa PAO1 [21]. In addition, new structural and functional defects such as changes in colony morphology, LPS structure and cellular division are reported in this communication. Finally, to study the proteomic changes that occurred during polyP deficiency recombinant strains were compared under different growth conditions and phases of growth. Interesting proteins related to energetic metabolism were overexpressed during polyP scarcity, such as three enzymes from the tricarboxylic acid (TCA) cycle, and one ATP synthase subunit. Protein folding, fatty acid catabolism and amino acid biosynthesis were other gene onthology (GO) categories overrepresented during polyP deficit. On the other hand, motility and transport proteins were the only categories underrepresented in this condition.
The proteomics results suggest a link between polyP and central metabolism that can be further explored to clarify the multiple structural and functional defects found during the lack of polyP in bacteria.
Results
Structural and functional defects in polyphosphate deficient bacteria
Overexpression of PPX resembled the functional defects found in motility and biofilm formation in a ppk1 mutant from P. aeruginosa PAO [21]. Despite several functional and structural defects have been reported in P. aeruginosa PAO1 ppk1 mutant [15,21,22], our polyP deficient cells showed new functional and structural phenotypes not previously reported. PPK1 is essential for biofilm development and virulence of P. aeruginosa PAO1. Considering that lipopolysaccharide (LPS) is also very important in both cellular processes; the electrophoretic profile of LPS from recombinants Pseudomonas sp. B4 were analyzed. Interestingly, changes in the core of the LPS were observed in Tricine/SDS-polyacrylamide gel electrophoresis (Figure 1). To our knowledge, the structure of the LPS core from Pseudomonas sp. B4 has not yet been elucidated and consequently it is difficult to determine the structural nature of the change found in the LPS core. It would be interesting to determine the structure of LPS in both strains [control and polyP(-)] to reveal the change in the LPS and its probable link with polyP.
Figure 1.
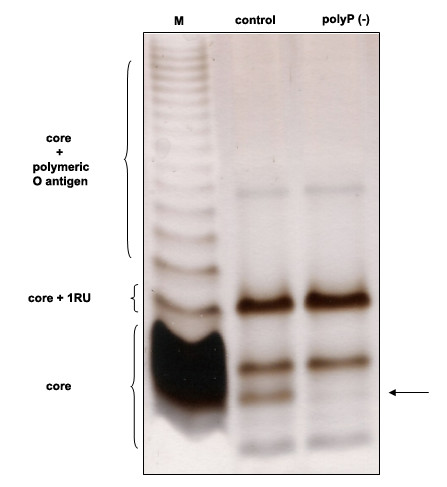
LPS profiles of polyP-deficient cells of Pseudomonas sp. B4. Equal numbers of Pseudomonas sp. B4 polyP-deficient and control cell samples were loaded in each lane and analysed by 12% (w/v) PAGE by using a Tricine-SDS buffer system. LPS from Salmonella serovars Typhi was used as LPS control (lane M). The arrow indicates the change seen in a band of the inner core. RU: repetitive units.
It was found that inorganic polyP influences not only biofilm formation but also colony morphology phenotype. Changes were seen when the colony phenotype of control cells was compared with that of polyP-deficient ones. Wild type and control cells were highly motile forming a rough colony with an irregular border (Figure 2A). In contrast, polyP-deficient cells displayed a round regular smooth colony (Figure 2A). The change observed in colony morphology could be directly a consequence of the absence of exopolymer production observed in the cells (Figure 2B) and in a P. aeruginosa PAO1 ppk1 mutant [22] but also due to the variation in the LPS core reported here. Altogether, the results suggest that biofilm formation capabilities of polyP-deficient mutants, may not only be attributed to the defect in exopolymer formation, but also to their altered LPS structure.
Figure 2.
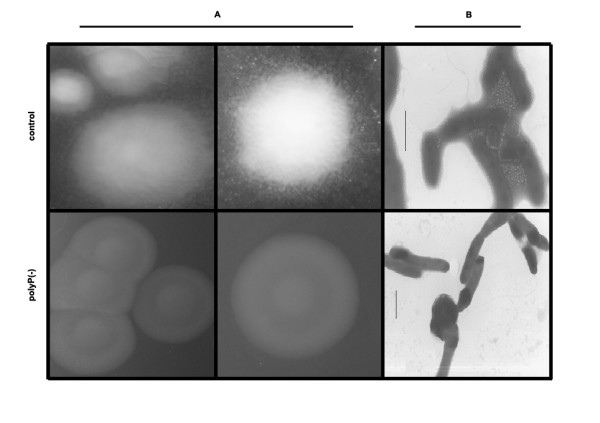
Colony morphology of polyP-deficient cells of Pseudomonas sp. B4. Pseudomonas sp. B4 polyP-deficient and control cells were grown in LB plates for 48 h and the colonies were photographed by using a magnifying glass (A). Unstained cells were analyzed by transmission electron microscopy (B).
Finally, during the entrance in stationary phase of growth in rich medium (LB) it was observed that polyP-deficient cells became highly filamentous compared to control cells most likely reflecting a cell division malfunction (Figure 3). Different defined media supplemented with various carbon sources were tested and this behaviour was found only during the entry into the stationary phase of growth in LB medium.
Figure 3.
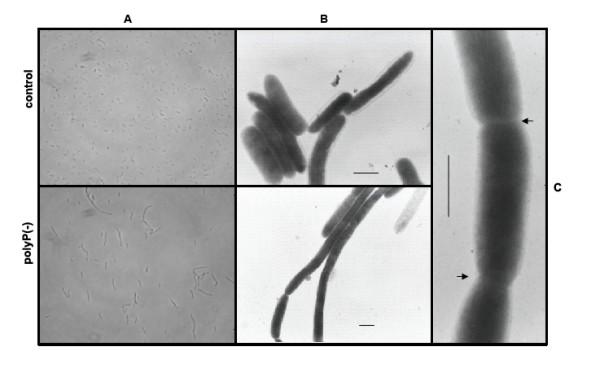
PolyP-deficient cells become filamentous during stationary phase of growth. Pseudomonas sp. B4 polyP-deficient and control cells were grown in LB medium and observed by using phase contrast-optical microscopy (A) and transmission electron microscopy of unstained cells (B). Magnified view of polyP-deficient cells (C). Arrows indicate the septum.
Differential proteomics of polyP-deficient Pseudomonas sp. B4
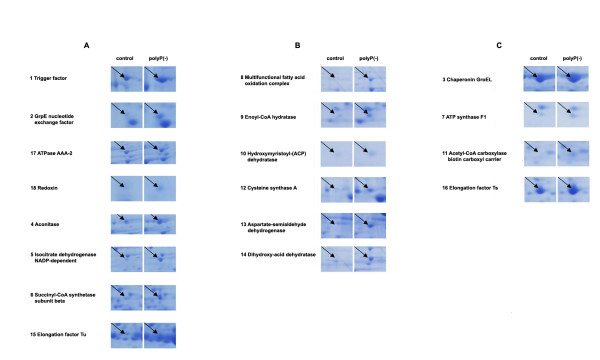
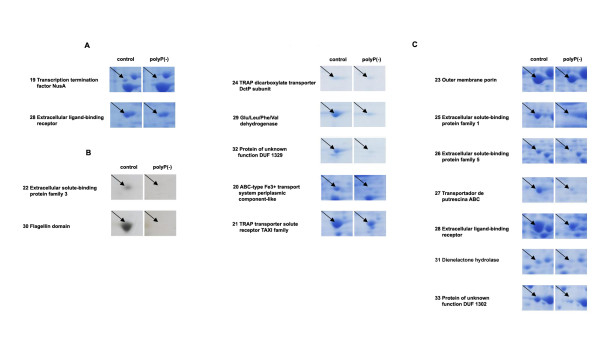
To gain insight into the effect of polyP deficiency and the metabolic adjustments taking place during the cellular response, the proteomes of Pseudomonas sp. B4 polyP-deficient and control cells were compared by two-dimensional gel electrophoresis (2D-PAGE) (Figure 4). We analyzed extracellular and total cell-free proteomes from planctonic cells grown in LB medium during exponential and stationary phase of growth and also analyzed the total cell-free proteome of the colony biofilm. These 8 samples were analyzed by using biological and experimental duplicates. This procedure yielded 81 spots of interest (proteins differentially expressed under polyP-deficiency) that were analysed by mass spectrometry resulting in 78 proteins that could be identified. Thirty-five different proteins whose expression consistently changed between the control and polyP-deficient cells in the conditions assayed are listed in Tables 1 and 2. Gel spots details are seen in Figures 5 and 6. Next, a summary of some relevant functional categories over- and under-represented during polyP deficiency is presented.
Figure 4.
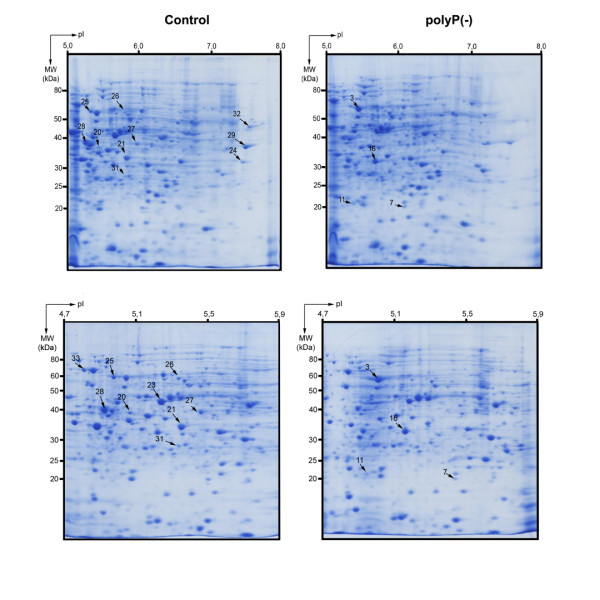
2D-PAGE gel electrophoresis of polyP-deficient and control Pseudomonas sp. B4 cell. Colonies from Pseudomonas sp. B4 polyP-deficient and control cells were grown in LB medium for 48 h. Samples were prepared and analyzed as described in Methods. The upper panels show the separation of proteins in the 5-8 pH range. To have a better resolution of some protein spots a 4.7-5.9 pH range was used (lower panels). Numbers with arrows indicate the spot numbers used for MS/MS analyses (Tables 1 and 2).
Table 1.
Summary of Gene Ontology categories of overrepresented proteins whose expressions increase during polyP deficiency in Pseudomonas sp. B4.
| GO Term Annotation | Spot | Protein Name IPR |
NCBI Accession |
Theo. Mr (kDa)/PI |
Exp. Mr (kDa)/PI |
Species/Coverage | Mascot Score |
|---|---|---|---|---|---|---|---|
| Biological Process | |||||||
| Protein folding GO:0006457 |
1 e, l | Trigger factor IPR008881 |
gi: 145575278 | 48.3/4.78 | 55/5.1 | Pseudomonas mendocina ymp/44% | 1359 |
| 2 e, l | GrpE nucleotide exchange factor IPR000740 |
gi: 60549562 | 20.4/4.9 | 24/5.1 | Pseudomonas putida/29% | 267 | |
| 3 st, a | Chaperonin GroEL IPR012723 |
gi: 146308703 | 56.8/5.02 | 55/5.2 | Pseudomonas mendocina ymp/35% | 674 | |
| Tricarboxylic acid cycle GO:0006099 |
4 e, l | Aconitase IPR004406 |
gi: 145575802 | 94.2/5.24 | 95/5.8 | Pseudomonas mendocina ymp/32% | 1715 |
| 5 e, l | Isocitrate dehydrogenase, NADP-dependent IPR004436 |
gi: 146307420 | 82.1/5.63 | 90/6.3 | Pseudomonas mendocina ymp/24% | 1130 | |
| Metabolic process GO:0008152 |
6 e, l | Succinyl-CoA synthetase IPR005809 |
gi: 146307523 | 41.8/5.5 | 49/6.5 | Pseudomonas mendocina ymp/34% | 654 |
| ATP synthesis proton transport GO:0015986 |
7 st, a | ATP synthase F1, delta subunit IPR000711 |
gi: 146309623 | 19/5.87 | 20/5.6 | Pseudomonas mendocina ymp/40% | 310 |
| Fatty acid metabolic process GO:0006631 |
8 st, l | Fatty acid oxidation complex IPR006180 |
gi: 146306611 | 77.5/5.58 | 70/6.5 | Pseudomonas mendocina ymp/51% | 159 |
| Metabolic process GO:0008152 |
9 st, l | Enoyl-CoA hydratase IPR001753 |
gi: 146307097 | 29.8/5.67 | 27/6.3 | Pseudomonas mendocina ymp/54% | 61 |
| Fatty acid biosynthetic process GO:0006633 |
10 st, l | Hydroxymyristoyl-(ACP) dehydratase IPR010084 |
gi: 146308063 | 16.8/6.3 | 15/7.5 | Pseudomonas mendocina ymp/67% | 106 |
| 11 st, a | Acetyl-CoA carboxylase biotin carboxyl carrier IPR001249 |
gi: 26987297 | 16.2/4.95 | 20/4.8 | Pseudomonas putida KT2440/20% | 415 | |
| Cysteine biosynthetic process serine GO:0006535 |
12 st, l | Cysteine synthase IPR005859 |
gi: 146306821 | 34.4/5.89 | 37/6.5 | Pseudomonas mendocina ymp/32% | 451 |
| Amino acid biosynthetic process GO:0008652 |
13 st, l | Aspartate-semialdehyde dehydrogenase IPR012280 |
gi: 146307742 | 40.5/5.33 | 40/6 | Pseudomonas mendocina ymp/63% | 128 |
| Branched chain family amino acid biosynthetic process GO:0009082 |
14 st, l | Dihydroxy-acid dehydratase IPR004404 |
gi: 146309219 | 66.2/5.69 | 60/6.5 | Pseudomonas mendocina ymp/44% | 114 |
| Translational elongation GO:0006414 |
15 e, l | Elongation factor Tu IPR004541 |
gi: 146308925 | 43.9/5.38 | 45/5.8 | Pseudomonas mendocina ymp/43% | 847 |
| 16 st, a | Elongation factor Ts IPR001816 |
gi: 146308073 | 30.5/5.22 | 30/5.2 | Pseudomonas mendocina ymp/52% | 895 | |
| Molecular function | |||||||
| ATP binding GO:0005524 |
17 e, l | ATPase AAA-2 | gi: 146308654 | 95/5.32 | 90/5.9 | Pseudomonas mendocina ymp/40% | 2404 |
| Antioxidant activity GO:0016209 |
18 e, l | Alkyl hydroperoxide reductase IPR000866 |
gi: 119860085 | 17.6/5.02 | 17/5.1 | Pseudomonas putida W619/24% | 149 |
GO: Gene Ontology Term Annotation; Spot numbers correspond to spots in 2D-PAGE; Growth Phase (e:exponential; st: stationary); Culture Medium LB (l: liquid; a: agar plate); IPR: InterPro entry; NCBI accession number from NCBI database; Theo. Mr (kDa)/PI: theoretical molecular mass and isoelectric point; Exp. Mr (kDa)/PI, experimental molecular mass and isoelectric point estimated from the 2D-PAGE gels.
Table 2.
Summary of Gene Ontology categories of overrepresented proteins whose expressions decrease during polyP deficiency in Pseudomonas sp. B4.
| GO Term Annotation | Spot | Protein Name IPR |
NCBI Accession |
Theo. Mr (kDa)/PI |
Exp. Mr (kDa)/PI |
Species/Coverage | Mascot Score |
|---|---|---|---|---|---|---|---|
| Biological Process | |||||||
| Regulation of transcription termination GO:0031554 |
19 e, l | Transcription termination factor NusA IPR010213 |
gi: 146308624 | 54.6/4.52 | 70/5.0 | Pseudomonas mendocina ymp/16% | 508 |
| Transport GO:0006810 |
20 st, a | ABC-type Fe3+ transport system periplasmic component-like IPR011587 |
gi: 146306364 | 38.1/5.27 | 38/5.3 | Pseudomonas mendocina ymp/50% | 627 |
| 21 st, a | TRAP transporter solute receptor, TAXI family IPR011852 |
gi: 146309574 | 33.3/5.74 | 35/6 | Pseudomonas mendocina ymp/26% | 808 | |
| 22 st, l | Extracellular solute-binding protein, family 3 IPR001638 |
gi: 146309284 | 27.6/4.79 | 27/5 | Pseudomonas mendocina ymp/66% | 545 | |
| 23 st, a | Outer membrane porin IPR005318 |
gi: 146309320 | 46.6/6.03 | 45/5.2 | Pseudomonas mendocina ymp/22% | 411 | |
| 24 st, a | TRAP dicarboxylate transporter, DctP subunit IPR004682 |
gi: 146307449 | 37.6/7.04 | 35/7.5 | Pseudomonas mendocina ymp/30% | 292 | |
| 25 st, a | Extracellular solute-binding protein, family 1 IPR006059 |
gi: 146307075 | 64.8/4.98 | 60/5 | Pseudomonas mendocina ymp/44% | 1080 | |
| 26 st, a | Extracellular solute-binding protein, family 5 IPR000914 |
gi: 146305880 | 59.3/5.72 | 55/5.3 | Pseudomonas mendocina ymp/16% | 354 | |
| Polyamine transport GO:0015846 |
27 st, a | Transportador de putrescina ABC IPR005893 |
gi: 70730588 | 42/6.67 | 40/5.4 | Pseudomonas fluorescens Pf-5/14% | 122 |
| Transport GO:0006810 |
28 e/l, st/a |
Extracellular ligand-binding receptor IPR001828 |
gi: 146306419 | 39.4/5.12 | 40/5.3 | Pseudomonas mendocina ymp/20% | 585 |
| Amino acid metabolic process GO:0006520 |
29 st, a | Glu/Leu/Phe/Val dehydrogenase IPR006097 |
gi: 146307897 | 37.1/5.85 | 40/7.5 | Pseudomonas mendocina ymp/21% | 366 |
| Ciliary or flagellar motility GO:0001539 |
30 st, l | Flagellin domain IPR001492 |
gi: 146307857 | 49.9/5.04 | 50/5 | Pseudomonas mendocina ymp/6% | 280 |
| Molecular function | |||||||
| Hydrolase activity GO:0016787 |
31 st, a | Dienelactone hydrolase IPR002925 |
gi: 146307513 | 27.8/5.45 | 30/5.3 | Pseudomonas mendocina ymp/24% | 411 |
| Unknown function | 32 st, a | Protein of unknown function DUF1329 | gi: 146308674 | 51.4/8.3 | 50/7.8 | Pseudomonas mendocina ymp/50% | 1200 |
| 33 st, a | Protein of unknown function DUF1302 | gi: 77457132 | 64.1/5.15 | 65/4.9 | Pseudomonas fluorescens PfO/13% | 340 | |
Conditions and abbreviations are the same as those in Table1.
Figure 5.
Summary of protein spots identified whose expression increases during polyP deficiency. A- Planktonic cultures, exponential phase. B- Planktonic cultures, stationary phase. C- Colonies grown on LB agar plates.
Figure 6.
Summary of protein spots identified whose expression decreases during polyP deficiency. A, Planktonic cultures from exponential phase. B, Planktonic cultures from stationary phase. C, Colonies grown on LB agar plates.
Energy metabolism
The polyP-deficient strain overexpressed three TCA cycle enzymes during exponential phase: aconitase, isocitrate dehydrogenase and succinyl-CoA synthetase. The last two proteins are directly involved in producing NADH and GTP (or ATP) respectively. Additionally, in solid medium, this strain overexpressed ATP synthase F1 (delta subunit) that synthesizes ATP coupled to an electrochemical protons gradient in the respiratory chain [23]. Several catabolic pathways converge on the TCA cycle and particularly; beta-oxidation is the process by which fatty acids are broken down to generate acetyl-CoA, the entry molecule for the TCA cycle. Curiously, during stationary phase of planktonic polyP(-) cultures, cells overexpressed two proteins belonging to the mutifunctional fatty acid oxidation complex that generates acetyl-CoA species: enoyl-CoA hydratase and 3-hydroxyacyl-CoA dehydrogenase. Both enzymes catalyze successive reactions, and their substrates are also related to polyhydroxyalkanoates (PHA) biosynthesis [24]. This polymer is accumulated in anaerobic cultures during stages in which polyPs are degraded [25], and perhaps low polyP levels may enhance PHA accumulation. It would be interesting to find out if the absence of polyP affected other storage biopolymers such as triacylglycerols (TAG), wax esters, polyhydroxyalkanoates (PHA) and glycogen.
Protein folding and stress response
Three proteins involved in protein folding were overexpressed during exponential phase by the polyP(-) strain: trigger factor, GrpE and ClpB. Additionally, GroEL was increased in the same strain during stationary phase. All of them are considered chaperones that prevent inappropriate molecular interactions by binding to hydrophobic regions in non-native proteins and allow proper protein folding acting as a molecular network [26]. Trigger factor is a ribosome-associated bacterial chaperone that begins nascent protein folding in an ATP-independent manner [27,28]. On the other hand, GrpE is a co-chaperone that works as a nucleotide exchange factor on a DnaK domain, whereas ClpB rescues stress-damaged proteins from an aggregated state asissted by DnaK [27,29]. GroEL interacts with recently synthesized proteins after their release from the ribosome [26]. With the exception of trigger factor, the other three chaperones form an ATP-dependent network.
Also, an alkyl hydroperoxide reductase (peroxiredoxin) was overexpressed in exponential phase of polyP-deficient cells. This enzyme reduces peroxides to water or alcohols and prevents oxidative stress in bacteria coupled to the TCA cycle and respiratory chain. Additionally, it regenerates the NAD pool and keeps oxidative and reducing balance [30,31]. Peroxiredoxin could act as protection factor against ROS generated by the stress caused by low polyP levels. Finally, increased levels of the translational factors EF-Tu and EF-Ts were found during polyP scarcity. This response has also been described in E. coli during acid stress and heavy metal (cobalt) exposure. It is suggested that these elongation factors could fold proteins in a way similar to that of stress chaperones [32]. Finally, as the GTP hydrolysis step is catalysed by EF-Tu, which binds to the large ribosomal subunit, it has been proposed that the interaction between polyphosphate and the large ribosomal subunit promotes translation fidelity by influencing the EF-Tu GTPase reaction [33].
Altogether, these results suggest that during polyP scarcity a general stress state occurred and cells succeeded by overexpressing protein-folding chaperones.
Transport proteins
From the 17 total proteins identified whose expressions decreased during lack of polyP, 10 were identified as transporters. Energy consuming ABC-type transporters responsible for carrying different solutes such as sugars, peptides, polyamines, amino acids and Fe3+ were identified. Also, C4-dicarboxylates TRAP transporters and outer membrane protein OprE, which has been involved in virulence process in the genus Pseudomonas [34], were reduced in polyP(-) cells.
Other processes and hypothetical proteins
The present study also yielded some results that appear to be conflicting. We, and others, have demonstrated that despite the lack of motility of polyP-deficient cells, the flagellum was intact (as seen by using transmission electron microscopy). Nevertheless, we found flagellin, the major component of flagella filaments, diminished in the total and extracelullar proteome of polyP-deficient cells.
Finally two protein spots present in the total proteome matched ORF sequences designated 'hypothetical' or 'conserved hypothetical" proteins. These hypothetical proteins identified here should be subjected to further characterization to confirm their possible role in polyP metabolism and to ascertain their true biological function.
Discussion and Conclusions
PolyP has numerous and diverse biological functions that have been discovered mainly by studying ppk1 mutants in bacteria. A P. aeruginosa PAO1 ppk1 null mutant exhibits pleiotropic phenotypes including decreased virulence, defective in motility, quorum sensing, biofilm formation and failure in responses to various stresses [13,15,22]. Many of these features were also observed in ppk1 mutants of other bacteria such as Vibrio cholerae, Salmonella, Shigella and others [35,36]. Nevertheless, new functional and structural defects were found in our recombinant cells that overexpressed the exopolyphosphatase, the enzyme in charge of polyP degradation to Pi. These results might be explained by the higher extent of polyP depletion when using this approach. In the genus Pseudomonas, despite the lack of detectable PPK1 activity (<1% of wild type), these mutants still possess as much as 20% of the wild-type levels of poly P as is the case of P. aeruginosa PAO1 [22]. We previously reported that the overexpression of exopolyphosphatase removed more than 95% of cellular polyP [21].
The changes observed in the colony morphology are not surprising taking into account that polyP deficient P. aeruginosa PAO1 cells fails to produce extracellular polysaccharide [22]. Similar results and an additional change in the LPS profile were seen in our polyP-deficient cells. Although, the LPS structure of Pseudomonas sp. B4 is not known in detail it can be speculated that the change seen in the LPS could be due to an alteration in the phosphate moiety of the LPS core or that polyP regulates some enzyme able to modify the LPS. Further experiments should be done to clarify this finding but it will be interesting to find out if some of the LPS kinases reported in the genus Pseudomonas (such as WaaP [37]) could use polyP instead of ATP during phosphorylation of Heptose I in the inner core of LPS. Furthermore, taking into account the role of LPS during pathogenesis development in many bacteria, this change might explain some dysfunction during virulence of polyP-deficient bacteria.
Bacterial cell division occurs through the formation of an FtsZ ring (Z ring) at the site of division. The ring is composed of the tubulin-like FtsZ protein that has GTPase activity and the ability to polymerize in vitro (reviewed in [38]). Our observation of cell division failure in polyP-deficient cells during entry into the stationary phase is in agreement with the finding that during polyP-deficiency energy metabolism, and particularly nucleoside triphosphate (NTP) formation, was affected (see below). As seen in Figure 3, the cells were apparently able to form the septum, but did not complete the separation process. It is possible that polyP scarcity affects the function of FtsZ, since its GTPase activity needs both, GTP and a bivalent ion. Considering that polyP can provide both, phosphate for the generation of GTP ([16,17] and bivalent metals [35], the absence of this biopolymer could block indirectly the polymerisation of Z ring, which would explain the observed phenotype. Curiously, the enzyme in charge of GTP synthesis from polyP in P. aeruginosa (PPK2), was induced 100-times in the stationary phase [16]. In this phase of growth GTP is necessary for the synthesis of alginate and other functions such as cellular division. At present, we cannot discard that other proteins from the divisome, that also employ GTP for their activity, are affected by the absence of polyP.
Living cells constantly require energy for maintaining their highly organized structures, synthesize cellular components, generate electric currents, and many other processes. PolyP acts as a reserve for high energy Pi and regulates intracellular ATP in combination with oxidative and substrate level phosphorylation. Our proteomic data support the hypothesis that polyP is an important component for energy regulation, and particularly in ATP regeneration [39]. During polyP deficiency, cells would prevail by increasing the flux of important energy generating pathways such as β-oxidation, citric acid cycle and oxidative phosphorylation as proposed in Figure 7. We found eight different proteins related to these pathways increased during polyP deficiency and in the case of the TCA cycle enzymes two of them are directly involved in the generating NADH and GTP by their activity (see Table 1). Interestingly, a previous link between polyP and the TCA cycle was reported in P. aeruginosa. AlgR2, a global transcriptional factor, positively regulates nucleoside diphosphate kinase (Ndk) and succinyl-CoA synthetase, enzymes critical in nucleoside triphosphate (NTP) formation [40]. Thus, AlgR2 positively regulates the production of alginate, GTP, ppGpp and inorganic polyP in P. aeruginosa [41]. It is possible then that polyP-deficiency induces AlgR2 expression to increase GTP and polyP production. This could explain the increase of succinyl-CoA synthetase in our polyP deficient cells.
Figure 7.

Working model proposed for the metabolic adjustment of bacterial cells during polyP deficiency. In red, metabolic pathways in which several of its components are overexpressed during polyP scarcity.
Active transport of ion and molecules across the membrane consumes energy and ATP. We found that the majority of protein spots decreasing their levels in polyP(-) cells belong to the transport protein category (see Table 2). It is possible that diminishing energy-consuming processes such as active transport can help the cells to overcome this polyP deficiency.
The defects in the ppk1 mutant described in P. aeruginosa [22,42], and those seen in the same E. coli mutant [10], suggest a failure to respond to a variety of stresses. We found that the levels of many important chaperones and enzymes related to stress response are increased in polyP deficient cells. It is suggested that a general stress response occurs during polyP deficiency and cells prevail by augmenting the levels of general chaperones and enzymes that would remove reactive oxygen species. In fact, our previous results showed that growth of Pseudomonas sp. B4 in certain conditions generates an oxidative stress and produced a massive increase of polyP [43].
Altogether the results presented in this communication demonstrate the usefulness of proteomics to study the effect of polyP deficiency in order to generate new hypothesis to clarify its role in bacteria. New suggestions such as the possible link between the central metabolic pathways and polyP metabolism proposed here should be the focus of future metabolic flux experiments. The integrations of OMICS technologies will definitely help in elucidating the role of polyP in bacteria and its importance during pathogenic process.
In summary, polyP has numerous and varied biological functions in bacteria that have been discovered mainly by studying its deficiency. To better understand the function of polyP we used broad-host-range constitutive and regulated vectors to deplete cellular polyP and found new functional and structural changes. In addition, it is generally accepted that energy supply of the cells could be severely compromised in the absence of polyP. We confirmed this evidence by using differential proteomic studies and suggested that during polyP scarcity energy metabolism and particularly nucleoside triphosphate (NTP) formation were affected, generating a general stress condition. We propose that bacterial cells prevail by increasing the flux of energy-generating metabolic pathways such as tricarboxilic acid (TCA) cycle and β-oxidation and by reducing energy-consuming ones such as active transporters and amino acid biosynthesis.
Methods
Bacterial strains and growth conditions
Pseudomonas sp. B4 wt, control (pMLS7) and polyP-deficient (pS7PPX1) recombinant strains were previously obtained [21] and grown aerobically at 37°C on Luria-Bertani (LB) rich medium supplemented with trimetropim (50 μg/ml). When required, LB plates (1,5% (w/v) of Bacto-agar) were used for obtaining cells from the colonies after 48 h of growth.
Optical and Electron microscopy
Unstained cells from the different cultures were routinely examined for the presence of polyP granules by transmission electron microscopy [43]. Cells were mixed and dispersed in distilled water and added onto carbon-coated nickel grids. The drops containing the microorganisms were drained off with filter paper and air dried during 30-50 s. Electron microscopy was performed with a Philips Tecnai 12 electron microscope using 80 kV accelerating voltage (Electron Microscopy Laboratory, Pontificia Universidad Católica de Chile). Optical microscopy was routinely performed in an Olympus BX50 microscope (Olympus Corporation, Japan).
LPS analysis
Culture samples were adjusted to an optical density at 600 nm of 2.0 in a final volume of 100 μl. Then, proteinase K-digested whole-cell lysates were prepared as described previously [44], and LPS was separated on 14% acrylamide gels using a Tricine-sodium dodecyl sulfate (SDS) buffer system [45]. Gel loadings were normalized so that each sample represented the same number of cells. Gels were silver stained by a modification of the procedure of Tsai and Frasch [46,47].
Samples preparation and 2D-PAGE
Cells (200 mg) were harvested by centrifugatio n (7,000 × g for 15 min at 25°C) from liquid cultures or were collected with an inoculation loop from agar plates. Pellets were washed four times in sonication buffer (40 mM Tris pH 8.15; 1 mM PMSF). Cells were disrupted by sonication (Misonix XL2020) and the sample was incubated on ice for 10 min with 50 μg/ml of DNAase. Cell debris was removed by centrifugation and then the sample was washed and concentrated (to half of the total volume) by using a Microcon® YM-3 filter unit (10,000 × g, 4°C). Protein quantitation was performed using Bio-Rad Protein Assay® system. A total of 500 μg of proteins was precipitated through Ready-Prep 2D Cleanup Bio-Rad® kit.
Precipitated proteins were resuspended in 300 μL IEF buffer (7 M urea, 2 M thiourea, 4% CHAPS, 0.0002% bromophenol blue) followed by the addition of DTT to 100 mM and 0.2% Bio-Rad ampholytes and the sample mix was incubated for 1 h at 25°C. The entire volume was loaded in the Protean® IEF focusing tray (17 cm) using the following strips pH ranges: 4.7-5.9/5-8/3-10NL (ReadyStrip™ IPG) that were actively rehydrated at 50 V for 12 h. The focusing step was performed at 250 V for 15 min; 2,000 V for 2 h; 8,000 V for 4 h and finally 10,000 V for 11 h, all the steps at 20°C. Focused proteins in the strip were then incubated at 25°C with gentle agitation for 15 min in equilibrium buffer (6 M urea; 2% SDS; 0,05 M Tris/Cl pH 8.8; 20% glycerol) containing 2% DTT and then 15 min in equilibrium buffer containing 2.5% iodoacetamide. Finally, the strip was placed onto a 12.5% polyacrilamide gel for the second dimension in Protean® II (Bio-Rad) system at 50 V for 23 h. The gels were fixed for 1 h (50% ethanol; 2% phosphoric acid), stained for 3 h (0,12% CBB G-250; 10% phosphoric acid; 10% ammonium sulphate; 20% methanol) and then washed three times with 15% methanol.
Digital images of the gels were analyzed and spots quantified using Delta2D v.3.6 software. Spot volume was normalized as a percentage of the total volume of all spots on the corresponding gel and also manually confirmed. The threshold for accepting a meaningful variation was a factor of 2.0 (p < 0,05). A total of 81 proteins spots showing differences in the expression pattern between control and polyP(-) strains (three independent replicates) were selected for further MS analysis.
In-gel protein digestion and sample preparation
Spots of interest from Coomassie blue-stained 2D gels were excised manually, deposited in 96-well plates and processed automatically in a Proteineer DP (Bruker Daltonics, Bremen, Germany). The digestion protocol used was based on Schevchenko et al. [48] with minor variations: gel plugs were washed firstly with 50 mM ammonium bicarbonate and secondly with acetonitrile (ACN) prior to reduction with 10 mM DTT in 25 mM ammonium bicarbonate solution, and alkylation was carried out with 55 mM IAA in 50 mM ammonium bicarbonate solution. Gel pieces were then rinsed with 50 mM ammonium bicarbonate and with ACN, and then dried under a stream of nitrogen. Modified porcine trypsin (sequencing grade; Promega, Madison WI) was added at a final concentration of 16 ng/μl in 25% ACN/50 mM ammonium bicarbonate solution and the digestion took place at 37°C for 6 h. The reaction was stopped by adding 0.5% TFA for peptide extraction. Tryptic peptides were dried by speed-vacuum centrifugation and resuspended in 4 μl of MALDI solution. A 0.8 μl aliquot of each peptide mixture was deposited onto a 386-well OptiTOF™ Plate (Applied Biosystems, Framingham, MA, USA) and allowed to dry at room temperature. A 0.8 μl aliquot of matrix solution (3 mg/mL CHCA in MALDI solution) was then added onto dried digest and allowed to dry at room temperature.
MALDI peptide mass fingerprinting, MS/MS analysis and database searching
For MALDI-TOF/TOF analysis, samples were automatically acquired in an ABi 4800 MALDI TOF/TOF mass spectrometer (Applied Biosystems, Framingham, MA, USA) in positive ion reflector mode (ion acceleration voltage was 25 kV for MS acquisition and 1 kV for MSMS) and the spectra were stored into the ABi 4000 Series Explorer Spot Set Manager. PMF and MSMS fragment ion spectra were smoothed and corrected to zero baseline using routines embedded in ABi 4000 Series Explorer Software v3.6. Each PMF spectrum was internally calibrated with the mass signals of trypsin autolysis ions to reach a typical mass measurement accuracy of <25 ppm. Known trypsin and keratin mass signals, as well as potential sodium and potassium adducts (+21 Da and +39 Da) were removed from the peak list. To submit the combined PMF and MS/MS data to MASCOT software v.2.1 (Matrix Science, London, UK), GPS Explorer v4.9 was used, searching in the non-redundant NCBI protein database.
LC-ESI MS/MS analysis
In some specific cases, alternative proteomic techniques were employed to confirm and improve protein identifications. For this purpose, we made use of liquid chromatography coupled to electrospray ion-trap mass spectrometry tandem MS (LC ESI-MS/MS). This was done using an Ultimate 3000 nano LC (Dionex, Amsterdam, The Netherland) and a 75 micrometer I.D, 100 mm reversed-phase column, at a 300 nL/min flow, coupled to a Bruker HCT Ultra ion-trap mass spectrometer (Bruker Daltonics, Bremen, Germany) working in dynamic exclusion mode.
Database Search
For protein identification, LC ESI MS/MS spectra were transferred to BioTools 2.0 interface (Bruker Daltonics) to search in the NCBInr database using a licensed version of Mascot v.2.2.04 search engine (http://www.matrixscience.com; Matrix Science, London, UK). Search parameters were set as follows: carbamidomethyl cystein as fixed modification by the treatment with iodoacetamide, oxidized methionines as variable modification, peptide mass tolerance of 0.5 Da for the parental mass and fragment masses and 1 missed cleavage site. In all protein identifications, the probability Mowse scores were greater than the minimum score fixed as significant with a p-value minor than 0.05. Selected proteins were based on that who exhibited higher Mascot score and sequence coverage. A total of thirty-three different proteins showing differential expression pattern between polyP+ and polyP- strains (three independent replicates) were selected. Furthermore, theoretical isoelectric points and molecular masses were compared to experimental values of thirty-three proteins were grouped by gene ontology (GO) categories [49].
Authors' contributions
FPC y CAJ conceived and designed the study; FPC performed some experiments and wrote the manuscript. CV performed proteomic experiments. CM carried out cellular experiments. AP y JPA carried out MS/MS protein identification. CAJ participated in coordination and critical evaluation of the manuscript. All authors read and approved the final manuscript.
Contributor Information
Cristian Varela, Email: cristian.varela.cl@gmail.com.
Cecilia Mauriaca, Email: cmauriaca@gmail.com.
Alberto Paradela, Email: alberto.paradela@cnb.csic.es.
Juan P Albar, Email: jpalbar@cnb.csic.es.
Carlos A Jerez, Email: cjerez@uchile.cl.
Francisco P Chávez, Email: fpchavez@uchile.cl.
Acknowledgements
This research was supported by Grants 1070986 and 11070180 from Fondecyt and ICM P05-001-F from MIDEPLAN.
References
- Brown M, Kornberg A. The long and short of it - polyphosphate, PPK and bacterial survival. Trends Biochem Sci. 2008;33(6):284–290. doi: 10.1016/j.tibs.2008.04.005. [DOI] [PubMed] [Google Scholar]
- Kornberg A. Inorganic polyphosphate: a molecule of many functions. Prog Mol Subcell Biol. 1999;23:1–18. doi: 10.1007/978-3-642-58444-2_1. [DOI] [PubMed] [Google Scholar]
- Blum J. Changes in orthophosphate, pyrophosphate and long-chain polyphosphate levels in Leishmania major promastigotes incubated with and without glucose. J Protozool. 1989;36(3):254–257. doi: 10.1111/j.1550-7408.1989.tb05358.x. [DOI] [PubMed] [Google Scholar]
- Kuroda A, Tanaka S, Ikeda T, Kato J, Takiguchi N, Ohtake H. Inorganic polyphosphate kinase is required to stimulate protein degradation and for adaptation to amino acid starvation in Escherichia coli. Proc Natl Acad Sci USA. 1999;96(25):14264–14269. doi: 10.1073/pnas.96.25.14264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda A, Nomura K, Ohtomo R, Kato J, Ikeda T, Takiguchi N, Ohtake H, Kornberg A. Role of inorganic polyphosphate in promoting ribosomal protein degradation by the Lon protease in **E. coli. Science. 2001;293(5530):705–708. doi: 10.1126/science.1061315. [DOI] [PubMed] [Google Scholar]
- Reusch R. Polyphosphate/poly-(R)-3-hydroxybutyrate) ion channels in cell membranes. Prog Mol Subcell Biol. 1999;23:151–182. doi: 10.1007/978-3-642-58444-2_8. [DOI] [PubMed] [Google Scholar]
- Reusch R. Transmembrane ion transport by polyphosphate/poly-(R)-3-hydroxybutyrate complexes. Biochemistry (Mosc) 2000;65(3):280–295. [PubMed] [Google Scholar]
- Crooke E, Akiyama M, Rao N, Kornberg A. Genetically altered levels of inorganic polyphosphate in Escherichia coli. J Biol Chem. 1994;269(9):6290–6295. [PubMed] [Google Scholar]
- Kim K, Rao N, Fraley C, Kornberg A. Inorganic polyphosphate is essential for long-term survival and virulence factors in Shigella and Salmonella spp. Proc Natl Acad Sci USA. 2002;99(11):7675–7680. doi: 10.1073/pnas.112210499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao N, Kornberg A. Inorganic polyphosphate supports resistance and survival of stationary-phase Escherichia coli. J Bacteriol. 1996;178(5):1394–1400. doi: 10.1128/jb.178.5.1394-1400.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao N, Liu S, Kornberg A. Inorganic polyphosphate in Escherichia coli: the phosphate regulon and the stringent response. J Bacteriol. 1998;180(8):2186–2193. doi: 10.1128/jb.180.8.2186-2193.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao N, Kornberg A. Inorganic polyphosphate regulates responses of Escherichia coli to nutritional stringencies, environmental stresses and survival in the stationary phase. Prog Mol Subcell Biol. 1999;23:183–195. doi: 10.1007/978-3-642-58444-2_9. [DOI] [PubMed] [Google Scholar]
- Rashid M, Kornberg A. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2000;97(9):4885–4890. doi: 10.1073/pnas.060030097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid M, Rao N, Kornberg A. Inorganic polyphosphate is required for motility of bacterial pathogens. J Bacteriol. 2000;182(1):225–227. doi: 10.1128/JB.182.1.225-227.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid M, Rumbaugh K, Passador L, Davies D, Hamood A, Iglewski B, Kornberg A. Polyphosphate kinase is essential for biofilm development, quorum sensing, and virulence of Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2000;97(17):9636–9641. doi: 10.1073/pnas.170283397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishige K, Zhang H, Kornberg A. Polyphosphate kinase (PPK2), a potent, polyphosphate-driven generator of GTP. Proc Natl Acad Sci USA. 2002;99(26):16684–16688. doi: 10.1073/pnas.262655299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Ishige K, Kornberg A. A polyphosphate kinase (PPK2) widely conserved in bacteria. Proc Natl Acad Sci USA. 2002;99(26):16678–16683. doi: 10.1073/pnas.262655199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seufferheld M, Alvarez H, Farias M. Role of polyphosphates in microbial adaptation to extreme environments. Appl Environ Microbiol. 2008;74(19):5867–5874. doi: 10.1128/AEM.00501-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kell D. Metabolomics and systems biology: making sense of the soup. Curr Opin Microbiol. 2004;7(3):296–307. doi: 10.1016/j.mib.2004.04.012. [DOI] [PubMed] [Google Scholar]
- Joyce A, Palsson B. The model organism as a system: integrating 'omics' data sets. Nat Rev Mol Cell Biol. 2006;7(3):198–210. doi: 10.1038/nrm1857. [DOI] [PubMed] [Google Scholar]
- Chávez F, Mauriaca C, Jerez C. Constitutive and regulated expression vectors to construct polyphosphate deficient bacteria. BMC Res Notes. 2009;2(1):50. doi: 10.1186/1756-0500-2-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraley C, Rashid M, Lee S, Gottschalk R, Harrison J, Wood P, Brown M, Kornberg A. A polyphosphate kinase 1 (ppk1) mutant of Pseudomonas aeruginosa exhibits multiple ultrastructural and functional defects. Proc Natl Acad Sci USA. 2007;104(9):3526–3531. doi: 10.1073/pnas.0609733104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi-Matsui M, Kashiwagi S, Ubukata T, Iwamoto-Kihara A, Wada Y, Futai M. Rotational catalysis of Escherichia coli ATP synthase F1 sector. Stochastic fluctuation and a key domain of the beta subunit. J Biol Chem. 2007;282(28):20698–20704. doi: 10.1074/jbc.M700551200. [DOI] [PubMed] [Google Scholar]
- Aldor I, Keasling J. Process design for microbial plastic factories: metabolic engineering of polyhydroxyalkanoates. Curr Opin Biotechnol. 2003;14(5):475–483. doi: 10.1016/j.copbio.2003.09.002. [DOI] [PubMed] [Google Scholar]
- Wilmes P, Wexler M, Bond P. Metaproteomics provides functional insight into activated sludge wastewater treatment. PLoS ONE. 2008;3(3):e1778. doi: 10.1371/journal.pone.0001778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuerling E, Bukau B. Chaperone-assisted folding of newly synthesized proteins in the cytosol. Crit Rev Biochem Mol Biol. pp. 261–277. [DOI] [PubMed]
- Lee S, Choi J, Tsai F. Visualizing the ATPase cycle in a protein disaggregating machine: structural basis for substrate binding by ClpB. Mol Cell. 2007;25(2):261–271. doi: 10.1016/j.molcel.2007.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merz F, Boehringer D, Schaffitzel C, Preissler S, Hoffmann A, Maier T, Rutkowska A, Lozza J, Ban N, Bukau B. Molecular mechanism and structure of Trigger Factor bound to the translating ribosome. EMBO J. 2008;27(11):1622–1632. doi: 10.1038/emboj.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsell D, Kowal A, Singer M, Lindquist S. Protein disaggregation mediated by heat-shock protein Hsp104. Nature. 1994;372(6505):475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- Nishiyama Y, Yamamoto H, Allakhverdiev S, Inaba M, Yokota A, Murata N. Oxidative stress inhibits the repair of photodamage to the photosynthetic machinery. EMBO J. 2001;20(20):5587–5594. doi: 10.1093/emboj/20.20.5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seib K, Wu H, Kidd S, Apicella M, Jennings M, McEwan A. Defenses against oxidative stress in Neisseria gonorrhoeae: a system tailored for a challenging environment. Microbiol Mol Biol Rev. 2006;70(2):344–361. doi: 10.1128/MMBR.00044-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Sundaram C, Luthra P, Singh Y, Sirdeshmukh R, Gade W. Role of proteins in resistance mechanism of Pseudomonas fluorescens against heavy metal induced stress with proteomics approach. J Biotechnol. 2006;126(3):374–382. doi: 10.1016/j.jbiotec.2006.04.032. [DOI] [PubMed] [Google Scholar]
- McInerney P, Mizutani T, Shiba T. Inorganic polyphosphate interacts with ribosomes and promotes translation fidelity in vitro and in vivo. Mol Microbiol. 2006;60(2):438–447. doi: 10.1111/j.1365-2958.2006.05103.x. [DOI] [PubMed] [Google Scholar]
- Jaouen T, Coquet L, Marvin-Guy L, Orange N, Chevalier S, Dé E. Functional characterization of Pseudomonas fluorescens OprE and OprQ membrane proteins. Biochem Biophys Res Commun. 2006;346(3):1048–1052. doi: 10.1016/j.bbrc.2006.06.013. [DOI] [PubMed] [Google Scholar]
- Kornberg A. Inorganic polyphosphate: toward making a forgotten polymer unforgettable. J Bacteriol. 1995;177(3):491–496. doi: 10.1128/jb.177.3.491-496.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg A, Rao N, Ault-Riché D. Inorganic polyphosphate: a molecule of many functions. Annu Rev Biochem. 1999;68:89–125. doi: 10.1146/annurev.biochem.68.1.89. [DOI] [PubMed] [Google Scholar]
- Zhao X, Lam J. WaaP of Pseudomonas aeruginosa is a novel eukaryotic type protein-tyrosine kinase as well as a sugar kinase essential for the biosynthesis of core lipopolysaccharide. J Biol Chem. 2002;277(7):4722–4730. doi: 10.1074/jbc.M107803200. [DOI] [PubMed] [Google Scholar]
- Lutkenhaus J, Addinall S. Bacterial cell division and the Z ring. Annu Rev Biochem. 1997;66:93–116. doi: 10.1146/annurev.biochem.66.1.93. [DOI] [PubMed] [Google Scholar]
- Harold F. Inorganic polyphosphates in biology: structure, metabolism, and function. Bacteriol Rev. 1966;30(4):772–794. doi: 10.1128/br.30.4.772-794.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledgham F, Soscia C, Chakrabarty A, Lazdunski A, Foglino M. Global regulation in Pseudomonas aeruginosa: the regulatory protein AlgR2 (AlgQ) acts as a modulator of quorum sensing. Res Microbiol. 2003;154(3):207–213. doi: 10.1016/S0923-2508(03)00024-X. [DOI] [PubMed] [Google Scholar]
- Kim H, Schlictman D, Shankar S, Xie Z, Chakrabarty A, Kornberg A. Alginate, inorganic polyphosphate, GTP and ppGpp synthesis co-regulated in Pseudomonas aeruginosa: implications for stationary phase survival and synthesis of RNA/DNA precursors. Mol Microbiol. 1998;27(4):717–725. doi: 10.1046/j.1365-2958.1998.00702.x. [DOI] [PubMed] [Google Scholar]
- Parks Q, Hobden J. Polyphosphate kinase 1 and the ocular virulence of Pseudomonas aeruginosa. Invest Ophthalmol Vis Sci. 2005;46(1):248–251. doi: 10.1167/iovs.04-0340. [DOI] [PubMed] [Google Scholar]
- Chávez F, Lünsdorf H, Jerez C. Growth of polychlorinated-biphenyl-degrading bacteria in the presence of biphenyl and chlorobiphenyls generates oxidative stress and massive accumulation of inorganic polyphosphate. Appl Environ Microbiol. 2004;70(5):3064–3072. doi: 10.1128/AEM.70.5.3064-3072.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchcock P, Brown T. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J Bacteriol. 1983;154(1):269–277. doi: 10.1128/jb.154.1.269-277.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesse A, Campagnari A, Bittner W, Apicella M. Increased resolution of lipopolysaccharides and lipooligosaccharides utilizing tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis. J Immunol Methods. 1990;126(1):109–117. doi: 10.1016/0022-1759(90)90018-Q. [DOI] [PubMed] [Google Scholar]
- Tsai C, Frasch C. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal Biochem. 1982;119(1):115–119. doi: 10.1016/0003-2697(82)90673-X. [DOI] [PubMed] [Google Scholar]
- Marolda C, Lahiry P, Vinés E, Saldías S, Valvano M. Micromethods for the characterization of lipid A-core and O-antigen lipopolysaccharide. Methods Mol Biol. 2006;347:237–252. doi: 10.1385/1-59745-167-3:237. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68(5):850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball C, Blake J, Botstein D, Butler H, Cherry J, Davis A, Dolinski K, Dwight S, Eppig J. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]