Analysis of the crystal structure of a phosphotransfer complex from the Rhodobacter sphaeroides chemotaxis pathway allowed reengineering of molecular recognition in a two-component signalling system.
Abstract
Two-component signal transduction pathways comprising histidine protein kinases (HPKs) and their response regulators (RRs) are widely used to control bacterial responses to environmental challenges. Some bacteria have over 150 different two-component pathways, and the specificity of the phosphotransfer reactions within these systems is tightly controlled to prevent unwanted crosstalk. One of the best understood two-component signalling pathways is the chemotaxis pathway. Here, we present the 1.40 Å crystal structure of the histidine-containing phosphotransfer domain of the chemotaxis HPK, CheA3, in complex with its cognate RR, CheY6. A methionine finger on CheY6 that nestles in a hydrophobic pocket in CheA3 was shown to be important for the interaction and was found to only occur in the cognate RRs of CheA3, CheY6, and CheB2. Site-directed mutagenesis of this methionine in combination with two adjacent residues abolished binding, as shown by surface plasmon resonance studies, and phosphotransfer from CheA3-P to CheY6. Introduction of this methionine and an adjacent alanine residue into a range of noncognate CheYs, dramatically changed their specificity, allowing protein interaction and rapid phosphotransfer from CheA3-P. The structure presented here has allowed us to identify specificity determinants for the CheA–CheY interaction and subsequently to successfully reengineer phosphotransfer signalling. In summary, our results provide valuable insight into how cells mediate specificity in one of the most abundant signalling pathways in biology, two-component signal transduction.
Author Summary
The ability to respond to environmental stimuli is a universal feature of living cells. Evolution has created a vast array of signalling mechanisms that enable cells to react in many ways to extracellular changes. In bacteria, two-component signalling mechanisms, comprising a sensor protein kinase paired with its a cognate response regulator, are used widely to sense and respond to environmental changes. Some species of bacteria have over 150 different two-component pairs in a single cell, so the specificity between these pairs has to be tightly controlled to prevent “crossed wires” between signalling pathways. In this study, we have identified the determinants of this specificity in a two-component complex that controls the movement of Rhodobacter sphaeroides along a chemical gradient. By solving and analysing the crystal structure of this complex, we were able to pinpoint the amino acid residues that are crucially involved in formation of the complex. Knowledge of these crucial residues allowed us to convert noncognate response regulators into cognate response regulators simply by changing two amino acids. This reengineering of two-component signalling pathways paves the way for producing custom-designed circuits for applications in synthetic biology.
Introduction
Bacteria, Archaea, and some eukaryotes use two-component signalling pathways to detect environmental conditions and bring about appropriate changes in cellular behaviour [1],[2]. Two-component pathways comprise sensor histidine kinases (HPK) and response regulators (RRs). Environmental stimuli control the rate at which the HPK autophosphorylates on a conserved histidine residue. Once phosphorylated, the HPK transfers the phosphoryl group to an aspartate residue within the receiver domain of the cognate RR. The phosphorylated RR (RR-P), often a transcriptional regulator, then effects a response appropriate to the original stimulus. Some bacteria have over 150 different HPK and RR pairs, and the specificity of the phosphorylation reactions between them needs to be tightly controlled to prevent HPKs from inappropriately phosphorylating and activating noncognate RRs. A number of mechanisms contribute to this specificity [3],[4], although the primary one is molecular recognition, in which a HPK shows a strong kinetic preference for its cognate RRs [5],[6]. Understanding the mechanisms involved in molecular recognition will not only allow prediction of interacting pairs, but potentially allow the rewiring of bacterial sensory pathways for use in synthetic biology.
Many two-component systems utilize an additional element, the Hpt (histidine-containing phosphotransfer) domain in phosphotransfer; these include multistep phosphorelays and chemotaxis signalling pathways. In multistep phosphorelays, the Hpt domain serves as an intermediate in the transfer of phosphoryl groups from the receiver domain phosphorylated by the HPK and the output RR. In contrast, the histidine within the Hpt domain is the initial site of phosphorylation in the chemotaxis HPK, CheA, and is phosphorylated using ATP as the phosphodonor [7]. Subsequently, the phosphoryl group is transferred from the histidine residue in the Hpt (P1) domain of CheA to an aspartate residue on either of two RRs, CheY or CheB [8]. In this study, we present the structure of the Hpt (P1) domain of CheA3 from R. sphaeroides in complex with its cognate RR CheY6, which, to our knowledge, is the first structure of a Hpt domain of a CheA protein in complex with its RR.
The chemotaxis pathway of Escherichia coli has been extensively characterized [9]–[12]. However, many bacteria have more complicated chemosensory pathways, employing multiple homologues of each of the chemosensory proteins [13]–[15]. R. sphaeroides has four CheA homologues and eight chemotaxis RR proteins (six CheYs and two CheBs) plus multiple homologues of the other E. coli chemotaxis proteins [13],[16]–[18]. Interestingly, the different CheAs show different phosphotransfer specificities for the CheY and CheB homologues [19]–[22]. CheA3 and CheA4 form a cluster with the soluble chemoreceptors in the cytoplasm, which is believed to sense the metabolic state of the cell [13],[23],[24]. CheA3 and CheA4 are unusual CheAs that lack some of the domains found in E. coli CheA [20],[25]. CheA4 has a P3 (dimerization) domain, a P4 (kinase) domain, and a P5 (regulatory) domain, whereas CheA3 has a P1 (Hpt) domain and a P5 (regulatory) domain separated by a 794-amino acid sequence that encodes a novel CheY-P phosphatase activity [26]. Neither CheA3 nor CheA4 is capable of autophosphorylation; instead, CheA4 phosphorylates CheA3 on H51 of the P1 (Hpt) domain. Once phosphorylated, CheA3-P serves as a specific phosphodonor for the chemotaxis RRs, CheY1, CheY6, and CheB2. However, it is unable to phosphorylate the other chemotaxis RRs, CheY2, CheY3, CheY4, CheY5, and CheB1 [20],[21],[27]. This phosphotransfer specificity must be determined by the interactions between the RRs and the P1 domain of CheA3 since the isolated P1 domain of CheA3 (CheA3P1) shows the same phosphotransfer specificity as full-length CheA3 [26].
In groundbreaking work, the Laub group used a mutual information bioinformatics approach to identify coevolving residues in HPKs and their cognate RRs [28]. They reasoned that many of the coevolving residues would be the specificity determinants for the phosphotransfer reaction and went on to show that it is possible to switch the RR substrate specificity of the HPK EnvZ by mutating these residues [28]. In this study, we use a structure-based approach to identify specificity determinants for the phosphotransfer reaction between CheA3P1-P and CheY6. By introducing these residues into the noncognate RRs, we have been able to change their kinase specificity, allowing them to be phosphorylated by CheA3P1-P.
Results
Structure of the CheA3P1.CheY6 Complex
In order to elucidate the molecular details of the interaction between CheA3 and CheY6, the complex structure of CheY6 and the unphosphorylated Hpt domain of CheA3 (residues 1–135) has been solved to 1.40 Å using Seleno single-wavelength anomalous dispersion (SAD) for phasing. Data collection and refinement statistics can be found in Table 1.
Table 1. Data collection and refinement statistics.
| Data Collection and Refinement | Statistics | Subcategory | SeMet | Unphosphorylated | Phosphorylated |
| Data collection | Resolution (Å) | 2.3 (2.38–2.30)a | 1.40 (1.45–1.40) | 2.80 (2.90–2.80) | |
| Space group | P21 | P21 | P1 | ||
| Cell dimension | a, b, c (Å) | 43.8, 62.7, 49.1 | 43.7, 62.0, 48.9 | 33.2, 43.4, 48.7 | |
| α, β, γ (°) | 90, 101.1, 90 | 90, 101.3, 90 | 78.8, 86.6, 80.9 | ||
| Redundancy | 9.5 (10.1) | 6.3 (3.7) | 1.8 (1.8) | ||
| Completeness (%) | 99.2 (99.0) | 99.7 (98.9) | 96.1 (94.5) | ||
| Rsym (%) | 8.9 (19.7) | 5.3 (14.8) | 12.0 (61.6) | ||
| Avg I/σ | 18.4 (12.9) | 31.6 (9.8) | 6.3 (1.2) | ||
| Refinement | Resolution | 38–1.40 | 42–2.80 | ||
| No. reflections | 50287 | 6232 | |||
| R work/R free | 17.0/20.7 | 24.1b/28.4b | |||
| No. atoms | Protein | 2040 | 1743 | ||
| Water | 272 | — | |||
| B-factors (Å2) | Protein | 28.5 | 86.5c | ||
| Water | 38.9 | — | |||
| r.m.s. deviations | Bond lengths (Å) | 0.010 | 0.008 | ||
| Bond angles (°) | 1.220 | 0.950 |
Numbers in parenthesis are for the highest resolution shell.
Rxpct, as described elsewhere [43].
Standard deviation of ±18 Å2.
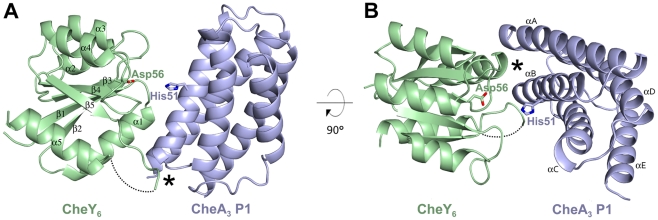
CheY6 has the typical (α/β)5 topology seen for E. coli CheY [29] and other structurally characterized RRs (Figure 1). Comparison with E. coli CheY reveals a high degree of structural conservation with a root mean square deviation (rmsd) of 1.4 Å over 115 Cα atoms (26% sequence identity). Although crystallized in the presence of Mn2+, there was no additional density in the CheY6 divalent cation binding site. However, the conformation of the metal coordinating residues Asp56, Asp9, and Asp10 is most similar to that found in the structure of Mg2+ bound CheY [30]. Only the backbone carbonyl of Glu58 is facing away rather than pointing towards the potential metal binding site (Figure S1). CheY6 has an elongated loop region connecting β5 and α5 (Figure 1). This loop comprises 13 residues (residues 107–119) in CheY6 compared to only three residues (109–111) in E. coli CheY. In the crystal, this loop is only partially ordered, suggesting that it is highly flexible. Residues 113–118 could not be traced and were omitted from the final model. The N-terminal region of this β5-α5 loop in combination with α1 of CheY6 form the vast majority of contacts to CheA3P1 (Figure 1).
Figure 1. Structure of the CheA3P1.CheY6 complex.
CheA3P1 is shown in light blue whereas CheY6 is in pale green. The phosphorylatable residue of CheA3P1, His51, and the phosphoacceptor on CheY6, Asp56, are shown in stick representation. Secondary structure elements are labelled in black. Residues 113–118 of CheY6 could not be traced and are depicted as a dotted line. (A) Side view onto the CheA3P1.CheY6 complex with the interaction between β5 and α5 in CheY6 and αB, and the following loop connecting αB and αC on CheA3P1, marked with an asterisk. (B) Top view showing the major site of interaction between the N-terminal end of α1 on CheY6 and αA/αB on CheA3P1 marked by an asterisk.
CheA3P1 forms a four-helix bundle (αA–αD) with an additional C-terminal helix (αE) (Figure 1). Despite low sequence identity, it is structurally very similar to previously determined CheA P1 structures [31]–[33]. Comparison with CheA P1 from Salmonella enterica serovar Typhimurium [32] and Thermotoga maritima [33] gives an rmsd of 1.2 Å over 116 Cα atoms (21% sequence identity) and 1.5 Å over 101 Cα (18% sequence identity), respectively. The site of phosphorylation, His51 in CheA3P1, is located on αB, in close proximity to the active site of the RR (Figure 1) with the phosphoacceptor Asp56 on CheY6 being 7.5Å from His51 on CheA3P1. Helices αA and αB face the RR and together with the αB–αC loop form the interface with CheY6 (Figure 1).
Structure of Phosphorylated CheA3P1 in Complex with CheY6(D56N, S83A)
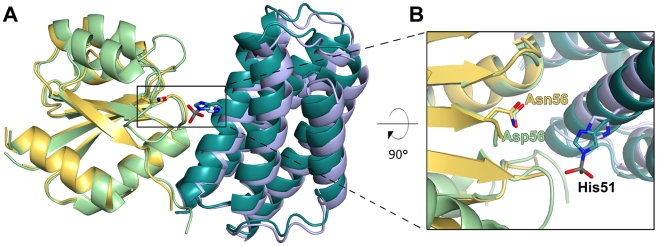
To show that the conclusions drawn from the unphosphorylated structure are also valid for the physiologically relevant, phosphorylated complex, we solved the structure of phosphorylated CheA3P1 in complex with CheY6. Since the rapid rate of phosphotransfer between CheA3P1-P and CheY6 does not allow crystallization of the wild-type, phosphorylated complex, we formed a stable, complex by introducing two substitutions, D56N and S83A, in the active site of CheY6 (see Materials and Methods for details). These substitutions have previously been shown to abolish phosphotransfer from CheA3P1-P [19]. The structure was solved to 2.8 Å by molecular replacement using the unphosphorylated structure as model. Data collection and refinement statistics can be found in Table 1. The structure shows clear additional density adjacent to Nε2 of His51 of CheA3P1 in agreement with phosphorylation of this residue (Figure S2). Due to the moderate resolution of the analysis, residues 60–65, 85–97, and 111–121 on CheY6 could not be traced and were not included in the final model.
Compared to the unphosphorylated complex, CheA3P1-P undergoes a rigid body translation of 2.1 Å relative to the RR (Figure 2A). This realignment of CheA3P1-P positions the phosphorylated His51 closer to the phosphoacceptor on CheY6, Asp56 (Figure 2B). The phosphorylated His51 is facing away from the active site of CheY6; however, a 180° flip of this side chain would put it in near linear geometry and place the phosphoryl group within 4.5 Å of Asp56 of CheY6 (Figure 2B). The orientation of the phosphorylated His51 seen in the crystal structure is likely caused by the lack of a divalent cation in the metal binding site of the RR leading to electrostatic repulsion from the CheY6 active site.
Figure 2. Superposition of the CheA3P1.CheY6 complex in the phosphorylated and unphosphorylated conformations.
Structures were aligned onto the RR. Colour coding for the unphosphorylated conformation as in Figure 1; for the phosphorylated conformation, CheY6 is shown in yellow and CheA3 P1 in teal. The active site residues His51 (CheA3 P1) and Asp56/Asn56 (CheY6 unphosphorylated/phosphorylated complex conformation) are shown in stick representation. (A) Overview highlighting the 2.1 Å rigid body translation of CheA3 P1 towards CheY6. (B) Close-up view onto the active site showing the movement of His51 towards Asp56/Asn56.
The difference in binding affinity for CheY6 between CheA3 and CheA3-P is not known. In E. coli, the difference in Kd values for CheY between CheA and CheA-P is relatively small, with both values being in the low micromolar range [34]. Consistent with this, we find very little change in the total buried surface area between the phosphorylated (605 Å2) and unphosphorylated (530 Å2) structures of CheA3P1 complexed with CheY6. The only difference is a hydrogen bond formed between Glu58 of CheY6 and His51 of CheA3P1 in the unphosphorylated complex that is released in the phosphorylated complex. As this is not contributing to the interface of the physiologically important complex, it is therefore not discussed further. Because the interfaces in the phosphorylated and unphosphorylated complex are highly similar, the following discussion of the interactions between CheA3P1 and CheY6 are based on the high-resolution, unphosphorylated structure.
Interactions between CheA3P1 and CheY6
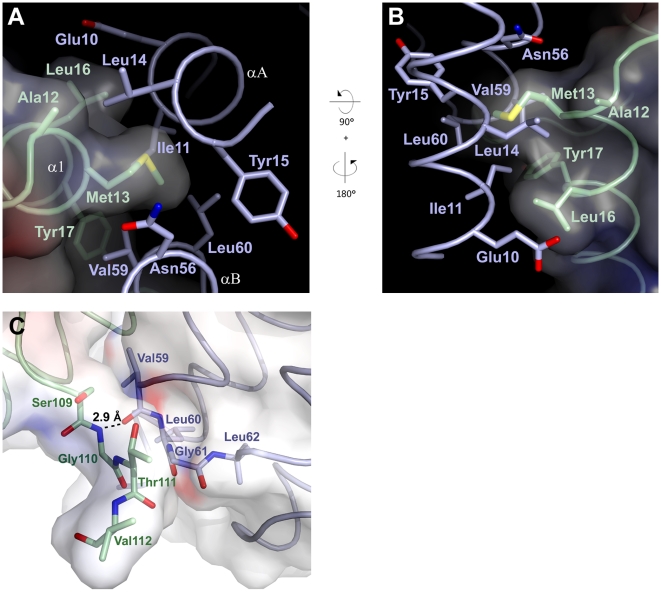
The interface between CheA3P1 and CheY6 is dominated by hydrophobic interactions with only one hydrogen bond and no salt bridges formed between the two proteins. The buried surface area of the interface is small (530 Å2), indicative of a weak interaction, consistent with the transient nature of the complexes [35]. A continuous interface is formed by two sites of interaction complemented by a hydrogen bond formed between Ser83 on CheY6 and Arg58 on CheA3P1. One of these sites lies between the elongated loop region between β5 and α5 in CheY6 and αB and the following loop connecting αB and αC on CheA3P1 (Figure 1A). The other site lies between the N-terminal end of α1 on CheY6 and αA/αB on CheA3P1 (Figure 1B). The latter is the major binding site including ∼70% of the total buried surface area (358 Å2).
In this region, helices αA and αB of CheA3P1 form a hydrophobic pocket comprising Ile11, Leu14, and Tyr15 on αA, and Asn56, Val59, and Leu60 on αB (Figure 3A and 3B). This pocket is situated adjacent to α1 on CheY6 and in ideal position to accommodate Met13 on the N-terminal region of this helix. This Met finger is protruding from α1, and its side chain fits snugly into the hydrophobic pocket, with 99.7% of its accessible surface area being buried (Figure 3B). The hydrophobic interaction is complemented by Ala12, Leu16, and Tyr17 of α1 on CheY6, all facing the same side on CheA3P1 as Met13 (Figure 3A). Ala12 of CheY6 extends the hydrophobic interface by interaction with Leu14 of CheA3P1, whereas Leu16 of CheY6 interacts with Ile11 and the aliphatic part of Glu10 on αA of CheA3P1. Tyr17 extends the binding surface towards the β5-α5 loop by interacting with Val59 at the C-terminus of αB. The elongated loop between β5 and α5 of CheY6 forms van der Waals interactions with Ser109, Gly110, and Thr111 of CheY6 contacting Ile59, Gly61, and Ser63 on αB and the following loop region on CheA3P1 (Figure 3C). A main chain hydrogen bond is formed between Gly110 and Val59.
Figure 3. Close-up view of the two binding sites between CheA3P1 and CheY6.
Colour coding as in Figure 1. Residues that are involved in the interaction are shown in stick representation. (A) Detailed view onto the major binding site between CheY6 and CheA3P1. Orientation as in Figure 1B, residues 19–53 of CheA3 P1 are omitted for clarity. (B) Solvent accessible surface of CheY6 coloured by electrostatic potential contoured at ±10 kT. Met13 fits snugly into the hydrophobic pocket on CheA3P1. (C) Close-up view onto the second binding site. Orientation as in Figure 1A. The solvent-accessible surface area is coloured by electrostatic potential contoured at ±10 kT. The hydrogen bond between Gly110 of CheY6 and Val59 of CheA3P1 is marked with a dotted line.
Of these interactions, the Met13 finger has the largest contribution towards the binding interface on CheY6, accounting for almost a third (156 Å2) of the total buried surface area. Together with Ala12, Leu16, and Tyr17, it is accounting for over 60% (321 Å2) of the interface. On CheA3P1, the residues involved in the interaction with CheY6 are slightly less clustered and spatially farther apart. Yet the three residues with the biggest contribution towards the total buried surface area, Val59 (103 Å2, 20% of total buried surface area), Leu14 (64 Å2, 12%), and Ile11 (48 Å2, 9%), together account for almost half of the overall binding surface.
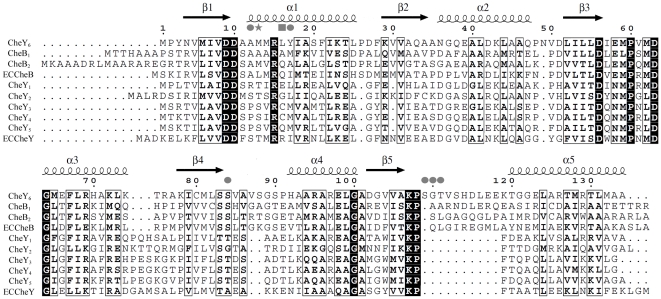
This analysis shows that only a small number of residues are necessary to form the majority of interactions within the complex. On CheY6, these are all located on α1. Within this helix, Met13 has the most crucial role in the hydrophobic interaction. A sequence alignment of all identified RRs in R. sphaeroides shows that only CheY6 and CheB2 have a Met residue at position 13, whereas all others either have a Ser, Thr, or Ala at this position (Figure 4). As CheA3-P serves as a phosphodonor for both CheY6 and CheB2 this suggests that Met13 has an important role in determining the specificity for binding to CheA3P1.
Figure 4. Structure-guided sequence alignment of the chemotaxis RRs from E. coli and R. sphaeroides.
Alignment is based on the structures of E. coli CheY (PDB code: 3CHY), E. coli CheB (1A2O), R. sphaeroides CheY3 (C. H. Bell, unpublished data), and R. sphaeroides CheY6. Secondary structure is shown for CheY6. Residues involved in binding of CheY6 to CheA3P1 are marked with a star (contributes >30% to total buried surface area), square (>15%), or circle (>5%).
Substitution of M13 in CheY6 Reduces Interaction and the Rate of Phosphotransfer from CheA3P1-P
Residues 11–17 of CheY6 collectively account for ∼67% of the buried surface area of CheY6 in the CheA3P1-CheY6 complex, with M13, L16, and Y17 each contributing ∼30%, ∼16%, and ∼8%, respectively. To confirm that this surface of CheY6 is involved in the interactions with CheA3P1-P that lead to phosphotransfer in solution, we substituted residues at this surface to mimic those found in the noncognate RRs, CheY3 and CheY4. M13 was changed to S, and Y17 was changed to M as found in both CheY3 and CheY4. CheY3 and CheY4 both have C at position 16; however, it has previously been shown that CheY3(C16S) and CheY4(C16S) behave indistinguishably from CheY3 and CheY4 in phosphotransfer assays (S. L. Porter, unpublished data), and therefore, to avoid any potential problems from disulphide bond formation during the purification, we changed L16 to S rather than C. Two mutant proteins were produced, CheY6(M13S) and CheY6(M13S,L16S,Y17M).
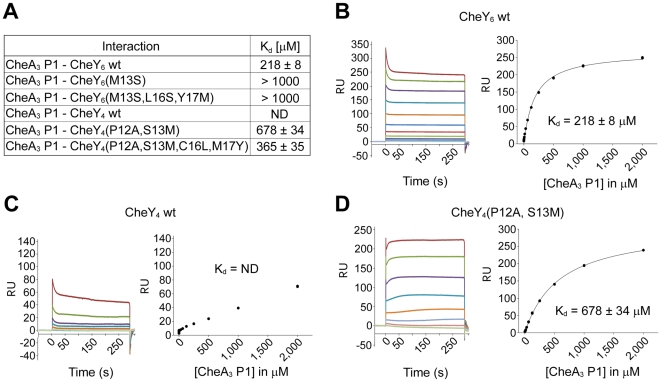
Surface plasmon resonance (SPR) assays showed that wild-type CheY6 binds to CheA3P1 with an affinity of 218 µM (Figure 5). This weak interaction is in agreement with the transient nature of the complex and the small buried surface area between the two proteins as observed in the crystal structure. Both the single-mutant protein, CheY6(M13S), and the triple-mutant protein CheY6(M13S,L16S,Y17M) showed a remarkable decrease in affinity, with a Kd of more than 1 mM (Figure 5 and Figure S3).
Figure 5. Binding of CheA3P1 to the response regulators.
(A) Table of binding constants (Kd) measured by SPR between CheA3P1 and CheY4, CheY6 and their mutant versions. Data are expressed as mean ± standard error of the mean (s.e.m.). ND, not determinable. (B–D) Binding of CheA3P1 to CheY6 wild type (wt), CheY4 wt, and CheY4(P12A,S13M). Left, representative sets of experimental sensorgrams from typical equilibrium-based binding experiments, with reference subtraction. Different concentrations of CheA3P1 were injected over surfaces coupled with the respective RR. For all injections, the experimental traces reached equilibrium and returned to baseline after the injection. Right, plot of the equilibrium binding response (response units [RU]) against CheA3P1 concentration ranging from 120 nM to 2 mM. Within one experiment, each concentration was measured twice. All experiments were performed in duplicate. Best-fit binding curves corresponding with a 1∶1 binding model are shown as lines.
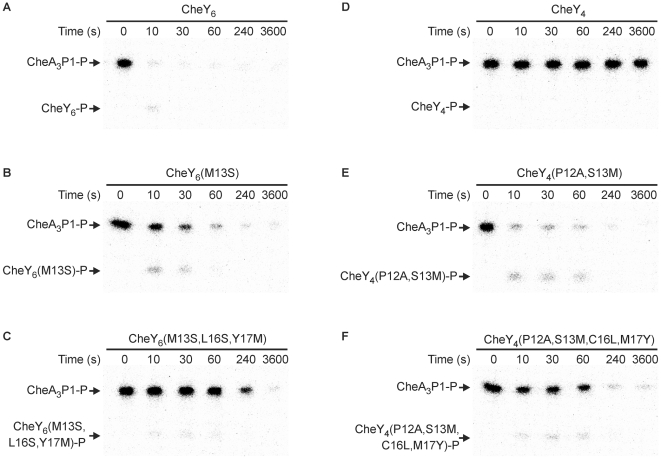
Consistent with the SPR binding assays, the rate of phosphotransfer from CheA3P1-P to each of the two mutant CheY6 proteins was much slower than to wild-type CheY6 (Figure 6A–6C), with phosphotransfer to the triple-mutant protein CheY6(M13S,L16S,Y17M) being slowest. These results underline the conclusions drawn from the structural characterization of the complex and stress the importance of the Met finger at position 13 for binding of CheY6 to CheA3P1. In addition, the hydrophobic interaction mediated by Leu16 and Tyr17 also adds significantly to recognition of CheY6 by CheA3P1.
Figure 6. Changing the phosphotransfer specificity of CheY6 and CheY4.
Phosphorimages of SDS-PAGE gels measuring phosphotransfer from CheA3P1-P to (A) CheY6, (B) CheY6(M13S), (C) CheY6(M13S,L16S,Y17M), (D) CheY4, (E) CheY4(P12A,S13M), and (F) CheY4(P12A,S13M,C16L,M17Y). CheY (5 µM) was added to 1 µM CheA3P1-32P. Ten-microlitre reaction samples were taken at the time points indicated and quenched in 20 µl of 1.5× SDS/EDTA loading dye. The quenched samples were analyzed by SDS-PAGE and detected by phosphorimaging.
Reengineering CheY4 into a Cognate RR of CheA3P1-P
The sequence of CheY4 is 36% identical to CheY6; however, unlike CheY6, CheY4 is not a cognate RR of CheA3P1-P (Figure 6D). Consistent with this, SPR assays failed to detect a significant interaction between CheA3P1 and CheY4 (Figure 5). In an attempt to alter the phosphotransfer specificity of CheY4 to allow phosphotransfer from CheA3P1-P, we substituted the CheY4 residues corresponding to the positions shown to interact in the CheA3P1-CheY6 structure, so that they matched CheY6. Two mutant proteins were produced: CheY4(P12A,S13M) and CheY4(P12A,S13M,C16L,M17Y). Ala12 was included in both mutant proteins since CheY4 has a Pro at position 12 that might influence the orientation of α1 and thus interfere with the proper positioning of Met13 for interaction with the hydrophobic pocket. SPR assays showed that both mutant proteins bind CheA3P1 (Figure 5 and Figure S3), and in phosphotransfer assays, both mutant proteins were phosphorylated by CheA3P1-P (Figure 6E and 6F). Phosphotransfer was fastest from CheA3P1-P to CheY4(P12A,S13M), with most of the initial CheA3P1-P dephosphorylated within 10 s (Figure 6E).
Interestingly, although the rate of phosphotransfer from CheA3P1-P to CheY4(P12A,S13M) was faster than to CheY4(P12A,S13M,C16L,M17Y), the SPR results show that CheA3P1 interacts slightly more strongly with CheY4(P12A,S13M,C16L,M17Y) than with CheY4(P12A,S13M) (Figure 5 and Figure S3). This apparent discrepancy could be explained by the alignment of the phosphorylated histidine and the phosphorylatable aspartate in the CheY4(P12A,S13M,C16L,M17Y).CheA3P1-P complex, which might be slightly less optimal for catalysis than in the CheY4(P12A,S13M).CheA3P1-P complex. Nevertheless, both methods show that mutating CheY4 so that it resembles CheY6 at the contact sites with CheA3P1 enhances both binding affinity and phosphotransfer rate. These results demonstrate that substitution of just two residues is sufficient to change the phosphotransfer specificity of CheY4.
Reengineering Other CheYs
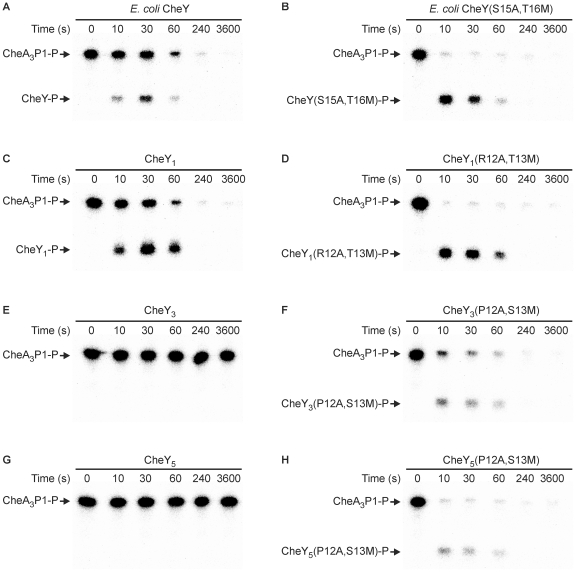
Having successfully reengineered the phosphotransfer specificity of CheY4 by introducing A12 and M13, we used the same approach to change the specificity of CheY1, CheY3, CheY5, and E. coli CheY (Figure 7). These proteins share between 30% and 33% sequence identity with CheY6. In all cases, the mutant proteins containing the alanine and methionine substitutions were phosphorylated more rapidly by CheA3P1-P than were their wild-type counterparts. Although CheY1 and E. coli CheY both lack a methionine residue at the position corresponding to M13 of CheY6, they are both phosphorylated by CheA3P1-P (Figure 7A and 7C); however, phosphotransfer to their corresponding alanine and methionine substitution mutant proteins proceeded even faster, with almost complete dephosphorylation of CheA3P1-P within 10 s (Figure 7B and 7D). Similar to CheY4, wild-type CheY3 and CheY5 were not phosphorylated by CheA3P1-P (Figure 7E and 7G), and the effect of the alanine and methionine substitutions was to allow rapid phosphotransfer from CheA3P1-P (Figure 7F and 7H). These results demonstrate that the RR residues at the positions equivalent to A12 and M13 of CheY6 play a major role in determining phosphotransfer specificity.
Figure 7. Changing the phosphotransfer specificity of other CheYs by introduction of A12 and M13.
Phosphorimages of SDS-PAGE gels measuring phosphotransfer from CheA3P1-P to (A) E. coli CheY, (B) E. coli CheY(S15A,T16M), (C) CheY1, (D) CheY1(R12A,T13M), (E) CheY3, (F) CheY3(P12A,S13M), (G) CheY5, and (H) CheY5(P12A,S13M). CheY (5 µM) was added to 1 µM CheA3P1-32P. Ten-microlitre reaction samples were taken at the time points indicated and quenched in 20 µl of 1.5× SDS/EDTA loading dye. The quenched samples were analyzed by SDS-PAGE and detected by phosphorimaging.
Discussion
Two-component signalling systems depend on a high level of selectivity between histidine kinases and their cognate RRs to prevent cross-talk between different pathways. Here, we present structural and functional data that elucidate how this specificity is mediated on a molecular level. We identified the key residues for molecular recognition and thereby were able to reengineer phosphotransfer specificity. Considering the high level of structural homology within the receiver and Hpt domain families, the presented structure together with complementary information, e.g., analysis of amino acid covariation [28],[36],[37], is likely to facilitate the rational reengineering of CheA-like histidine kinase–RR pairs for use, for example, in synthetic signalling circuits.
Conserved Features of the CheA3P1.CheY6 Complex
The structure of CheA3P1 in complex with CheY6 is, to our knowledge, the first structure of a Hpt domain of a CheA protein in complex with a RR. To date, there are only two other structures of a Hpt domain in complex with a RR, namely Spo0B/Spo0F [38],[39] from Bacillus subtilis and YPD1/SLN1 [40],[41] from Saccharomyces cerevisiae. Although the overall structure of the RR is well conserved in all three complexes, there are remarkable differences in the Hpt domains. Spo0B exists as a dimer in which the two α-helices of the helical hairpin domain of each protomer associate to form a four-helix bundle. In addition, Spo0B has a C-terminal domain with an α/β fold, which is involved in binding of the RR. YPD1 is a monomeric Hpt protein consisting of a four-helix bundle with an additional short helix at the N-terminus. This N-terminal helix is involved in RR binding and complements the interface formed by helices αB and αC. When the three complexes were superimposed with respect to the receiver domains, CheA3P1 aligned well with YPD1 but only poorly with Spo0B (Figure S4).
Phosphotransfer Specificity Determinants
CheA3P1.CheY6 shows the smallest binding interface amongst the three complexes solved so far, with 530 Å2 compared to 953 Å2 in YPD1/SLN1 and 1,200 Å2 in Spo0F/Spo0B. The interface is smaller since CheA3P1 has neither the C-terminal α/β fold domain seen in Spo0B nor the additional N-terminal helix seen in YPD1. Despite this small interface, CheA3P1 shows binding and efficient phosphotransfer to CheY6. It was previously proposed that the P2 domain of CheA-like proteins might be necessary for binding of the RR to CheA by increasing the binding interface [40]. However, CheA3 does not have a P2 domain, and the experimental data presented here and elsewhere [26] show that the P1 domain alone is sufficient for binding of and specific phosphotransfer to the cognate RR. Moreover, the structural and mutational analysis suggests that their interaction is mediated by very distinct residue clusters on the RR and the Hpt domain, the former being located on the N-terminal region of α1 and the loop region between β5 and α5, and the latter being located on the N- and C-terminal region of αA and αB, respectively, and the loop region connecting αB and αC. This is in good agreement with a recently published computational analysis of amino acid coevolution of cognate histidine kinase–RR pairs [28] and the binding site found in the YPD1/SLN1 [40],[41] or Spo0B/Spo0F [38],[39] complex.
Reengineering Phosphotransfer Specificity
The specificity of protein–protein interactions is essential for most cellular processes. Despite the vast number of these interactions, our understanding of the molecular basis of their specificity is limited. The structure presented here has allowed us to identify specificity determinants for the CheA–CheY interaction. We successfully used this information to redesign noncognate RRs to allow them to be rapidly phosphorylated by CheA3P1-P (Figures 6 and 7). Whereas the Laub group have reengineered a HPK to specifically phosphorylate non-cognate RR substrates [28], we have now shown that it is possible to rationally reengineer a RR so that it can be phosphorylated by a noncognate HPK. The changing of phosphotransfer specificity described here represents, to our knowledge, the first example of the redesign of the intracellular part of the chemotaxis pathway and provides valuable insight into how cells mediate specificity in one of the most abundant signal transduction pathways, two-component signalling. The ability to reengineer phosphotransfer specificity coupled with recent work from the Bourret group [42], which has shown that RR autodephosphorylation rate can be manipulated, provide a platform for the future design of synthetic two-component circuits with customizable kinetics.
Materials and Methods
Protein Overexpression and Purification
CheA3P1 (residues 1–135 of CheA3, GenBank ID 3720125), CheY6 (GenBank ID 3720126), and CheY4 (GenBank ID 3722004) were cloned into the bacterial expression vector pQE80 (Qiagen), which includes an N-terminal His6-tag. Sequence verified plasmids were transformed into M15pREP4 cells (Qiagen) and cultivated in Terrific Broth to an absorbance at 600 nm (A600) of 0.8. Cultures were cooled to 20°C, induced with 0.25 mM IPTG, and then grown for ∼15 h before harvesting. The bacterial pellets were resuspended in 25 mM sodium phosphate (pH 8.0), 500 mM NaCl, 0.5 mM β-mercaptoethanol, and EDTA-free protease inhibitor cocktail (Roche). Cells were lysed using a Basic Z model cell disruptor (Constant Systems) and fractionated by centrifugation (45,000g, 4°C, 60 min). CheA3P1, CheY6, and CheY4 were purified from the supernatant by immobilized metal-affinity chromatography [19]–[21]. The samples were dialysed against standard buffer (30 mM Hepes [pH 7.5], 150 mM NaCl, 5 mM sodium acetate, 2 mM manganese chloride, 2 mM Tris(2-carboxyethyl)phosphine [TCEP]) and further purified by size-exclusion chromatography. Protein purity and concentration was measured as described [43]. Purified proteins were stored at −80°C. Seleno-methionine (SeMet)-labelled CheY6 was produced essentially as described [44]. The complex between CheY6 and CheA3P1 was formed by mixing equimolar amounts of both proteins. The sample was incubated for 30 min at room temperature (20°C), and the complex was purified by size-exclusion chromatography. CheA3P1 was phosphorylated using ATP and CheA4, and purified as described previously [26]. The final preparation of CheA3P1-P was free of ATP and CheA4. Phosphorylation was verified by mass spectrometry. Equimolar amounts of phosphorylated CheA3P1 and CheY6(D56N,S83A) were mixed and set up for crystallization.
Site-Directed Mutagenesis
Mutations were introduced using the Quikchange Site-Directed Mutagenesis Kit (Stratagene).
Crystallization and Data Collection
Crystal trials were set up with the purified CheA3P1.CheY6 complex at a concentration of 20 mg/ml. We set up nanoliter crystallization trials using a Cartesian Technologies robot (100 nl of protein solution plus 100 nl of reservoir solution) in 96-well Greiner plates [45], placed them in a TAP (The Automation Partnership) Homebase storage vault maintained at 295 K, and imaged them via a Veeco visualization system. Crystals of the phosphorylated, unphosphorylated and SeMet-labelled complex between CheY6 and CheA3P1 could be obtained in 100 mM Bicine (pH 9), 1 M LiCl, 20% (w/v) PEG6000. Crystals were optimized using a dilution series of the initial condition. Crystals reached their final size of 400×100×100 µm3 after 12 h.
Diffraction data were collected at 100 K, crystals being flash-cooled in a cryo N2 gas stream. Before flash-freezing, crystals were cryo-protected with perfluoropolyether oil PFO-X125/03 (Lancaster Synthesis). The native dataset was collected at beamline I03 (λ = 0.9757 Å) and the SeMet dataset at beamline I04 (λ = 0.979 Å) at the Diamond Light Source. Data for the phosphorylated complex were collected on a MAR345 detector (Marresearch) mounted on a Miromax007 generator (Rigaku), equipped with Varimax HR Osmic mirrors (Rigaku). X-ray data were processed and scaled with the HKL suite [46]. Data collection statistics are shown in Table 1.
Structure Determination and Refinement
The CheA3P1-CheY6 complex structure was determined by SAD analysis. The positions of 12 selenium atoms were determined by using SHELXD [47]. This solution was put into AUTOSHARP [48] for phase calculation, improvement, and phase extension using the high-resolution native data to 1.4 Å resolution. The resulting map was of high quality and allowed tracing of the whole polypeptide chain. An initial model was built automatically using Arp/wARP [49] and manually adjusted using COOT [50]. The structure was refined using autoBUSTER [51], REFMAC [52], and Phenix [53]. The phosphorylated complex was solved by molecular replacement using Phaser [54] with the unphosphorylated complex as model.
Refinement statistics are given in Table 1; all data within the indicated resolution range were included. Stereochemical properties were assessed by MOLPROBITY [55] and PROCHECK [56]. Ramachandran statistics are as follows (favored/disallowed, %): CheA3P1-CheY6 unphosphorylated 98.8/0, CheA3P1-CheY6 phosphorylated 99.1/0. Superpositions were calculated using lsqkab [57] implemented in the CCP4 suite and electrostatic potentials were generated using APBS [58]. Buried surface areas of protein–protein interactions were calculated using the PISA Webserver (http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html). Coordinates are deposited in RSCB Data Bank under 3KYI and 3KYJ.
Surface Plasmon Resonance Binding Studies
SPR experiments were performed using a Biacore T100 machine (GE Healthcare) at 25°C in standard buffer supplemented with 0.05% (v/v) Tween 20. Protein concentrations were determined from the absorbance at 280 nm using calculated molar extinction coefficients. Proteins for surface attachment were enzymatically biotinylated within an engineered C-terminal tag. These proteins were then attached to surfaces on which 3,000 response units (RU) of streptavidin were coupled via primary amines [59] yielding a density of 200–1,500 RU of biotinylated protein. All experiments were done in duplicates with independently purified proteins. The signal from experimental flow cells was corrected by subtraction of a blank and reference signal from a mock or irrelevant protein-coupled flow cell. In all experiments analyzed, the experimental trace returned to baseline after each injection and the data fitted to a simple 1∶1 Langmuir model of binding. K d values were obtained by nonlinear curve fitting of the Langmuir binding isotherm (bound = Cmax/(K d + C), where C is analyte concentration and max is the maximum analyte binding) evaluated using the Biacore Evaluation software (GE Healthcare).
Detection of Phosphotransfer from CheA3P1-P to the RRs
Assays were performed at 20°C in TGMNKD buffer (50 mM Tris HCl, 10% [v/v] glycerol, 5 mM MgCl2, 150 mM NaCl, 50 mM KCl, 1 mM DTT [pH 8.0]). CheA3P1 was phosphorylated using [γ-32P] ATP and CheA4, and purified as described previously [26]. The final preparation of CheA3P1-32P was free of ATP and CheA4. CheA3P1-32P (1 µM) was mixed with 5 µM RR in a final reaction volume of 100 µl. Following the addition of RR, reaction aliquots of 10 µl were taken at the indicated time points and quenched immediately in 20 µl of 1.5× SDS-PAGE loading dye (3.75% [w/v] SDS, 45 mM EDTA, 18.75 mM Tris HCl, 18.75% [v/v] glycerol, 1.5% [v/v] β-mercaptoethanol [pH 6.8]). Quenched samples were analyzed using SDS-PAGE and phosphorimaging as described previously [21].
Supporting Information
Stereoview of a superposition of the metal binding site of CheY6 and E. coli CheY. CheY6 is shown in pale green, E. coli CheY with Mg2+ bound (PDB-code: 1CHN) in yellow, and E.coli CheY without Mg2+ (3CHY) in teal. Structures were aligned on their secondary structure elements using secondary structure matching (SSM) implemented in COOT [50]. Residues involved in the coordination of Mg2+ in E. coli CheY are shown in stick representation for all three structures. Residues in E. coli structures are only labelled for the Mg2+ bound form. Mg2+ from the E. coli structure is shown as a grey sphere. CheY6 resembles the Mg2+ bound form of E. coli CheY despite not having a divalent cation bound to its metal binding site.
(6.24 MB TIF)
Electron density of phosphorylated His51 in CheA3P1 in the active complex structure. The orientation is similar to Figure 2B. The density represents a 2Fobs-Fcalc map contoured at 1.5 σ and calculated after initial rigid body refinement in Phaser [54] and one round of positional refinement in autoBUSTER [51], both omitting the phosphate group from the model.
(0.64 MB TIF)
Binding of CheA3P1 to substitution mutants of CheY4 and CheY6. (A–C) Binding of CheA3P1 to CheY4(P12A,S13M,C16L,M17Y), CheY6(M13S), and CheY6(M13S,L16S,Y17M). Left, representative sets of experimental sensorgrams from typical equilibrium-based binding experiments, with reference subtraction. Different concentrations of CheA3P1 were injected over surfaces coupled with the respective RR. For all injections, the experimental traces reached equilibrium and returned to baseline after the injection. Right, plot of the equilibrium binding response (response units [RU]) against CheA3P1 concentration ranging from 120 nM to 2 mM. Within one experiment, each concentration was measured twice. All experiments were performed in duplicate. Best-fit binding curves corresponding with a 1∶1 binding model are shown as lines. Experiments with CheY6(M13S) and CheY6(M13S,L16S,Y17M) did not reach saturation due to very low affinity, thus the Kd value is estimated.
(0.52 MB TIF)
Superposition of the CheA3P1-CheY6 complex structure with the Spo0B-Spo0F complex and YPD1-SLN1 complex. (A) Superposition of CheA3P1-CheY6 with Spo0B-Spo0F (PDB-code: 1F51). (B) Superposition of CheA3P1-CheY6 with YPD1-SLN1 (1OXB). All structures were superimposed on the RRs using SSM as implemented in COOT [50]. CheY6 is shown in pale green, CheA3P1 in light blue, Spo0F in orange, Spo0B in teal, SLN1 in purple, and YPD1 in aquamarine. Only the four-helix bundle of the Spo0B dimer is shown; the C-terminal domain is omitted for clarity. The orientation is similar to Figure 1. CheA3P1 is structurally more similar to the monomeric YPD1 than to Spo0B.
(2.64 MB TIF)
Acknowledgments
We thank the staff of Diamond beamlines I03 and I04 for assistance with data collection, T. Walter for help with crystallization, and K. Harlos for help with data collection. We acknowledge the use of the crystallization facilities provided by the Medical Research Council (MRC)–funded Oxford Protein Production Facility (OPPF).
Abbreviations
- RR
response regulator
- SPR
surface plasmon resonance
Footnotes
The authors have declared that no competing interests exist.
This study was funded by the Wellcome Trust and the BBSRC (Biotechnology and Biological Sciences Research Council). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.West A. H, Stock A. M. Histidine kinases and response regulator proteins in two- component signaling systems. Trends Biochem Sci. 2001;26:369–376. doi: 10.1016/s0968-0004(01)01852-7. [DOI] [PubMed] [Google Scholar]
- 2.Stock A. M, Robinson V. L, Goudreau P. N. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 3.Laub M. T, Goulian M. Specificity in two-component signal transduction pathways. Ann Rev Genet. 2007;41:121. doi: 10.1146/annurev.genet.41.042007.170548. [DOI] [PubMed] [Google Scholar]
- 4.Siryaporn A, Goulian M. Cross-talk suppression between the CpxA-CpxR and EnvZ-OmpR two-component systems in E. coli. Mol Microbiol. 2008;70:494–506. doi: 10.1111/j.1365-2958.2008.06426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Skerker J. M, Prasol M. S, Perchuk B. S, Biondi E. G, Laub M. T. Two-component signal transduction pathways regulating growth and cell cycle progression in a bacterium: a system-level analysis. PLoS Biology. 2005;3:e334. doi: 10.1371/journal.pbio.0030334. doi: 10.1371/journal.pbio.0030334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casino P, Rubio V, Marina A. Structural insight into partner specificity and phosphoryl transfer in two-component signal transduction. Cell. 2009;139:325–336. doi: 10.1016/j.cell.2009.08.032. [DOI] [PubMed] [Google Scholar]
- 7.Hess J. F, Bourret R. B, Simon M. I. Histidine phosphorylation and phosphoryl group transfer in bacterial chemotaxis. Nature. 1988;336:139–143. doi: 10.1038/336139a0. [DOI] [PubMed] [Google Scholar]
- 8.Bourret R. B, Hess J. F, Simon M. I. Conserved aspartate residues and phosphorylation in signal transduction by the chemotaxis protein CheY. Proc Natl Acad Sci U S A. 1990;87:41–45. doi: 10.1073/pnas.87.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tindall M. J, Maini P. K, Porter S. L, Armitage J. P. Overview of mathematical approaches used to model bacterial chemotaxis II: bacterial populations. Bull Math Biol. 2008;70:1570–1607. doi: 10.1007/s11538-008-9322-5. [DOI] [PubMed] [Google Scholar]
- 10.Tindall M. J, Porter S. L, Maini P. K, Gaglia G, Armitage J. P. Overview of mathematical approaches used to model bacterial chemotaxis I: the single cell. Bull Math Biol. 2008;70:1525–1569. doi: 10.1007/s11538-008-9321-6. [DOI] [PubMed] [Google Scholar]
- 11.Hazelbauer G. L, Falke J. J, Parkinson J. S. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem Sci. 2008;33:9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baker M. D, Wolanin P. M, Stock J. B. Signal transduction in bacterial chemotaxis. Bioessays. 2006;28:9–22. doi: 10.1002/bies.20343. [DOI] [PubMed] [Google Scholar]
- 13.Porter S. L, Wadhams G. H, Armitage J. P. Rhodobacter sphaeroides: complexity in chemotactic signalling. Trends Microbiol. 2008;16:251–260. doi: 10.1016/j.tim.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 14.Wadhams G. H, Armitage J. P. Making sense of it all: bacterial chemotaxis. Nat Rev Mol Cell Bio. 2004;5:1024–1037. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- 15.Hamer R, Chen P-Y, Armitage J. P, Reinert G, Deane C. M. Deciphering chemotaxis pathways using cross species comparisons. BMC Syst Biol. 2010;4:3. doi: 10.1186/1752-0509-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.del Campo A. M, Ballado T, de la Mora J, Poggio S, Camarena L, et al. Chemotactic control of the two flagellar systems of Rhodobacter sphaeroides is mediated by different sets of CheY and FliM proteins. J Bacteriol. 2007;189:8397–8401. doi: 10.1128/JB.00730-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shah D. S, Porter S. L, Harris D. C, Wadhams G. H, Hamblin P. A, et al. Identification of a fourth cheY gene in Rhodobacter sphaeroides and interspecies interaction within the bacterial chemotaxis signal transduction pathway. Mol Microbiol. 2000;35:101–112. doi: 10.1046/j.1365-2958.2000.01680.x. [DOI] [PubMed] [Google Scholar]
- 18.Shah D. S. H, Porter S. L, Martin A. C, Hamblin P. A, Armitage J. P. Fine tuning bacterial chemotaxis: analysis of Rhodobacter sphaeroides behaviour under aerobic and anaerobic conditions by mutation of the major chemotaxis operons and cheY genes. EMBO J. 2000;19:4601–4613. doi: 10.1093/emboj/19.17.4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Porter S. L, Wadhams G. H, Martin A. C, Byles E. D, Lancaster D. E, et al. The CheYs of Rhodobacter sphaeroides. J Biol Chem. 2006;281:32694–32704. doi: 10.1074/jbc.M606016200. [DOI] [PubMed] [Google Scholar]
- 20.Porter S. L, Armitage J. P. Chemotaxis in Rhodobacter sphaeroides requires an atypical histidine protein kinase. J Biol Chem. 2004;279:54573–54580. doi: 10.1074/jbc.M408855200. [DOI] [PubMed] [Google Scholar]
- 21.Porter S. L, Armitage J. P. Phosphotransfer in Rhodobacter sphaeroides chemotaxis. J Mol Biol. 2002;324:35–45. doi: 10.1016/s0022-2836(02)01031-8. [DOI] [PubMed] [Google Scholar]
- 22.Martin A. C, Wadhams G. H, Shah D. S. H, Porter S. L, Mantotta J. C, et al. CheR- and CheB-dependent chemosensory adaptation system of Rhodobacter sphaeroides. J Bacteriol. 2001;183:7135–7144. doi: 10.1128/JB.183.24.7135-7144.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wadhams G. H, Warren A. V, Martin A. C, Armitage J. P. Targeting of two signal transduction pathways to different regions of the bacterial cell. Mol Microbiol. 2003;50:763–770. doi: 10.1046/j.1365-2958.2003.03716.x. [DOI] [PubMed] [Google Scholar]
- 24.Wadhams G. H, Martin A. C, Porter S. L, Maddock J. R, Mantotta J. C, et al. TlpC, a novel chemotaxis protein in Rhodobacter sphaeroides, localizes to a discrete region in the cytoplasm. Mol Microbiol. 2002;46:1211–1221. doi: 10.1046/j.1365-2958.2002.03252.x. [DOI] [PubMed] [Google Scholar]
- 25.Porter S. L, Warren A. V, Martin A. C, Armitage J. P. The third chemotaxis locus of Rhodobacter sphaeroides is essential for chemotaxis. Mol Microbiol. 2002;46:1081–1094. doi: 10.1046/j.1365-2958.2002.03218.x. [DOI] [PubMed] [Google Scholar]
- 26.Porter S. L, Roberts M. A. J, Manning C. S, Armitage J. P. A bifunctional kinase-phosphatase in bacterial chemotaxis. Proc Natl Acad Sci U SA. 2008;105:18531–18536. doi: 10.1073/pnas.0808010105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ind A. C, Porter S. L, Brown M. T, Byles E. D, de Beyer J. A, et al. An inducible expression plasmid for Rhodobacter sphaeroides and Paracoccus denitrificans. Appl Environ Microbiol. 2009;75:6613–6615. doi: 10.1128/AEM.01587-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Skerker J. M, Perchuk B. S, Siryaporn A, Lubin E. A, Ashenberg O, et al. Rewiring the specificity of two-component signal transduction systems. Cell. 2008;133:1043–1054. doi: 10.1016/j.cell.2008.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Volz K, Matsumura P. Crystal structure of Escherichia coli CheY refined at 1.7Å resolution. J Biol Chem. 1991;266:15511. doi: 10.2210/pdb3chy/pdb. [DOI] [PubMed] [Google Scholar]
- 30.Stock A. M, Martinez-Hackert E, Rasmussen B. F, West A. H, Stock J. B, et al. Structure of the Mg2+-bound form of CheY and mechanism of phosphoryl transfer in bacterial chemotaxis. Biochemistry. 1993;32:13375–13380. doi: 10.1021/bi00212a001. [DOI] [PubMed] [Google Scholar]
- 31.Zhou H, Dahlquist F. W. Phosphotransfer site of the chemotaxis-specific protein kinase CheA as revealed by NMR. Biochemistry. 1997;36:699–710. doi: 10.1021/bi961663p. [DOI] [PubMed] [Google Scholar]
- 32.Mourey L, Da Re S, Pedelacq J. D, Tolstykh T, Faurie C, et al. Crystal structure of the CheA histidine phosphotransfer domain that mediates response regulator phosphorylation in bacterial chemotaxis. J Biol Chem. 2001;276:31074–31082. doi: 10.1074/jbc.M101943200. [DOI] [PubMed] [Google Scholar]
- 33.Quezada C. M, Gradinaru C, Simon M. I, Bilwes A. M, Crane B. R. Helical shifts generate two distinct conformers in the atomic resolution structure of the CheA phosphotransferase domain from Thermotoga maritima. J Mol Biol. 2004;341:1283–1294. doi: 10.1016/j.jmb.2004.06.061. [DOI] [PubMed] [Google Scholar]
- 34.Stewart R. C, Jahreis K, Parkinson J. S. Rapid phosphotransfer to CheY from a CheA protein lacking the CheY-binding domain. Biochemistry. 2000;39:13157–13165. doi: 10.1021/bi001100k. [DOI] [PubMed] [Google Scholar]
- 35.Nooren I. M. A, Thornton J. M. Structural characterisation and functional significance of transient protein-protein interactions. J Mol Biol. 2003;325:991–1018. doi: 10.1016/s0022-2836(02)01281-0. [DOI] [PubMed] [Google Scholar]
- 36.Weigt M, White R. A, Szurmant H, Hoch J. A, Hwa T. Identification of direct residue contacts in protein–protein interaction by message passing. Proc Natl Acad Sci U S A. 2009;106:67–72. doi: 10.1073/pnas.0805923106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szurmant H, Bobay B. G, White R. A, Sullivan D. M, Thompson R. J, et al. Co-evolving motions at protein–protein interfaces of two-component signaling systems identified by covariance analysis. Biochemistry. 2008;47:7782–7784. doi: 10.1021/bi8009604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zapf J, Sen U, Madhusudan, Hoch J. A, Varughese K. I. A transient interaction between two phosphorelay proteins trapped in a crystal lattice reveals the mechanism of molecular recognition and phosphotransfer in signal transduction. Structure. 2000;8:851–862. doi: 10.1016/s0969-2126(00)00174-x. [DOI] [PubMed] [Google Scholar]
- 39.Varughese K. I, Tsigelny I, Zhao H. The crystal structure of Beryllofluoride Spo0F in complex with the phosphotransferase Spo0B represents a phosphotransfer pretransition state. J Bacteriol. 2006;188:4970–4977. doi: 10.1128/JB.00160-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu O. P, Porter S. W, West A. H. The yeast YPD1/SLN1 complex: insights into molecular recognition in two-component signaling systems. Structure. 2003;11:1569–1581. doi: 10.1016/j.str.2003.10.016. [DOI] [PubMed] [Google Scholar]
- 41.Zhao X, Copeland D. M, Soares A. S, West A. H. Crystal structure of a complex between the phosphorelay protein YPD1 and the response regulator domain of SLN1 bound to a phosphoryl analog. J Mol Biol. 2008;375:1141–1151. doi: 10.1016/j.jmb.2007.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thomas S. A, Brewster J. A, Bourret R. B. Two variable active site residues modulate response regulator phosphoryl group stability. Mol Microbiol. 2008;69:453–465. doi: 10.1111/j.1365-2958.2008.06296.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Porter S. L, Wadhams G. H, Armitage J. P. In vivo and in vitro analysis of the Rhodobacter sphaeroides chemotaxis signaling complexes. Method Enzymol. 2007;423:392–413. doi: 10.1016/S0076-6879(07)23018-6. [DOI] [PubMed] [Google Scholar]
- 44.Hendrickson W. A, Horton J. R, Lemaster D. M. Selenomethionyl proteins produced for analysis by Multiwavelength Anomalous Diffraction (Mad): a vehicle for direct determination of 3-dimensional structure. EMBO J. 1990;9:1665–1672. doi: 10.1002/j.1460-2075.1990.tb08287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walter T. S, Diprose J. M, Mayo C. J, Siebold C, Pickford M. G, et al. A procedure for setting up high-throughput nanolitre crystallization experiments. Crystallization workflow for initial screening, automated storage, imaging and optimization. Acta Crystallogr Sect D-Biol Crystallogr. 2005;61:651–657. doi: 10.1107/S0907444905007808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 47.Schneider T. R, Sheldrick G. M. Substructure solution with SHELXD. Acta Crystallogr Sect D-Biol Crystallogr. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 48.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 49.Langer G, Cohen S. X, Lamzin V. S, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protocols. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr Sect D-Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 51.Blanc E, Roversi P, Vonrhein C, Flensburg C, Lea S. M, et al. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr Sect D-Biol Crystallogr. 2004;60:2210–2221. doi: 10.1107/S0907444904016427. [DOI] [PubMed] [Google Scholar]
- 52.Murshudov G. N, Vagin A. A, Dodson E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr Sect D-Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 53.Adams P. D, Grosse-Kunstleve R. W, Hung L. W, Ioerger T. R, Mccoy A. J, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr Sect D-Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 54.McCoy A. J, Grosse-Kunstleve R. W, Storoni L. C, Read R. J. Likelihood-enhanced fast translation functions. Acta Crystallogr Sect D-Biol Crystallogr. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 55.Davis I. W, Leaver-Fay A, Chen V. B, Block J. N, Kapral G. J, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucl Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Laskowski R. A, Macarthur M. W, Moss D. S, Thornton J. M. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 57.Kabsch W. Solution for best rotation to relate 2 sets of vectors. Acta Crystallogr Sect A. 1976;32:922–923. [Google Scholar]
- 58.Baker N. A, Sept D, Joseph S, Holst M. J, McCammon J. A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.O'Callaghan C. A, Byford M. F, Wyer J. R, Willcox B. E, Jakobsen B. K, et al. BirA enzyme: production and application in the study of membrane receptor-ligand interactions by site-specific biotinylation. Anal Biochem. 1999;266:9–15. doi: 10.1006/abio.1998.2930. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Stereoview of a superposition of the metal binding site of CheY6 and E. coli CheY. CheY6 is shown in pale green, E. coli CheY with Mg2+ bound (PDB-code: 1CHN) in yellow, and E.coli CheY without Mg2+ (3CHY) in teal. Structures were aligned on their secondary structure elements using secondary structure matching (SSM) implemented in COOT [50]. Residues involved in the coordination of Mg2+ in E. coli CheY are shown in stick representation for all three structures. Residues in E. coli structures are only labelled for the Mg2+ bound form. Mg2+ from the E. coli structure is shown as a grey sphere. CheY6 resembles the Mg2+ bound form of E. coli CheY despite not having a divalent cation bound to its metal binding site.
(6.24 MB TIF)
Electron density of phosphorylated His51 in CheA3P1 in the active complex structure. The orientation is similar to Figure 2B. The density represents a 2Fobs-Fcalc map contoured at 1.5 σ and calculated after initial rigid body refinement in Phaser [54] and one round of positional refinement in autoBUSTER [51], both omitting the phosphate group from the model.
(0.64 MB TIF)
Binding of CheA3P1 to substitution mutants of CheY4 and CheY6. (A–C) Binding of CheA3P1 to CheY4(P12A,S13M,C16L,M17Y), CheY6(M13S), and CheY6(M13S,L16S,Y17M). Left, representative sets of experimental sensorgrams from typical equilibrium-based binding experiments, with reference subtraction. Different concentrations of CheA3P1 were injected over surfaces coupled with the respective RR. For all injections, the experimental traces reached equilibrium and returned to baseline after the injection. Right, plot of the equilibrium binding response (response units [RU]) against CheA3P1 concentration ranging from 120 nM to 2 mM. Within one experiment, each concentration was measured twice. All experiments were performed in duplicate. Best-fit binding curves corresponding with a 1∶1 binding model are shown as lines. Experiments with CheY6(M13S) and CheY6(M13S,L16S,Y17M) did not reach saturation due to very low affinity, thus the Kd value is estimated.
(0.52 MB TIF)
Superposition of the CheA3P1-CheY6 complex structure with the Spo0B-Spo0F complex and YPD1-SLN1 complex. (A) Superposition of CheA3P1-CheY6 with Spo0B-Spo0F (PDB-code: 1F51). (B) Superposition of CheA3P1-CheY6 with YPD1-SLN1 (1OXB). All structures were superimposed on the RRs using SSM as implemented in COOT [50]. CheY6 is shown in pale green, CheA3P1 in light blue, Spo0F in orange, Spo0B in teal, SLN1 in purple, and YPD1 in aquamarine. Only the four-helix bundle of the Spo0B dimer is shown; the C-terminal domain is omitted for clarity. The orientation is similar to Figure 1. CheA3P1 is structurally more similar to the monomeric YPD1 than to Spo0B.
(2.64 MB TIF)