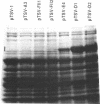
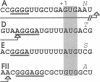
Abstract
The late operon of bacteriophage lambda contains the genes encoding the morphogenetic proteins of the phage. These genes are transcribed equally from the single late promoter. Although the functional half-lives of the mRNA for the various genes of this operon vary less than 2-fold, their relative rates of expression have been shown to vary by nearly 1000-fold. This variation could result from differing rates of translation initiation, from overlapping upstream translation, or from differential elongation rates due to the presence of codons for which the corresponding tRNAs are rare. To distinguish between these possibilities, we have cloned sequences surrounding the initiator codons of several of these genes and measured their ability to drive synthesis of hybrid lambda-beta-galactosidase proteins. The rates of expression of the hybrid genes thus produced correlate very well with the natural rates of expression of the corresponding phage genes, suggesting that the rate of initiation of translation controls the relative expression rates of these genes.
Full text
PDF
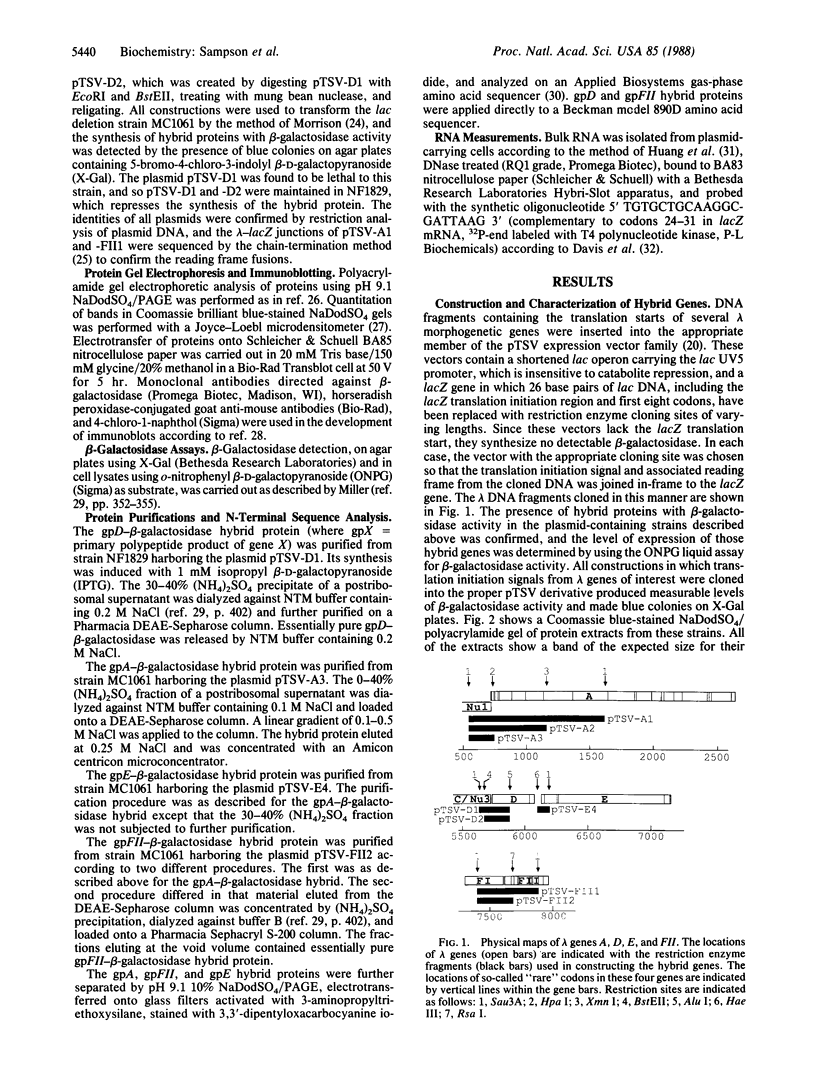



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aebersold R. H., Teplow D. B., Hood L. E., Kent S. B. Electroblotting onto activated glass. High efficiency preparation of proteins from analytical sodium dodecyl sulfate-polyacrylamide gels for direct sequence analysis. J Biol Chem. 1986 Mar 25;261(9):4229–4238. [PubMed] [Google Scholar]
- Bonekamp F., Andersen H. D., Christensen T., Jensen K. F. Codon-defined ribosomal pausing in Escherichia coli detected by using the pyrE attenuator to probe the coupling between transcription and translation. Nucleic Acids Res. 1985 Jun 11;13(11):4113–4123. doi: 10.1093/nar/13.11.4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter P. W., Bartkus J. M., Calvo J. M. Transcription attenuation in Salmonella typhimurium: the significance of rare leucine codons in the leu leader. Proc Natl Acad Sci U S A. 1986 Nov;83(21):8127–8131. doi: 10.1073/pnas.83.21.8127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J., Chou J., Cohen S. N. In vitro gene fusions that join an enzymatically active beta-galactosidase segment to amino-terminal fragments of exogenous proteins: Escherichia coli plasmid vectors for the detection and cloning of translational initiation signals. J Bacteriol. 1980 Aug;143(2):971–980. doi: 10.1128/jb.143.2.971-980.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens S. Bacteriophage lambda FII gene protein: role in head assembly. J Mol Biol. 1974 Nov 25;90(1):1–20. doi: 10.1016/0022-2836(74)90252-6. [DOI] [PubMed] [Google Scholar]
- Casjens S., Hayden M., Jackson E., Deans R. Additional restriction endonuclease cleavage sites on the bacteriophage P22 genome. J Virol. 1983 Feb;45(2):864–867. doi: 10.1128/jvi.45.2.864-867.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan R. K., Botstein D. Genetics of bacteriophage P22. I. Isolation of prophage deletions which affect immunity to superinfection. Virology. 1972 Jul;49(1):257–267. doi: 10.1016/s0042-6822(72)80027-8. [DOI] [PubMed] [Google Scholar]
- Dreyfuss G., Adam S. A., Choi Y. D. Physical change in cytoplasmic messenger ribonucleoproteins in cells treated with inhibitors of mRNA transcription. Mol Cell Biol. 1984 Mar;4(3):415–423. doi: 10.1128/mcb.4.3.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flinta C., Persson B., Jörnvall H., von Heijne G. Sequence determinants of cytosolic N-terminal protein processing. Eur J Biochem. 1986 Jan 2;154(1):193–196. doi: 10.1111/j.1432-1033.1986.tb09378.x. [DOI] [PubMed] [Google Scholar]
- Georgopoulos C. P., Hendrix R. W., Casjens S. R., Kaiser A. D. Host participation in bacteriophage lambda head assembly. J Mol Biol. 1973 May 5;76(1):45–60. doi: 10.1016/0022-2836(73)90080-6. [DOI] [PubMed] [Google Scholar]
- Gold L., Pribnow D., Schneider T., Shinedling S., Singer B. S., Stormo G. Translational initiation in prokaryotes. Annu Rev Microbiol. 1981;35:365–403. doi: 10.1146/annurev.mi.35.100181.002053. [DOI] [PubMed] [Google Scholar]
- Gouy M., Gautier C. Codon usage in bacteria: correlation with gene expressivity. Nucleic Acids Res. 1982 Nov 25;10(22):7055–7074. doi: 10.1093/nar/10.22.7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H., Fiers W. Preferential codon usage in prokaryotic genes: the optimal codon-anticodon interaction energy and the selective codon usage in efficiently expressed genes. Gene. 1982 Jun;18(3):199–209. doi: 10.1016/0378-1119(82)90157-3. [DOI] [PubMed] [Google Scholar]
- Hawkes R., Niday E., Gordon J. A dot-immunobinding assay for monoclonal and other antibodies. Anal Biochem. 1982 Jan 1;119(1):142–147. doi: 10.1016/0003-2697(82)90677-7. [DOI] [PubMed] [Google Scholar]
- Hayden M., Adams M. B., Casjens S. Bacteriophage L: chromosome physical map and structural proteins. Virology. 1985 Dec;147(2):431–440. doi: 10.1016/0042-6822(85)90145-x. [DOI] [PubMed] [Google Scholar]
- Herskowitz I., Signer E. R. A site essential for expression of all late genes in bacteriophage lambda. J Mol Biol. 1970 Feb 14;47(3):545–556. doi: 10.1016/0022-2836(70)90321-9. [DOI] [PubMed] [Google Scholar]
- Huang W. M., Ao S. Z., Casjens S., Orlandi R., Zeikus R., Weiss R., Winge D., Fang M. A persistent untranslated sequence within bacteriophage T4 DNA topoisomerase gene 60. Science. 1988 Feb 26;239(4843):1005–1012. doi: 10.1126/science.2830666. [DOI] [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E. coli translational system. J Mol Biol. 1981 Sep 25;151(3):389–409. doi: 10.1016/0022-2836(81)90003-6. [DOI] [PubMed] [Google Scholar]
- Ish-Horowicz D., Burke J. F. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981 Jul 10;9(13):2989–2998. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson A. B., Good L., Simonetti J., Zuker M. Some simple computational methods to improve the folding of large RNAs. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):45–52. doi: 10.1093/nar/12.1part1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King J., Casjens S. Catalytic head assembling protein in virus morphogenesis. Nature. 1974 Sep 13;251(5471):112–119. doi: 10.1038/251112a0. [DOI] [PubMed] [Google Scholar]
- Klein R. D., Selsing E., Wells R. D. A rapid microscale technique for isolation of recombinant plasmid DNA suitable for restriction enzyme analysis. Plasmid. 1980 Jan;3(1):88–91. doi: 10.1016/s0147-619x(80)90037-2. [DOI] [PubMed] [Google Scholar]
- Konigsberg W., Godson G. N. Evidence for use of rare codons in the dnaG gene and other regulatory genes of Escherichia coli. Proc Natl Acad Sci U S A. 1983 Feb;80(3):687–691. doi: 10.1073/pnas.80.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison D. A. Transformation in Escherichia coli: cryogenic preservation of competent cells. J Bacteriol. 1977 Oct;132(1):349–351. doi: 10.1128/jb.132.1.349-351.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murialdo H., Davidson A., Chow S., Gold M. The control of lambda DNA terminase synthesis. Nucleic Acids Res. 1987 Jan 12;15(1):119–140. doi: 10.1093/nar/15.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murialdo H., Siminovitch L. The morphogenesis of bacteriophage lambda. IV. Identification of gene products and control of the expression of the morphogenetic information. Virology. 1972 Jun;48(3):785–823. doi: 10.1016/0042-6822(72)90162-6. [DOI] [PubMed] [Google Scholar]
- Müller-Hill B., Kania J. Lac repressor can be fused to beta-galactosidase. Nature. 1974 Jun 7;249(457):561–563. doi: 10.1038/249561a0. [DOI] [PubMed] [Google Scholar]
- Pedersen S. Escherichia coli ribosomes translate in vivo with variable rate. EMBO J. 1984 Dec 1;3(12):2895–2898. doi: 10.1002/j.1460-2075.1984.tb02227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purvis I. J., Bettany A. J., Santiago T. C., Coggins J. R., Duncan K., Eason R., Brown A. J. The efficiency of folding of some proteins is increased by controlled rates of translation in vivo. A hypothesis. J Mol Biol. 1987 Jan 20;193(2):413–417. doi: 10.1016/0022-2836(87)90230-0. [DOI] [PubMed] [Google Scholar]
- Ray P. N., Pearson M. L. Evidence for post-transcriptional control of the morphogenetic genes of bacteriophage lambda. J Mol Biol. 1974 May 5;85(1):163–175. doi: 10.1016/0022-2836(74)90135-1. [DOI] [PubMed] [Google Scholar]
- Ray P. N., Pearson M. L. Functional inactivation of bacteriophage lambda morphogenetic gene in RNA. Nature. 1975 Feb 20;253(5493):647–650. doi: 10.1038/253647a0. [DOI] [PubMed] [Google Scholar]
- Robinson M., Lilley R., Little S., Emtage J. S., Yarranton G., Stephens P., Millican A., Eaton M., Humphreys G. Codon usage can affect efficiency of translation of genes in Escherichia coli. Nucleic Acids Res. 1984 Sep 11;12(17):6663–6671. doi: 10.1093/nar/12.17.6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Hong G. F., Hill D. F., Petersen G. B. Nucleotide sequence of bacteriophage lambda DNA. J Mol Biol. 1982 Dec 25;162(4):729–773. doi: 10.1016/0022-2836(82)90546-0. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultz J., Silhavy T. J., Berman M. L., Fiil N., Emr S. D. A previously unidentified gene in the spc operon of Escherichia coli K12 specifies a component of the protein export machinery. Cell. 1982 Nov;31(1):227–235. doi: 10.1016/0092-8674(82)90422-6. [DOI] [PubMed] [Google Scholar]
- Stanssens P., Remaut E., Fiers W. Inefficient translation initiation causes premature transcription termination in the lacZ gene. Cell. 1986 Mar 14;44(5):711–718. doi: 10.1016/0092-8674(86)90837-8. [DOI] [PubMed] [Google Scholar]
- Walker J. E., Auffret A. D., Carne A., Gurnett A., Hanisch P., Hill D., Saraste M. Solid-phase sequence analysis of polypeptides eluted from polyacrylamide gels. An aid to interpretation of DNA sequences exemplified by the Escherichia coli unc operon and bacteriophage lambda. Eur J Biochem. 1982 Apr 1;123(2):253–260. doi: 10.1111/j.1432-1033.1982.tb19761.x. [DOI] [PubMed] [Google Scholar]
- Wyckoff E., Sampson L., Hayden M., Parr R., Huang W. M., Casjens S. Plasmid vectors useful in the study of translation initiation signals. Gene. 1986;43(3):281–286. doi: 10.1016/0378-1119(86)90217-9. [DOI] [PubMed] [Google Scholar]