Abstract
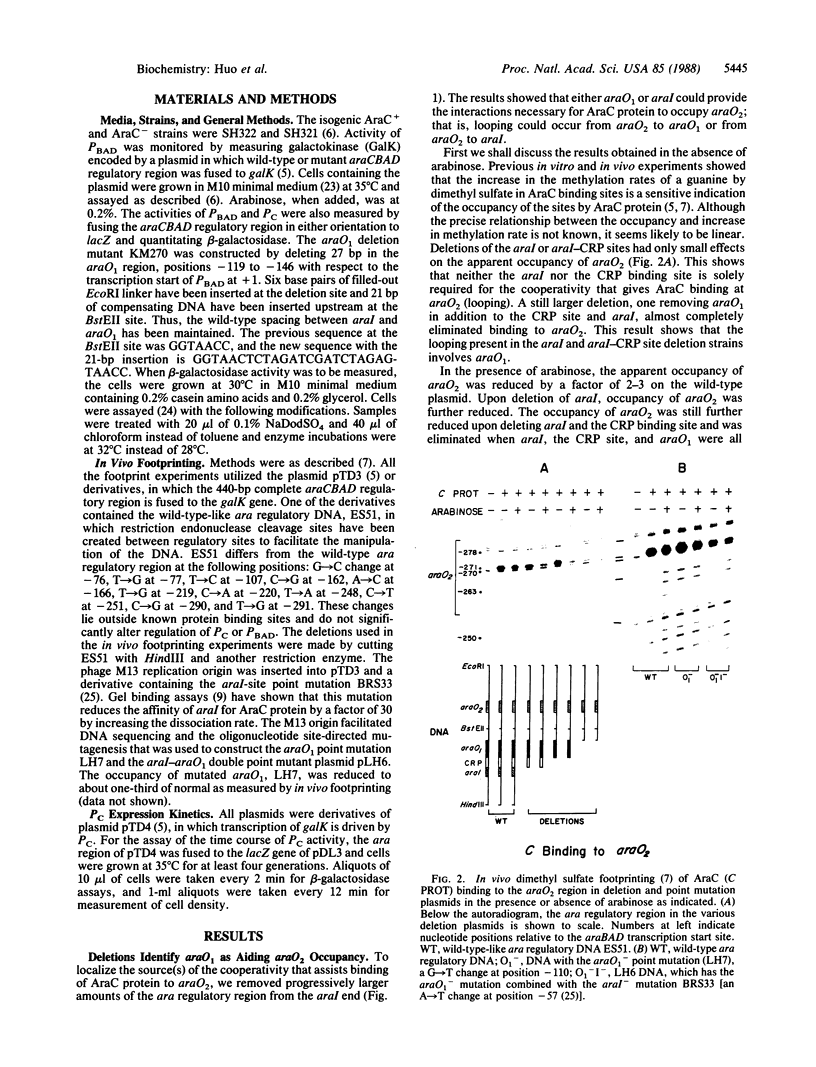
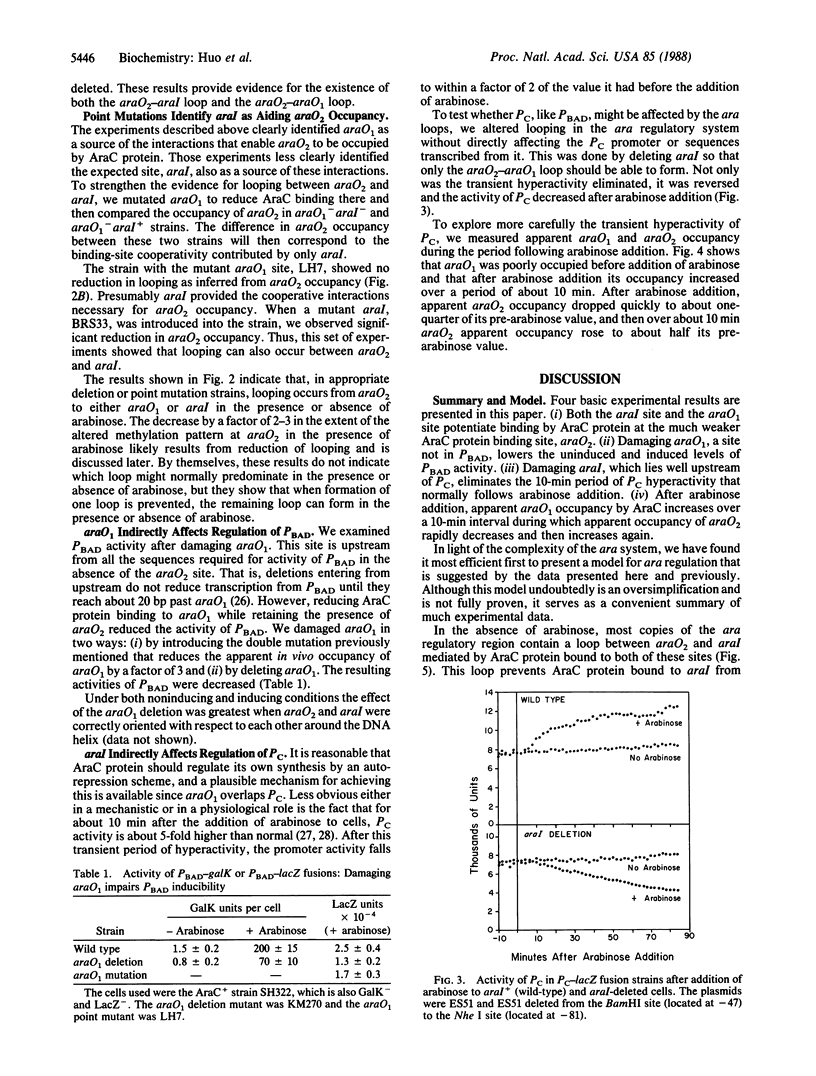
The araCBAD regulatory region of Escherichia coli contains two divergently oriented promoters and three sites to which AraC, the regulatory protein of the operon, can bind. This paper presents the results of in vivo dimethyl sulfate "footprinting" experiments to monitor occupancy of the three AraC sites and measurements of activity of the two promoters. These measurements were made both in the absence of the inducer arabinose and at various times after arabinose addition to growing cells containing the wild-type ara regulatory region or the regulatory region containing various deletions and point mutations. The data lead to the conclusion that two different DNA loops can form in the ara regulatory region. These loops are generated by AraC protein molecules binding to two different DNA sites and binding to each other. One of these loops predominates in the absence of arabinose and plays a major role in repressing activity of one of the promoters. Upon the addition of arabinose the amount of the first loop type, the repression loop, decreases and the amount of a second loop increases. Formation of this second loop precludes the counterproductive formation of the repression loop.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Borowiec J. A., Zhang L., Sasse-Dwight S., Gralla J. D. DNA supercoiling promotes formation of a bent repression loop in lac DNA. J Mol Biol. 1987 Jul 5;196(1):101–111. doi: 10.1016/0022-2836(87)90513-4. [DOI] [PubMed] [Google Scholar]
- Casadaban M. J. Regulation of the regulatory gene for the arabinose pathway, araC. J Mol Biol. 1976 Jul 5;104(3):557–566. doi: 10.1016/0022-2836(76)90120-0. [DOI] [PubMed] [Google Scholar]
- Dandanell G., Hammer K. Two operator sites separated by 599 base pairs are required for deoR repression of the deo operon of Escherichia coli. EMBO J. 1985 Dec 1;4(12):3333–3338. doi: 10.1002/j.1460-2075.1985.tb04085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandanell G., Valentin-Hansen P., Larsen J. E., Hammer K. Long-range cooperativity between gene regulatory sequences in a prokaryote. 1987 Feb 26-Mar 4Nature. 325(6107):823–826. doi: 10.1038/325823a0. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Travers A. A. DNA bending and its relation to nucleosome positioning. J Mol Biol. 1985 Dec 20;186(4):773–790. doi: 10.1016/0022-2836(85)90396-1. [DOI] [PubMed] [Google Scholar]
- Dunn T. M., Hahn S., Ogden S., Schleif R. F. An operator at -280 base pairs that is required for repression of araBAD operon promoter: addition of DNA helical turns between the operator and promoter cyclically hinders repression. Proc Natl Acad Sci U S A. 1984 Aug;81(16):5017–5020. doi: 10.1073/pnas.81.16.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn T. M., Schleif R. Deletion analysis of the Escherichia coli ara PC and PBAD promoters. J Mol Biol. 1984 Nov 25;180(1):201–204. doi: 10.1016/0022-2836(84)90437-6. [DOI] [PubMed] [Google Scholar]
- Griffith J., Hochschild A., Ptashne M. DNA loops induced by cooperative binding of lambda repressor. Nature. 1986 Aug 21;322(6081):750–752. doi: 10.1038/322750a0. [DOI] [PubMed] [Google Scholar]
- Hahn S., Dunn T., Schleif R. Upstream repression and CRP stimulation of the Escherichia coli L-arabinose operon. J Mol Biol. 1984 Nov 25;180(1):61–72. doi: 10.1016/0022-2836(84)90430-3. [DOI] [PubMed] [Google Scholar]
- Hahn S., Hendrickson W., Schleif R. Transcription of Escherichia coli ara in vitro. The cyclic AMP receptor protein requirement for PBAD induction that depends on the presence and orientation of the araO2 site. J Mol Biol. 1986 Apr 5;188(3):355–367. doi: 10.1016/0022-2836(86)90160-9. [DOI] [PubMed] [Google Scholar]
- Hahn S., Schleif R. In vivo regulation of the Escherichia coli araC promoter. J Bacteriol. 1983 Aug;155(2):593–600. doi: 10.1128/jb.155.2.593-600.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson W., Schleif R. F. Regulation of the Escherichia coli L-arabinose operon studied by gel electrophoresis DNA binding assay. J Mol Biol. 1984 Sep 25;178(3):611–628. doi: 10.1016/0022-2836(84)90241-9. [DOI] [PubMed] [Google Scholar]
- Hochschild A., Ptashne M. Cooperative binding of lambda repressors to sites separated by integral turns of the DNA helix. Cell. 1986 Mar 14;44(5):681–687. doi: 10.1016/0092-8674(86)90833-0. [DOI] [PubMed] [Google Scholar]
- Kolodrubetz D., Schleif R. Regulation of the L-arabinose transport operons in Escherichia coli. J Mol Biol. 1981 Sep 15;151(2):215–227. doi: 10.1016/0022-2836(81)90512-x. [DOI] [PubMed] [Google Scholar]
- Krämer H., Niemöller M., Amouyal M., Revet B., von Wilcken-Bergmann B., Müller-Hill B. lac repressor forms loops with linear DNA carrying two suitably spaced lac operators. EMBO J. 1987 May;6(5):1481–1491. doi: 10.1002/j.1460-2075.1987.tb02390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichenstein H. S., Hamilton E. P., Lee N. Repression and catabolite gene activation in the araBAD operon. J Bacteriol. 1987 Feb;169(2):811–822. doi: 10.1128/jb.169.2.811-822.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar A., Adhya S. Demonstration of two operator elements in gal: in vitro repressor binding studies. Proc Natl Acad Sci U S A. 1984 Oct;81(19):6100–6104. doi: 10.1073/pnas.81.19.6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin K., Huo L., Schleif R. F. The DNA loop model for ara repression: AraC protein occupies the proposed loop sites in vivo and repression-negative mutations lie in these same sites. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3654–3658. doi: 10.1073/pnas.83.11.3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossing M. C., Record M. T., Jr Upstream operators enhance repression of the lac promoter. Science. 1986 Aug 22;233(4766):889–892. doi: 10.1126/science.3090685. [DOI] [PubMed] [Google Scholar]
- Mukherjee S., Erickson H., Bastia D. Enhancer-origin interaction in plasmid R6K involves a DNA loop mediated by initiator protein. Cell. 1988 Feb 12;52(3):375–383. doi: 10.1016/s0092-8674(88)80030-8. [DOI] [PubMed] [Google Scholar]
- Ogden S., Haggerty D., Stoner C. M., Kolodrubetz D., Schleif R. The Escherichia coli L-arabinose operon: binding sites of the regulatory proteins and a mechanism of positive and negative regulation. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3346–3350. doi: 10.1073/pnas.77.6.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard D. E., Englesberg E. Further evidence for positive control of the L-arabinose system by gene araC. J Mol Biol. 1967 May 14;25(3):443–454. doi: 10.1016/0022-2836(67)90197-0. [DOI] [PubMed] [Google Scholar]
- Stoner C., Schleif R. The araE low affinity L-arabinose transport promoter. Cloning, sequence, transcription start site and DNA binding sites of regulatory proteins. J Mol Biol. 1983 Dec 25;171(4):369–381. doi: 10.1016/0022-2836(83)90035-9. [DOI] [PubMed] [Google Scholar]
- Théveny B., Bailly A., Rauch C., Rauch M., Delain E., Milgrom E. Association of DNA-bound progesterone receptors. Nature. 1987 Sep 3;329(6134):79–81. doi: 10.1038/329079a0. [DOI] [PubMed] [Google Scholar]
- Ulanovsky L., Bodner M., Trifonov E. N., Choder M. Curved DNA: design, synthesis, and circularization. Proc Natl Acad Sci U S A. 1986 Feb;83(4):862–866. doi: 10.1073/pnas.83.4.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitson P. A., Hsieh W. T., Wells R. D., Matthews K. S. Supercoiling facilitates lac operator-repressor-pseudooperator interactions. J Biol Chem. 1987 Apr 15;262(11):4943–4946. [PubMed] [Google Scholar]






