Abstract
The co-chaperone Hsp70-Hsp90 organizing protein (HOP) plays a central role in protein folding in vivo, binding to both Hsp70 and Hsp90 and bringing them together in a functional complex. Reports in the literature concerning the oligomeric state of HOP have been inconsistent—is it a monomer, dimer, or higher order oligomer? Knowing the oligomeric state of HOP is important, because it places limits on the number and types of multiprotein complexes that can form during the folding cycle. Thus, the number of feasible models is simplified. Here, we explicitly investigate the oligomeric state of HOP using three complementary methods: gel filtration chromatography, sedimentation equilibrium analytical ultracentrifugation (AUC), and an in vivo coexpression assay. We find that HOP does not behave like a monomeric globular protein on gel filtration. Rather its behavior is consistent with it being either an elongated monomer or a dimer. We follow-up on these studies using sedimentation equilibrium AUC, which separates on the basis of molecular weight (MW), independent of shape. Sedimentation equilibrium AUC clearly shows that HOP is a monomer, with no indication of higher MW species. Finally, we use an in vivo coexpression assay that also supports the conclusion that HOP is a monomer.
Keywords: heat shock organizing protein (HOP), tetratricopeptide repeat (TPR), analytical ultracentrifugation (AUC), gel filtration chromatography
Introduction
In eukaryotic cells, the molecular chaperone Hsp90 plays a critical role in the activation and conformational maturation of a diverse group of client proteins, including transcription factors, cell cycle regulators, and kinases.1–4 Hsp90 does not act alone, but rather functions in concert with a variety of other chaperones and co-chaperones.5,6 As its name implies, Hsp70-Hsp90 organizing protein (HOP) plays a key role by bringing the chaperones Hsp70 and Hsp90 together via two tetratricopeptide repeat (TPR) domains, TPR1 and TPR2A. The TPR1 domain binds to the C-terminal peptide of Hsp70, and the TPR2A binds to the C-terminal peptide of Hsp90.7 Hsp70 and Hsp90 are, thus, assembled into a functional complex, facilitating transfer of client proteins from Hsp70 to Hsp90 where the final steps of their maturation are completed.8 If protein folding by Hsp90 is inhibited, the client proteins are targeted for degradation. They are ubiquitinylated by C-terminus of Hsp interacting protein, an E3 ubiquitin ligase that also has a TPR domain that has been proposed to interact with the C-terminal peptide of either Hsp70 or Hsp90.9
It is well established that Hsp90 is a dimer10 and that Hsp70 is a monomer.11 The oligomeric state of HOP, whether it is a dimer or a monomer, thus has important implications for the types of folding and degradation complexes that can be formed. To propose reasonable and quantitative models for Hsp70/Hsp90-mediated folding, it is essential to know the oligomeric state of HOP.
Results
We present the results of using three different techniques to investigate the oligomeric state of HOP. We studied full-length human HOP (hHOP), Drosophila HOP (dHOP), and deletion fragments of these proteins. We studied stable fragments of hHOP, because certain regions of it have been proposed to mediate the putative dimerization. It was important, therefore, to test the effect on the measured properties by deleting these sections. It has been proposed, for example, that the TPR2A domain of HOP is a putative dimerization region.12,13 All the HOP fragments studied are shown in Figure 1. All are stable and folded, as assessed by Circular dichroism (CD) measurements (Fig. 2).
Figure 1.
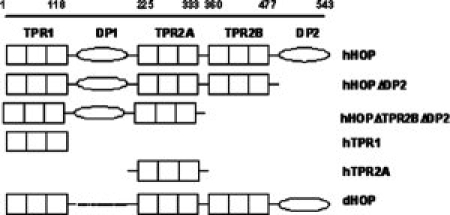
Schematic illustration of full-length human HOP (hHOP), drosophila HOP (dHOP), and the HOP deletion constructs used in this study.
Figure 2.
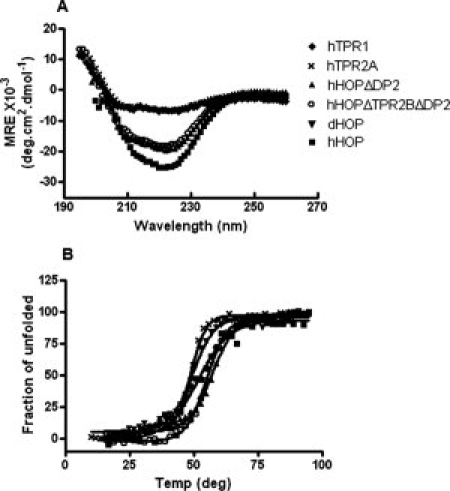
HOP deletion fragments remain properly folded. (A) Far ultraviolet region circular dichroism spectra for human TPR1 domain (solid diamond), TPR2A domain (cross), HOPΔDP2 fragment (solid upon triangle), and HOPΔTPR2B ΔDP2 (empty circle) were measured and compared with that of full-length Human HOP (solid square) and full-length Drosophila HOP (solid down triangle). Measurements were taken at protein concentrations of 2 μM at pH 7.4. (B) Thermal denaturation curves of HOP and its variants were measured by monitoring the ellipticity signals at 222 nm. A two-state denaturation model was fit to all the denaturation curves, with the estimated melting temperatures being 49°C, 57°C, 55°C, 50°C, and 52°C for TPR2A domain, HOPΔDP2, and HOPΔTPR2B ΔDP2 of hHOP, full-length dHOP, and hHOP, respectively.
Gel filtration chromatography
Gel filtration chromatography is a widely used technique, which separates on the basis of both mass and shape. When a gel filtration column is calibrated with globular proteins, it is a reasonable expectation that the elution volume of another globular protein can be used to determine the mass of that protein. For nonglobular proteins, however, this should not be the expectation, because their shape (hydrodynamic radius) will also influence the elution properties.
A Superdex S-200 16/60 clumn was calibrated using four globular proteins plus vitamin B12, spanning a molecular weight (MW) ranging from 1350 to 670,000 Da. Full-length hHOP, dHOP, and fragments of these proteins were chromatographed on this column. Each of the proteins was eluted with a single, symmetrical peak. The apparent MW of each protein was estimated by interpolation using the globular protein calibration curve (Fig. 3). The hHOP and dHOP migrated as if their MWs were approximately twice that their actual MW, as did hHOPΔDP2 and hHOPΔTPR2BΔDP2. By contrast, the single TPR1 domain and TPR2A domain of hHOP migrated according to their actual MW (Table I).
Figure 3.
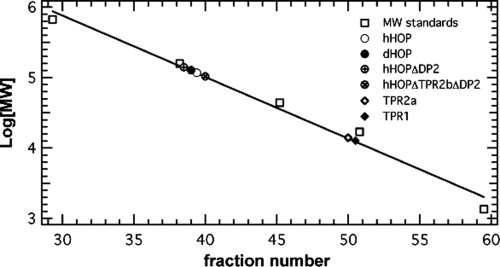
Gel filtration chromatography of globular molecular weight standards, HOP, and HOP fragments.
Table I.
Summary of Gel Filtration Chromatography Apparent MW of HOP Variants
| Calculated monomer MW (with histag) (kDa) | Experimental MW (S200) (kDa) | MW meas/ MW cal | |
|---|---|---|---|
| WT-hHOP | 67.5 | 117.3 | 1.7 |
| hHOP- ΔTPR2BΔDP2 | 46.9 | 104 | 2.2 |
| hHOP-ΔDP2 | 61.5 | 140.4 | 2.2 |
| hTPR2A | 19.9 | 14.0 | 0.7 |
| hTPR1 | 15.0 | 14.0 | 0.9 |
| dHOP | 57.9 | 127.1 | 2.2 |
MWmeas/MWcal is the ratio between the measured and the calculated molecuar weight.
From these results, we conclude that the individual TPR1 and TPR2A domains are globular and monomeric. We hypothesize that full-length HOP proteins are either globular and dimeric or elongated and monomeric. Gel filtration data alone cannot distinguish between these two possibilities. We also conclude that the deletion of the TPR2BDP2 region has no effect on the hydrodynamic properties of hHOP.
Analytical ultracentrifugation
We performed sedimentation equilibrium analytical ultracentrifugation (AUC), a method to determine the MW of a protein that is independent of its shape. Three different concentrations were analyzed for full-length hHOP (16, 8, and 4 μM), full-length dHOP (26, 13, and 6.5 μM), and hHOPΔTPR2BΔDP2 (20, 10, and 5 μM). The concentrations were chosen to cover the proposed dissociation constant for a dimer, which had been suggested in previous studies to be in the low micromolar range.14 We avoided protein concentrations greater than 1.5 mg/ml, to limit nonideal effects that can be an issue at high protein concentrations. We performed a global analysis of the data at each protein concentration and at all three speeds.
The ideal single species model fit the data best with a root mean square deviation (RMSD) of 0.01 (Fig. 4). Fitting a monomer-dimer equilibrium model to the data yielded a good fit (RMSD = 0.01) yet very high dissociation constant (Kd) values (weak association, >mM), suggesting a dimer formation unlikely at the conditions tested). Similarly, analyzing the data with monomer-dimer equilibrium model with a fixed monomer MW and a fixed Kd of 1 μM gave a poor fit (RMSD = 0.05). No concentration dependence of the apparent MW for any of the proteins was observed, suggesting no oligomerization at the high concentrations tested (Table II). Figure 4 shows a representative plot from the global analysis of the data for hHOP at 8 μM at 3 speeds using an ideal single species model. This analysis yields a MW for hHOP of 67,811 Da, close to the formula MW of a monomer (67591.7 Da). Performing the same analysis on hHOP at the other two concentrations yielded similar results. Global analysis of the truncation mutant hHOPΔTPR2BΔDP2 using an ideal single species model at each concentration also yielded apparent MW close to its monomer MW. dHOP, which differs from hHop in not having the DP1 domain, also behaved as a monomer (Table II).
Figure 4.
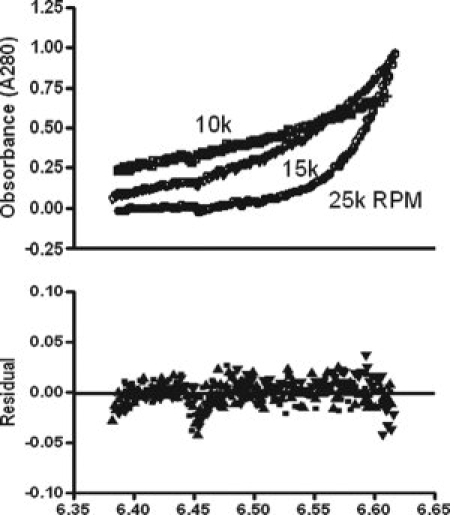
Sedimentation equilibrium AUC (SE-AUC) of hHOP. Displayed in the upper panel is the global analysis by fitting an ideal single species model to the SE-AUC data for hHOP at 8 μM at 10k, 15k and 25k RPM. The fitting yielded a single species with a molecular weight of 67,811 Da, with a root mean square deviation of 0.01. Data (symbols) and fit (solid line) are overlaid. Lower panel shows a superposition of the residuals for different speeds.
Table II.
Summary of Sedimentation Equilibrium AUC of HOP Variants at Different Concentrations
| Ideal one species model |
||||
|---|---|---|---|---|
| Theorectical MW (Da) | Concentration (μM) | Predicted MW (Da) | RMSD | |
| hHOP | 67591 | 16 | 65,860 | 0.01 |
| 8 | 67,811 | 0.01 | ||
| 4 | 64,320 | 0.009 | ||
| dHOP | 57861 | 26 | 46,455 | 0.01 |
| 13 | 55,921 | 0.006 | ||
| 6.5 | 55,227 | 0.01 | ||
| hHOPΔTPR2B ΔDP2 (1–368) | 47420 | 20 | 49,915 | 0.009 |
| 10 | 46,575 | 0.009 | ||
| 5 | 48,348 | 0.007 | ||
To summarize, the results of the sedimentation equilibrium AUC studies lead us to conclude that the reason for the nonideal behavior of HOP on the gel filtration column is because it has an elongated, nonglobular shape13 and not because it is a dimer.
An in vivo assay for protein dimerization
To investigate the dimerization potential of HOP by a completely different strategy, we devised an in vivo coexpression test. Two slightly different versions of hHOP—one with a 26 amino acid N-terminal tag that includes a hexahistidine (His6) sequence and one that is untagged—were coexpressed in Escherichia coli. We hypothesized that if HOP forms dimers, coexpression of tagged and untagged HOP would result in the formation of both homodimers (tagged-tagged or untagged-untagged) and heterodimers (tagged-untagged). Total protein extracts of cells expressing both the tagged and untagged forms of HOP were prepared and incubated with Ni-NTA resin. His6-tagged HOP can bind to the Ni-NTA resin directly, but the untagged HOP can only bind if it forms a heterodimer with tagged HOP. The tagged and untagged HOP can be resolved on a sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS PAGE) gel, because of their slight difference in size. It is evident from the data [Fig. 5(A)] that tagged and untagged HOPs are not able to form heterodimers, supporting the previous results that indicate HOP is a monomer.
Figure 5.
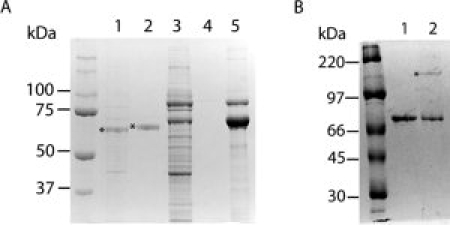
In vivo analysis of hHOP dimerization. (A) Lanes 1 and 2: total protein extract from cells expressing HOP and HOP-His6 tag were prepared and incubated with Ni-NTA resin. Lane 1: “flow through”—protein that did not bind to the Ni-NTA column, *indicates non-His6-tagged HOP. Lane 2: “bound protein”—protein that bound to the Ni-NTA column, *indicates His6-tagged HOP. Note that only His-tagged HOP is present in the bound protein fraction. Lanes 3, 4 and 5: validation of the coexpression assay using Hsp90 as a control dimer: performing the same experiment with Hsp90 and the His-tagged C-terminal (amino acid residues 177–724) domain of Hsp90. Total protein extract from cells coexpressing Hsp90 and His-tagged C-terminal domain of Hsp90 were prepared and incubated with Ni-NTA resin. Lane 3: “flow through”—protein that did not bind to the Ni-NTA column. Lane 4: washes from the Ni-NTA column after the “flow through” were collected. Lane 5: “bound protein”—protein that bound to the Ni-NTA column. Note the presence of both full-length Hsp90 and His-tagged C-terminal domain of Hsp90 in the bound protein fraction. (B) In vitro assay of HOP propensity to dimerize via disulfide bonds under nonreducing conditions. Lane 1: purified HOP stored in presence of reducing agent. Lane 2: purified HOP stored in the absence of reducing agent. Note the disulfide-linked HOP dimer in lane 2, indicated with *.
As a control, we performed a similar experiment with Hsp90, a protein that is known to be a dimer. It is known that the carboxy-terminal portion of Hsp90 (540–724) is necessary and sufficient for dimerization.10 A C-terminal fragment Hsp90 (177–724) was His6 tagged, whereas full-length Hsp90 (1–724) was untagged. When total cell-extract from cells expressing both the His-tagged C-terminal fragment of Hsp90 and untagged full-length Hsp90 were incubated with Ni-NTA resin, both proteins bound and were eluted together by imidazole (Fig. 5). This results is the expectation for a dimeric protein and demonstrates that the method works.
Discussion
HOP is a TPR containing co-chaperone of Hsp90 that has been most well known for its role as an Hsp70/Hsp90 adaptor protein.6 It plays an important role in both coordinating and modulating the activities of Hsp70 and Hsp90 in the Hsp90 chaperone cycle, despite the fact that it does not have chaperoning activity on its own. Physical linking of Hsp70 and Hsp90 by HOP to form the intermediate multichaperone complex of the Hsp90 chaperone cycle is essential for the proper function of the client protein folding pathway. HOP binds to the C-termini of Hsp70 and Hsp90 via its two distinct TPR domains, TPR1, TPR2A, respectively, forming the intermediate complex in the Hsp90 chaperoning pathway.7
There has been disparity in the literature regarding the stoichiometry of the Hsp70-HOP-Hsp90 complex, because of inconsistent reports of the oligomeric state of HOP. There have been reports that HOP is a monomer12 or dimer13–15 or both.16 We know that Hsp70 is a monomer and that Hsp90 is a dimer. To understand the complex formation between these chaperones and HOP, it is essential that we know the oligomeric state of HOP.
In this study, we consistently observe that hHOP, dHOP, and stable fragments of these proteins behave as monomers, and we see no indication of any propensity to form dimers or other higher MW species. Therefore, one must ask why some (but not all) researchers have suggested that HOP forms dimers. One possible explanation is if the experiments that report HOP is a monomer were performed at significantly lower concentrations than those that report it is a dimer. This is not the case. The AUC experiments in which HOP behaves exclusively as a monomer were performed at protein concentrations of more than 20 μM. The crystallization of the TRP2A and TPR2B domains of hHOP, with peptide ligand bound, was performed at protein concentrations of about 10 mg/mL (0.5 mM). The structures show that the isolated TPR domains are monomeric. AUC experiments on Sti1, the yeast homologue of HOP, were performed at concentrations up to 15 μM, and it was reported that there was a monomer-dimer equilibrium, with dimer at the highest concentration. In this example, whether these observations reflect an intrinsic difference between this protein and hHOP and dHOP is unclear. Onuoha et al.15 reported the results of a Small Angle X-Ray Scattering (SAXS) study, suggesting a model in which hHOP has a “butterfly-like” structure with TPR2A being the primary dimerization domain. In this example, HOP was reported to be dimeric at all concentrations studied (down to 5 μM).
We propose several reasons that may account for the discrepancies between different studies. The method of gel filtration experiment separates on the basis of hydrodynamic radius. One only expects a protein to migrate at an apparent MW that corresponds to its actual MW, and if it is a globular protein, that runs on a column that has been calibrated with globular proteins. Indeed, we also observe that HOP migrates as if it is not a monomeric globular protein. From gel filtration alone, one cannot distinguish between an elongated monomer, a dimer, or even an elongated dimer. Thus, for full-length HOP, it is in the interpretation of gel filtration data that we differ from other researchers, not in the observation. For the TPR2A domain, we see migration exclusively as a monomer (Fig. 3). We consistently observe that the isolated TPR1 and TPR2A domains are globular and monomeric in solution.
Sedimentation equilibrium AUC is a method to determine MW, independent of the shape of the molecule. In our equilibrium ultracentrifugation experiments, HOP and its fragments behave exclusively as monomers, over a range of concentrations and rotor speeds. There is no indication of dimer or higher MW oligomer formation.
Finally, we have noticed that if either the TRP2A domain or full-length HOP is not stored and used in the presence of fresh reducing agent, there is a tendency to form disulfide-linked dimers (Fig. 5). We suspect that disulfide-linked dimers might be present in some of the experiments of other researchers. HOP is active within the cytoplasm of eukaryotic cells, in a reducing environment, and therefore, we believe that disulfide-linked dimers are not relevant for biological function of HOP in vivo.
Let us consider the implications of HOP being a monomer on the assembly of Hsp70-HOP-Hsp90 complex. If HOP is a monomer, it has one binding site for monomeric Hsp70 and one for dimeric Hsp90. In such a complex, one of the Hsp90 monomers has a free C-terminus that is potentially available to interact with other co-chaperones.
By contrast, if HOP were a dimer, it has been proposed that a dimeric HOP could bind to dimeric Hsp90 with a bidentate dimer-dimer interaction. In such a complex, there would be no free Hsp90 C-terminus available to interact with other co-chaperones. It was proposed that the bidentate dimer-dimer HOP-Hsp90 interaction may lock the conformation of Hsp90 dimer, both preventing it binding to other co-chaperones and inhibiting its ATPase activity. Clearly, this model is not viable if HOP is a monomer.14
We favor a model in which one HOP monomer binds to one Hsp90 dimer. In the cell, if we assume HOP is monomeric, and we know that its endogenous levels are significantly lower than these of Hsp90, a model in which one Hsp90 dimer interacts with one monomeric HOP molecule seems the most reasonable. Indeed, in a study by the Murphy et al.17 using rabbit reticulocyate lysate demonstrated that all the HOP, ∼30% of Hsp90 and 9% of Hsp70, in the lysate is present in a Hsp70-HOP-Hsp90 complex with a stoichiometry of 1:1:2.
In summary, our results indicate that HOP exists exclusively as a monomer in solution, and therefore, it seems reasonable to hypothesize that HOP is monomeric when it performs its co-chaperone activities in the Hsp90 chaperon cycle.
Materials and Methods
Construction of the HOP truncation mutants
The bacterial expression plasmid of full-length hHOP in pProEx-HTa was used as the template DNA for generating truncated hHOP mutants. A stop codon (Fig. 1) was introduced into the hHOP sequence at specified positions by site directed mutagenesis to construct various truncated hHOP variants (hTPR1, hTPR2A, hHOPΔTPR2BΔDP2, and hHOPΔDP2) with domain boundaries defined similarly to that in the study of Chen and Smith.6 Another plasmid expressing TPR2A domain alone (position 222 to 349) was from a previous study.18
Construction of coexpression plasmids
For in vivo coexpression studies, full-length HOP was amplified by polymerase chain reaction (PCR), using hHOP gene in pProEx-HTa expression vector as a template, and cloned as EcoRI/HindIII and MfeI/AatII restriction fragments into the corresponding sites of Multiple Cloning Site (MCS)1 and MCS2, respectively, of pCOLADuet-1 (Novagen) expression vector. The hHOP expressed from MCS1 carries an amino-terminal His6 affinity tag (3010 Da) encoded by the vector, and the tag includes additional 12 amino acid residues carried over from the template used for PCR amplification of HOP gene. The genes encoding for carboxy-terminal fragment (amino acid residues 177–724) of Hsp90 and full-length Hsp90 were amplified by PCR and cloned as BamHI/NotI and MfeI/AatII restriction fragments into the corresponding sites of MCS1 and MCS2, respectively. Hsp90 (amino acid residues 177–724) carries an amino-terminal His6 affinity tag encoded by MCS1 of the expression vector. BamHI restriction site within the hsp90 gene was deleted by T2016C mutation using site-directed mutagenesis (QuickChange kit, Stratagene, La Jolla, CA), without altering the amino acid sequence of Hsp90. All constructs were verified by sequencing.
Protein expression and purification
Full-length hHOP, dHOP, hHOP truncation mutants, pCOLADuet1-HOP/MCS1-HOP/MCS2, and pCOLADuet1-hsp90c/MCS1-hsp90/MCS2 constructs were expressed in E. coli BL21(DE3) (Novagen). Cells were grown in Luria-Bertani (LB) medium at 37°C to an optical density at 600 nm (A600) of 0.6. Overexpression of target genes was induced with 0.6 mM and 1.0 mM isopropyl 1-thio-β-D-galactopyranoside for pCOLADuet1-HOP/MCS1-HOP/MCS2 and pCOLADuet1-hsp90c/MCS1-hsp90/MCS2, respectively, and the cell growth continued for 5 h at 30°C. The cells were pelleted by centrifugation, resuspended in 50 mM Tris (pH 8.0), 300 mM NaCl, 10 mM imidazole, 5% glycerol, and 5 mM β-mercaptoethanol (buffer A), incubated for 30 min on ice in the presence of 1 mg/mL lysozyme, DNAase, and protease inhibitors, and disrupted by sonication. The soluble fraction was separated from cell debris by centrifugation and incubated with Ni-NTA resin (Qiagen) for 40 min at 4°C. After collection of a flow-through fraction, bound proteins were washed with buffer A and eluted with 300 mM imidazole in buffer A containing 150 mM NaCl, followed by dialysis against 50 mM Tris, 100 mM NaCl, and 10 mM β-mecaptomethanol. The proteins expressed form pCOLADuet1-HOP/MCS1-HOP/MCS2 and pCOLADuet1-hsp90c/MCS1-hsp90/MCS2 constructs were analyzed using 10% SDS-PAGE gels and visualized by Coomassie Blue staining. All proteins were estimated to be >90% pure.
Circular dichroism
CD spectra were recorded using an AVIV spectrophotometer Model 215 (AVIV Instruments Inc.). Proteins were buffer exchanged and equilibrated with the assay buffer in 20 mM phosphate, pH 7.4, 100 mM NaCl, and 0.5 mM Tris(2-carboxyethyl) phosphine (TCEP). Protein secondary structures were measured in the far ultraviolet region (260 ∼ 200 nm) in a continuous mode with data point taken every 1 nm, with an average time of 6 seconds. Measurements were taken at 2-μM protein concentrations in a 0.1-cm path-length cuvette at 25°C. Molar ellipticity for each sample was calculated based on the net CD signals of each sample, after subtracting a buffer blank measured in the same cuvette.
Gel filtration chromatography
The apparent MWs of hHOP, dHOP, and their variants were determined using gel filtration chromatography. Samples with concentrations from 10 to 26 μM were loaded onto a Superdex S-200 16/60 column equilibrated with 50 mM Tris, pH 8.0, 100 mM NaCl, and 10 mM β-mercaptoethanol at a flow rate of 0.3 ml/min. Thyroglobulin (670 kDa), bovine γ-globulin (158 kDa), chicken ovalbumin (44 kDa), equine myoglobine (17 kDa), and vitamin B12 (1.35 kDa) were used as MW references (Bio-rad Laboratories, Hercules, CA).
Sedimentation equilibrium analytical ultracentrifugation
Sedimentation equilibrium experiments were performed on a Beckman XL-I analytical ultracentrifuge at 25°C by using a 60-Ti 4-hole rotor equipped with six-channel, carbon-epoxy composite centerpieces (Beckman Coulter). Purified His6-tagged hHOP, hHOPΔTPR2BΔDP2, and dHOP were dialyzed into the experimental buffers (50 mM Tris-HCl, 150 mM NaCl, and 1mM TCEP at pH 8.0), and the dialysate was used as an optical reference. Data were acquired at rotor speeds of 10k, 15k, 25k revolution per minute (RPM). For each protein, three different concentrations were used—4 μM, 8 μM, and 16 μM for hHOP; 5 μM, 10 μM, and 20 μM for hHOP(ΔTPR2BΔDP2); 6.5 μM, 13 μM, and 25.9 μM for dHOP. Samples were centrifuged for 24 h at each speed. Sedimentation equilibrium curves, measured by absorbance at 280 nm, were acquired every 2 h. The MATCH program was used to assess whether the run at each speed had achieved equilibrium by comparing the radial concentration profile in eight successive scans. The various data sets for each sample were fitted both individually for each speed and simultaneously at all speeds by using HeteroAnalysis software (V1.1.33, James Cole, Jeffery Larry, Analytical Ultracentrifugation Facility, Biotechnology and Bioservices Center, University of Connecticut, Storrs, CT,). Fits of both the ideal single species model and the monomer-dimer equilibrium model to the data were compared. Fixed monomer MWs of 67,591, 47,420 Da and 57,861 Da for hHOP, hHOPΔTPR2BΔAP2(1-368), and dHOP, respectively, as calculated from their amino acid sequences were used in the monomer-dimer model. By using the software SEDNTERP (V1.09, John Philo), the density of the buffer at 25°C was calculated to be 1.003 mg/ml, and the partial specific volumes for hHOP, hHOP (1–368), and dHOP were calculated on the basis of amino acid composition as 0.7327, 0,7322, and 0.7319 ml/mg, respectively.
Acknowledgments
We thank Dr. Michael Chinkers for the Drosophila HOP plasmid and Dr. Sophie Jackson for the hHOP plasmid. We also thank the Regan laboratory members for critically reviewing the manuscript.
References
- 1.Young J, Moarefi I, Hartl FU. Hsp90: a specialized but essential protein-folding tool. J Cell Biol. 2001;154:267–273. doi: 10.1083/jcb.200104079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pratt WB, Toft DO. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp Biol Med. 2003;228:111–133. doi: 10.1177/153537020322800201. [DOI] [PubMed] [Google Scholar]
- 3.Bishop SC, Burlison JA, Blagg BS. Hsp90: a novel target for the disruption of multiple signaling cascades. Curr Cancer Drug Targets. 2007;7:369–388. doi: 10.2174/156800907780809778. [DOI] [PubMed] [Google Scholar]
- 4.Koga F, Kihara K, Neckers L. Inhibition of cancer invasion and metastasis by targeting the molecular chaperone heat-shock protein 90. Anticancer Res. 2009;29:797–807. [PubMed] [Google Scholar]
- 5.Bose S, Weiki T, Bugl B, Buchner J. Chaperone function of Hsp90-associated proteins. Science. 1996;274:1715–1717. doi: 10.1126/science.274.5293.1715. [DOI] [PubMed] [Google Scholar]
- 6.Chen S, Smith DF. Hop as an adaptor in the heat shock protein 70 (Hsp70) and hsp90 chaperone machinery. J Biol Chem. 1998;273:35194–35200. doi: 10.1074/jbc.273.52.35194. [DOI] [PubMed] [Google Scholar]
- 7.Scheufler C, Brinker A, Bourenkov G, Bartunik H, Hartl FU, Moarefi I. Structure of TPR domain-peptide complexes: critical elements of the assembly of the Hsp70–Hsp90 multichaperone machine. Cell. 2000;101:199–210. doi: 10.1016/S0092-8674(00)80830-2. [DOI] [PubMed] [Google Scholar]
- 8.Wegele H, Wandinger SK, Schmid AB, Reinstein J, Buchner J. Substrate transfer from the chaperone Hsp70 to Hsp90. J Mol Biol. 2006;356:802–811. doi: 10.1016/j.jmb.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 9.Connell P, Ballinger CA, Jiang J, Wu Y, Thompson LJ, Höhfeld J, Patterson C. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat Cell Biol. 2001;3:93–96. doi: 10.1038/35050618. [DOI] [PubMed] [Google Scholar]
- 10.Minami Y, Kimura Y, Kawasaki H, Suzuki K, Yahara I. The carboxy-terminal region of mammalian HSP90 is required for its dimerization and function in vivo. Mol Cell Biol. 1994;14:1459–1464. doi: 10.1128/mcb.14.2.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.King C, Eisenberg E, Greene L. Polymerization of 70-kDa heat shock protein by yeast DnaJ in ATP. J Biol Chem. 1995;270:22535–22540. doi: 10.1074/jbc.270.38.22535. [DOI] [PubMed] [Google Scholar]
- 12.Young JC, Obermann MJ, Hartl FU. Specific binding of tetratricopeptide repeat proteins to the C-terminal 12-kDa domain of hsp90. J Biol Chem. 1998;273:18007–18010. doi: 10.1074/jbc.273.29.18007. [DOI] [PubMed] [Google Scholar]
- 13.Flom G, Behal RH, Rosen L, Cole DG, Johnson JL. Definition of the minimal fragments of Sti1 required for dimerization, interaction with Hsp70 and Hsp90 and in vivo functions. Biochem J. 2007;404:159–167. doi: 10.1042/BJ20070084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prodromou C, Siligardi G, O'Brien R, Woolfson DN, Regan L, Panaretou B, Ladbury JE, Piper PW, Pearl LH. Regulation of Hsp90 ATPase activity by tetratricopeptide repeat (TPR) domain co-chaperones. EMBO J. 1999;18:754–762. doi: 10.1093/emboj/18.3.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Onuoha SC, Coulstock ET, Grossmann JG, Jackson SE. Structural studies on the co-chaperone Hop and its complexes with Hsp90. J Mol Biol. 2008;379:732–744. doi: 10.1016/j.jmb.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 16.van der Spuy J, Cheetham ME, Dirr HW, Blatch GL. The cochaperone murine stress-inducible protein 1: overexpression, purification, and characterization, protein expression and purification. 2001;21:462–469. doi: 10.1006/prep.2001.1399. [DOI] [PubMed] [Google Scholar]
- 17.Murphy PJM, Kanelakis KC, Galigniana MD, Morishima Y, Pratt WB. Stoichiometry, abundance, and functional significance of the hsp90/hsp70-based multiprotein chaperone machinery in reticulocyte lysate. J Biol Chem. 2001;277:30092–30098. doi: 10.1074/jbc.M103773200. [DOI] [PubMed] [Google Scholar]
- 18.Yi F, Regan L. A novel class of small molecule inhibitors of Hsp90. ACS Chem Biol. 2008;3:645–654. doi: 10.1021/cb800162x. [DOI] [PMC free article] [PubMed] [Google Scholar]


