Abstract
A variety of proteins are capable of converting from their soluble forms into highly ordered fibrous cross-β aggregates (amyloids). This conversion is associated with certain pathological conditions in mammals, such as Alzheimer disease, and provides a basis for the infectious or hereditary protein isoforms (prions), causing neurodegenerative disorders in mammals and controlling heritable phenotypes in yeast. The N-proximal region of the yeast prion protein Sup35 (Sup35NM) is frequently used as a model system for amyloid conversion studies in vitro. Traditionally, amyloids are recognized by their ability to bind Congo Red dye specific to β-sheet rich structures. However, methods for quantifying amyloid fibril formation thus far were based on measurements linking Congo Red absorbance to concentration of insulin fibrils and may not be directly applicable to other amyloid-forming proteins. Here, we present a corrected formula for measuring amyloid formation of Sup35NM by Congo Red assay. By utilizing this corrected procedure, we explore the effect of different sodium salts on the lag time and maximum rate of amyloid formation by Sup35NM. We find that increased kosmotropicity promotes amyloid polymerization in accordance with the Hofmeister series. In contrast, chaotropes inhibit polymerization, with the strength of inhibition correlating with the B-viscosity coefficient of the Jones-Dole equation, an increasingly accepted measure for the quantification of the Hofmeister series.
Keywords: amyloid, Congo red assay, Hofmeister effect, prion, Sup35
Introduction
Amyloids and prions
Amyloids are highly ordered cross-β fibrous polymers associated with a variety of human diseases including Alzheimer's and Parkinson's diseases. Because of the debilitating nature of these diseases, considerable effort has been devoted to studying amyloid formation in hope of developing a cure. Amyloids can form as misfolded proteins self-assemble into a stable fibril with a characteristic cross β-repeat structure, and proteins or peptides of different sequences have been shown to produce amyloid fibrils that share similar characteristics, such as X-ray diffraction patterns and ability to bind certain dyes, for example, Congo Red or thioflavine T.1–3
Prion diseases, or transmissible spongiform encephalopathies (TSE), are infectious amyloid diseases caused by a protein-based agent, composed of a certain protein (called PrP) in an amyloidogenic forms. Human forms of prion illness include Creutzfeld-Jacob disease (CJD), Gerstmann-Sträussler-Scheinker disease (GSS), fatal familial insomnia (FFI) and Kuru, whereas the animal forms include scrapie (Sc) in sheep and goat, and bovine spongiform encephalopathy (BSE), or “mad cow” disease, in cattle, that is transmissible to humans.4 Recent studies show that prion infection and propagation occurs when an abnormally structured protein converts a normal protein molecules of the same sequence into the abnormal (prion) form,5 which then aggregates with other abnormal forms and accumulates as amyloid deposits in the brain of infected humans or animals.6
Prion-like proteins have also been found in fungi including yeast.7–9 Fungal prions are not homologous to mammalian PrP but control transmissible phenotypic traits. They do not ultimately kill fungi, and they certainly are not infectious to humans. The best studied fungal prion protein is the translation termination factor Sup35 protein of yeast Saccharomyces cerevisiae. Yeast cells containing the prion form of Sup35 are referred to [PSI+], as opposed to [psi−] cells bearing the nonprion form of this protein. Due to amyloid aggregation of Sup35, [PSI+] cells are partially defective in termination of translation. The amyloidogenic properties of the Sup35 protein are determined by its N-terminal region (Sup35N), also termed the “prion domain” which contains a QN-rich stretch and region of imperfect oligopeptide repeats.7–9 The adjacent middle region (Sup35M) contains a high concentration of charged residues. The fragment of Sup35 protein bearing N and M regions (Sup35NM) is dispensable for its function in translation termination, which is determined by the C-proximal release factor domain (Sup35C).8
In addition to in vivo studies, amyloid formation of Sup35 derivatives has also been widely studied in vitro.10,11 Protein fragments containing only the N-terminal or the N-terminal with middle region of Sup35 (Sup35NM) are sufficient for forming amyloid fibers.10,11 Due to the presence of M domain, the Sup35NM fragment of 253 amino acids behaves in a more reproducible and controlled fashion than the N-terminal region alone and is therefore most frequently employed for in vitro work. In vitro generated amyloids of Sup35NM are infectious and can confer the prion state to the cellular Sup35 protein upon transfection into a yeast cell.12,13
One of the most commonly used assays for detection of amyloid formation in vitro is based on the ability of Congo Red (CR) dye to bind amyloid fibers, resulting in a red shift of the characteristic absorbance spectrum of CR. This shift is stoichiometrically dependent upon the amount of amyloid, so that the amount of dye bound to amyloid protein can be readily calculated.14 Purified Sup35 or its fragments have been shown to form amyloid fibers that bind to CR in vitro. However, most studies using a CR assay for quantifying Sup35NM and other amyloids utilize formulas previously developed for insulin, which is not directly applicable to other proteins.14,15
Protein stability: Hofmeister series
The Hofmeister series was originally introduced as a series of salts ranked by their ability to precipitate hen egg white proteins.16 This ranking has since been observed to describe the relative effects of ions on a number of protein parameters, such as solubility and stability.16 The ions that increase solubility and decrease stability are commonly known as the chaotropes; conversely, the ions that decrease solubility and increase stability are known as the kosmotropes. Although both cations and anions can be ranked by the Hofmeister series, the anionic effects tend to dominate in aqueous solutions due to their ability to affect polarizable molecules and thus to hydrate more strongly than cations.16 Relative differences in ion hydration have been speculated to be the major cause of the Hofmeister effect,17 and an indication of the ion hydration can be found in the B-coefficient of the Jones-Dole equation, which expresses the relative viscosity of salt solutions as a function of salt concentration.18
The A term in this equation is used to describe electrostatic effects and becomes negligible at higher concentrations (>0.1 M). The D term is only relevant at very high concentrations (>1 M). Owing to their larger hydrated radii, kosmotropes have larger, positive B-viscosity coefficients while chaotropes have negative B-values (Fig. 1). Our previous research has demonstrated that the Jones-Dole B-viscosity coefficient can be used to quantitatively predict the effect of the Hofmeister salts on protein stability.19 Specifically, Broering and Bommarius have shown that the negative B-viscosity coefficient of chaotropes correlates logarithmically with the observed deactivation constants of different proteins, while there seems to be a maximum stabilization effect with kosmotropes (B > 0). Although previous work has shown qualitatively that the Hofmeister series salts correlates with the rate of fibril formation of α-synuclein, no quantitative relationships was demonstrated.20 One recent article on hen egg white lysozyme fibril formation in ionic liquids found no correlation with anion size/type, but the concentration used in the study was much lower than what is used typically to demonstrate Hofmeister effects.21
Figure 1.
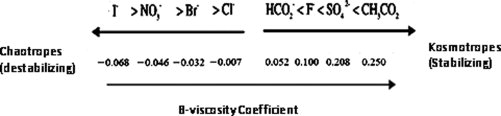
The Hofmeister salts are listed along with their corresponding B-viscosity coefficients. The relative kosmotropicity and chaotropicity of each salt can be followed by its position on the arrow.
Scope of present work
In this article, we adjusted the CR assay for use in studying Sup35NM aggregates. By using this improved assay, we expand the analysis of protein deactivation kinetics by Hofmeister salts to proteins that irreversibly aggregate via forming amyloids, such as Sup35NM. Specifically, we investigate if the simple linear relationship observed between the logarithm of the observed deactivation rate constants of other proteins and the B-viscosity coefficient of deactivating salt anions can also describe the effects of salts on the kinetics of Sup35NM amyloid formation.
Results
Adjustment of congo red assay for Sup35NM fibrils
Almost all previous work involving quantification of Sup35NM and most other amyloids via CR assay utilizes the method developed by Klunk et al.14 However, this method was developed using insulin fibers and has been adopted for Sup35NM and other amyloids without modification. Moreover, a later publication by the same group effectively invalidated the application of the original CR assay to quantification of noninsulin derived amyloid fibers.15 Nevertheless, the original insulin-derived equation linking spectrophotometric measurements to fibril concentrations is still utilized in published reports.10,22,23 We have performed an analysis of CR spectra in the presence of Sup35NM amyloid fibers to adjust the quantitative parameters of the CR assay to the properties of Sup35NM.
Upon binding to amyloid fibrils, the color of CR changes from orange-red to rose red, and the absorption spectra are changed correspondingly. The degree of color change has been shown to correspond quantitatively and linearly to the amount of amyloid present.15 Following Klunk's method, the appropriate parameters needed to adapt the CR assay to Sup35NM fibrils can be identified. Based on absorbance spectra of free CR [Fig. 2(A)] and amyloid-saturated CR [Fig. 2(B)], the wavelengths of maximum and minimum difference were identified as 543 nm and 415 nm, respectively. The molar absorptivities of free CR [Fig. 2(C)] and CR saturated by amyloids [Fig. 2(D)] at these two wavelengths are then obtained by plotting the concentration-dependent absorbance at these two wavelengths. [The numbers can be found in the Supporting Information.]
Figure 2.
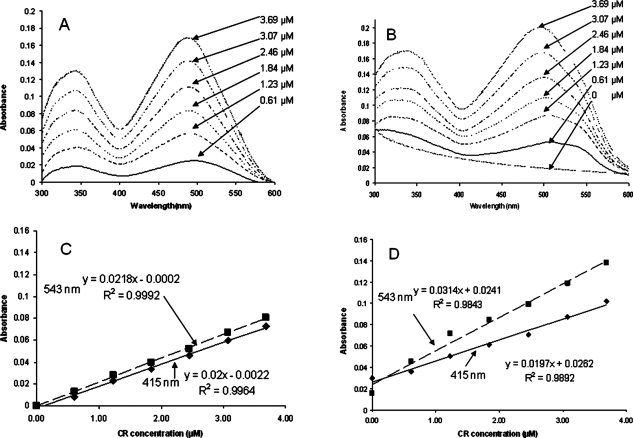
Spectral characteristics of Congo Red (CR) and Congo Red with amyloid fibrils. The legends in panel A and B indicates the concentration of CR. (A) Absorbance spectra of free CR at varying CR concentrations in (μM). (B) Absorbance spectra of a CR and varying amyloid concentrations (μM), including pure scattering without CR. (C) Molar absorptivity of free CR at the isosbestic point (415 nm) and point of maximum difference (543 nm). (D) Molar absorptivity of bound CR at isosbestic point (415 nm) and point of maximum difference (543 nm).
The total absorbance at a wavelength (w), wAt, is equal to the sum of the absorbance of bound CR (Ab) and free CR (Af), and using the Lambert-Beer law with an assumed path length of 1 cm, this total can be written as:
where wa is the molar absorptivity of the at wavelength w, and c is the concentration of bound or free CR.15 The only two unknowns that cannot be directly observed are cb and cf. However, they can be calculated from absorbance measurements at two wavelengths using Eqs. (1) and (2):
| (1) |
| (2) |
Then equating (1) to (2) and solving for cb:
| (3) |
In the spectrum of a mixture of free and amyloid-bound CR, the highest signal to noise is obtained at the point of maximum difference, which for Sup35NM is observed to occur at 543 nm. Using this wavelength, the detection limit is maximized when dealing with a low ratio of cb/cf. The second point is chosen somewhat arbitrarily to be a point of minimum difference in absorbance, which was chosen to be 415 nm. Therefore:
| (4) |
From Figure 2, we can obtain the molar absorptivities listed in Table I. Substituting these values into Eq. (4) gives:
| (5) |
Table I.
Molar Absorptivities of Congo Red Solution and Values Used for Calculation
| Wavelength (nm) | Unbound CR Ab (M cm)−1 | Bound CR Af (M cm)−1 | % difference |
|---|---|---|---|
| 543 | 21800 | 31400 | 44.04 |
| 415 | 20000 | 19700 | 1.50 |
Klunk et al.15 recognized the importance of correcting absorbance measurements for effects of light scattering due to the presence of amyloid fibrils. This correction is applied to the Sup35NM amyloids following Klunk's method using an “r” value of 0.538, which can be determined from Figure 2(B) as the ratio of 543A/415A of a solution of only fibrils, to obtain the final equation for the calculation of CR-amyloid complexes:
| (6) |
In this form, the CR-amyloid concentration in an undetermined sample can be calculated from the absorbance at 543 nm, 415 nm, and the absorbance of an amyloid-free control CR sample at 415 nm. Eq. (6) has the correct coefficients that have to be used to calculate the concentration of Sup35NM amyloids; they are different from the coefficients derived for insulin that are usually used in literature. The error when neglecting scattering by fibrils was found to be around 66%; when accounting for background scatter but employing the coefficients derived for insulin the error still is on the order of 50% (see Supporting Information).
Evaluating the effect of the Hofmeister series on amyloid polymerization
Experiments on Sup35NM polymerization were performed under three different protocols: (1) unseeded polymerization in quiescent solution; (2) unseeded polymerization in agitated solution; and (3) seeded polymerization (see “Material and Methods” section for the description of protocols).
The lag phase, defined as the period in which little or no polymerization is observed, is highly noticeable under quiescent conditions, and can last up to 20 hours.10 In solutions subjected to continuous rotation, the lag phase is almost eliminated and polymerization rate is increased in a concentration dependent manner (Supporting Information Fig. S-1). In the seeded experiments, sonicated preformed fibrils were added to the Sup35NM solution, thus eliminating the lag phase.10
In both unseeded and seeded experiments, an increase of Sup35NM polymerization rate was observed in the presence of kosmotropes (NaHCO2, NaF, Na2SO4, and NaCH3CO2). This increase was more pronounced solutions containing stronger kosmotropes [Fig. 3(A,B)]. In contrast, little to no polymerization occurred in with the presence of strongly chaotropic salts both with and without seeding [Fig. 3(C,D)].
Figure 3.
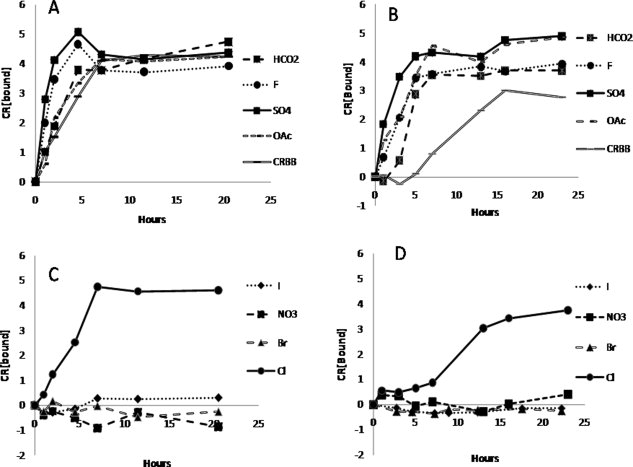
The effect of the Hofmeister salts on the polymerization of Sup35 NM under seeded and unseeded condition. Sodium salt anions are listed to the right. Seeded reactions contained 2% by volume sonicated preformed fibril samples. (A) Seeded condition and kosmotropic salts. (B) Unseeded condition and kosmotropic salts. (C) Seeded condition and chaotropic salts. (D) Unseeded condition and chaotropic salts. Units of CR[bound] are (M cm−1).
In agreement with the quiescent and seeded experiments, strongly kosmotropic salts also increased Sup35NM polymerization rate in the unseeded agitated solutions [Fig. 4(A)]. Surprisingly, the total amount of amyloids detected by the CR assay began to decrease after reaching the peak in the presence of three of the four kosmotropic salts studied, and eventually returned to the level observed at the outset of the experiment. Further research is required to elucidate the mechanism of this effect. The fibril concentration observed in sodium formate solutions did not show a decrease in polymer abundance even after 40 hrs. Chaotropes delayed polymerization in the agitated solutions and decreased the polymerization rate although they did not completely abolish amyloid formation [Fig. 4(B)].
Figure 4.
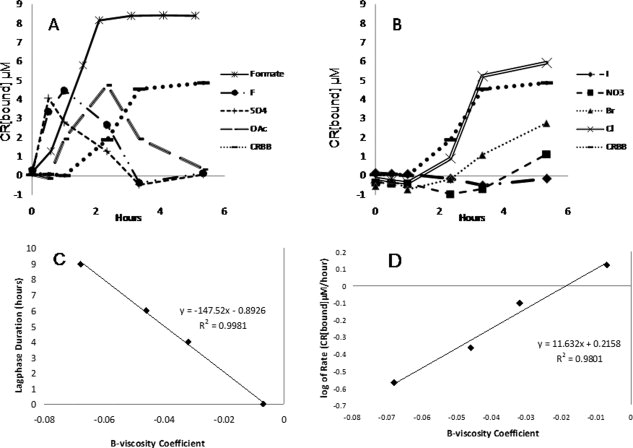
The effect of the Hofmeister salts on agitated Sup35 NM polymerization. (A) Agitated polymerization of 10 μM Sup35 NM in kosmotropic solutions. (B) Agitated polymerization of 10 μM Sup35NM in chaotropic solutions. (C) In a separately conducted agitated experiment with 15 μM Sup35 NM focusing on the chaotropic samples, a negative linear relationship was found between the lag phase of the solution (in hours) to the B-viscosity coefficient of the Hofmeister salt. The Figure with original data can be found in the Supporting Information. (D) In the same experiment as panel C, a linear correlation was found between the initial rate of aggregation to the B-viscosity coefficient of the Hofmeister salt.
In general, the increase in observed polymerization rates was roughly proportional to the kosmotropicity of the anion. In our experiments, the general ranking of polymerization rate has consistently shown to be sulfate > fluoride > acetate > formate > chloride in all experimental conditions, which is consistent with experimentally observed rankings of kosmotropic effects on protein properties such as solubility and stability.16 However, the effect of kosmotropes on amyloid polymerization did not correlate with the B-viscosity coefficient, as we originally hypothesized. Enzyme deactivation kinetics were previously demonstrated to correlate with the B-viscosity coefficient of chaotropic anions while deactivation kinetics appear relatively unaffected by kosmotropic salts. If ranked by B-viscosity coefficients, the general kosmotropicity ranking of the salts should be acetate > sulfate > formate > fluoride > chloride.19 Chloride, with a B-viscosity coefficient around −0.007, lies at the interface between chaotropes and the kosmotropes and seemed to somewhat inhibit polymerization but not nearly as efficiently as strongly chaotropic salts.
Quite interestingly, especially at higher concentrations of Sup35NM, we observe that reduced chaotropicity decreases lag time and accelerates fibril formation rates. The length of the lag phase decreased linearly with B-viscosity coefficient, while the logarithm of polymerization rate correlated linearly [Fig. 4(C,D)]. This suggests that, as noted with protein stability kinetics,19 amyloid fibril formation kinetics are sensitive to chaotropic anion hydration and ion hydration plays an important role in the formation of amyloidal fibrils.19
Finally, we tested whether the inhibitory effect of chaotropic ions on amyloid polymerization is dosage-dependent. Indeed, polymerization of Sup35NM was gradually inhibited as the concentration of NaNO3 was increased, with both length of lag phase [Fig. 5(B)] and maximal polymerization rate [Fig. 5(C)] linearly dependent on concentration of NaNO3.
Figure 5.
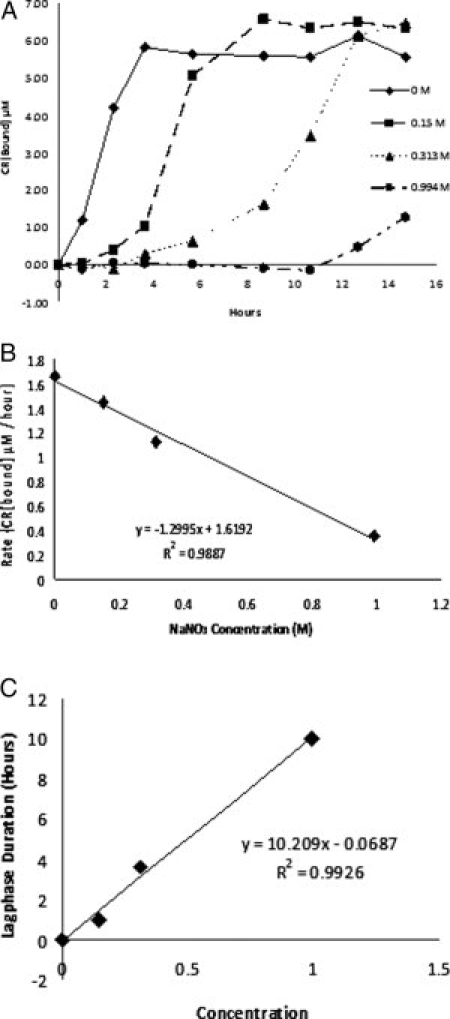
Concentration dependence of the inhibition effect of NaNO3 on Sup35NM polymerization. (A) Various concentrations of NaNO3 were tested under the agitated condition. The concentrations used were 0 M, 0.15 M, 0.313 M, and 0.994 M NaNO3 with 5 mM NaH2PO4 at pH 7.4. (B) Negative correlation between the initial polymerization rate and concentration of NaNO3 were observed. (C) Linear correlation between the length of polymerization lag phase and the concentration of NaNO3 was observed.
Discussion
There is considerable evidence that amyloid formation proceeds via a nucleated polymerization mechanism.7–9,24–26 In such a model, the amyloid-forming (prion) protein initially generates an oligomer capable of immobilizing the native protein molecules and converting them into a prion form, resulting in oligomer growth and, eventually, in formation of the amyloid fibril. While large fibrils may have low or no conversion-promoting activity, smaller pieces generated by fragmentation of the amyloid fibril are capable of restarting the cycle.11 Studies of the in vitro Sup35NM polymerization indicate that relatively small Sup35NM oligomers, between 20 and 80 molecules in size, are the molecular intermediate for amyloid formation.27,28 In a subsequent study, we intend to test if Sup35NM aggregation fits the nucleated polymerization model.
Our current experimental results clearly demonstrate that the chaotropic salts inhibit amyloid formation. As this effect is observed in both unseeded and seeded reactions, most likely chaotropes prevent protein conversion into an amyloidogenic state and disrupt both initial formation of oligomeric nuclei and extension of the pre-existing ones. Kosmotropic salts solutions, on the other hand, consistently increased the polymerization rate.
Because strong denaturants such as urea and GdnHCl are commonly used to maintain the amyloidogenic proteins in a soluble state, it is perhaps not too surprising to observe chaotropic inhibition of Sup35NM fibril formation. The physical basis for such a behavior, however, is less obvious. It is rather intriguing that anion hydration (as indicated by the B-viscosity coefficient) can be used to rank effects of chaotropic salts on fibril formation lag time and polymerization rate, though it does not accurately rank the effects of kosmotropic salts. Our recent analysis on globular proteins suggests that protein deactivation in the chaotropic region is thermodynamically controlled and proceeds through an unfolded protein state, while deactivation in kosmotropic solutions is more likely to be kinetically controlled.29 Similar consideration of the cosolute effects on the native and intermediate species during Sup35NM amyloid formation might also explain the differential effects of each salt. One possibility is that dissolved kosmotropes disrupt the stability of the native Sup35NM molecule. Sup35NM is naturally an unfolded protein in solution with a high degree of random coil content.22 This conformational flexibility is also found in other amyloidogenic proteins and is thought to be important for prion-forming capabilities.7 Interestingly, loosely packed, random-coil proteins have been shown to be thermodynamically destabilized with more accessible conformation states in kosmotropic solution.30 Therefore, dependence of the polymerization rate on the Hofmeister effect could be a reflection of the relative thermodynamic stability of native, loosely packed and amyloidic, more tightly-packed state induced by cosolute addition; then, the kosmotropic salts acting as destabilizers while the chaotropic salts act as thermodynamically stabilizing agents.
Alternatively, and more likely, the Hofmeister effect on prion polymerization could be considered as a result of the cosolute effect on the secondary structures in misfolded Sup35NM and its fibrils. Kosmotropic salt solutions has been shown to decrease the random coil content and subsequently increase beta sheet content of proteins while chaotropic salt solutions have the opposite effect.22 As amyloid formation depends on the cross-β interactions, and possibly on generation of the precursor with high β-sheet contents,31 strongly kosmotropic salts may promote amyloid formation by stabilizing the misfolded form and increasing the population of misfolded forms, while the chaotropic salts may inhibit amyloid formation by antagonizing β-sheet formation.
One more explanation could focus on kinetics. On the basis of solubility, it is not beyond reason to surmise that the kosmotropic salt solutions facilitate the formation of protein oligomers, while chaotropic salts can potentially inhibit the formation of oligomers. If relative concentration of oligomeric intermediates in the solution is increased, this would effectively increase the concentration of the substrates interacting with amyloidogenic nuclei. In such a model, the increased lag phase in the chaotropic solutions would be explained by low concentration of the oligomerizing intermediates joining the nuclei, rather than by low concentration of nuclei per se.
These models are not mutually exclusive and may simply reflect different parameters influenced by the Hofmeister effect. Further experiments are needed to decipher the molecular mechanism of the observed phenomena. It is also interesting to see whether effects detected for Sup35NM will be applicable to other amyloids. A number of recent studies have supported our observation of accelerated fibril formation in kosmotropic salts using different amyloid forming systems.32,33 If our observation on chaotropic salt inhibition holds true for other systems, chaotropic salt solutions may potentially serve as a basis for the treatments that would decrease amyloidogenic potential of the prion contaminants and in this way, prevent the spread of prion infections via medical mistreatment.
In conclusion, the results in this study clearly show that polymerization of the yeast prion protein Sup35NM into an amyloid is influenced by the Hofmeister effect. In particular, the kosmotropes accelerate polymerization while the chaotropes inhibit polymerization. The stringency of the acceleration effect of the kosmotropes is shown to follow the Hofmeister series, and the inhibition effect of chaotropes correlates with both the B-viscosity coefficient and chaotrope concentration. These findings suggest that water-protein interactions play a critical role in the processes of amyloid formation and propagation.
Material and Methods
Strains and Plasmids
HMS174 (pLysS) (Novagen) was used as a host E. coli strain for the pET20b expression system. The construction of plasmid pET-20b-SUP35NM-(His)6 was published previously.34
Expression and purification of Sup35NM
Sup35NM was expressed in HMS174 (DE3)/pLysS. In each case, 10 individual freshly obtained transformants were grown each in 5 mL of LB media with 7–52 μg/mL chloramphenicol and 100 μg/mL ampicillin, then pooled to inoculate a 1 L culture with a starting OD600 of about 0.01–0.05. The culture was grown to OD of 0.6–0.8 and induced with 1 mM IPTG for 4 hours. Cell pellets were collected by centrifugation and stored at −80°C until purification.
All steps of Sup35NM purification were carried out at room temperature (∼23°C). Cells were lysed by incubation in the denaturing buffer (10 mM TrisċHCl, pH 8, 8 M urea) for 30 min. Cell debris was removed by centrifugation at 10,000 g for 20 min. The supernatant was then applied to a 5 mL bed volume of Ni2+ NTA resin in a superflow column. The column was washed with ∼5 bed volumes of the Congo Red binding buffer (CRBB) (5 mM potassium phosphate, 150 mM sodium chloride, pH 7.4) containing 10 mM imidazole, and eluted with the same buffer containing 50 mM imidazole. The eluted fraction was then concentrated to 6–8 mg/mL and was used immediately after purification. Sup35NM concentrations were measured on a Beckmann-Coulter DU®800 spectrophotometer (Fullerton, CA) at 280 nm, using a protein extinction coefficient of 1.045 abs/(mg/mL)/cm−1.35
Congo red assay development
To create amyloid fibers for the Congo Red (CR) assay, an aliquot of the concentrated Sup35NM stock was diluted to 10 mL of 20 μM Sup35 NM with CRBB. If left for several days in these conditions at room temperature, Sup35NM spontaneously forms fibers. As residual urea from the original purification might interfere the Congo Red assay, special care was taken to remove the urea by dialysis. The entire volume of the Sup35 NM solution was sealed tightly in a 3500 molecular weight cut off dialysis membrane and dialyzed against 1 L CR binding buffer for 4 days, with a buffer being changed every day. At the end of the fourth day, the liquid inside the dialysis membrane became visibly turbid, and the entire contents were moved to a separate tube carefully. A small portion of the amyloid stock was tested for residual soluble Sup35NM: a 500 μL sample was centrifuged at 18,000g for 5 minutes, and the supernatant measured with A280. The A280 results (<0.05) indicate that soluble Sup35NM concentration was negligible. The Sup35NM amyloid stock solution was then mixed with various concentrations of CR ranging from 0 to 4 μM. These concentrations were chosen to ensure saturation of the CR by amyloids. A control sample set with the same concentrations of CR but with no amyloid samples was also created. After 15 minutes of incubation at room temperature, scans from 300 to 600 nm were taken for amyloid and CR and CR alone. The remaining derivation of the CR assay is shown in the “Results” section.
Congo red assay
To monitor Sup35NM amyloid fibril formation via CR assay, aliquots were taken from the protein sample at desired time points, diluted with CR buffer (12 μM CR, 5 μM potassium phosphate buffer, 150 mM sodium chloride, pH 7.4) to a final concentration of 2 μM protein and 10 μM CR, and incubated for 20 minutes at ambient temperature. Absorbance measurements were then taken at 543 nm and 415 nm. Congo Red binding is then calculated as described in “Results” section. Because the calculated CR value does not always begin at zero extinction due to the variation in the Hofmeister sample buffer, the value at the time point t = 0 is used to create a base line.
CR inhibition study
To test whether or not the salts used in our experiments can potentially interfere with the Congo Red Assay, fully polymerized 20 μM Sup35NM solutions in CRBB were mixed at a 1:1 ratio with twice-concentrated solutions of the eight Hofmeister salts at 5 mM potassium phosphate buffer pH 7.4 to create a final salt concentration of 0.97 water activity. The resulting mixtures were then assayed with the Congo Red Assay. The results clearly show that very little difference can be seen in the assay result of the eight different Hofmeister buffers when equal amount of amyloids are present (Fig. 6).
Figure 6.
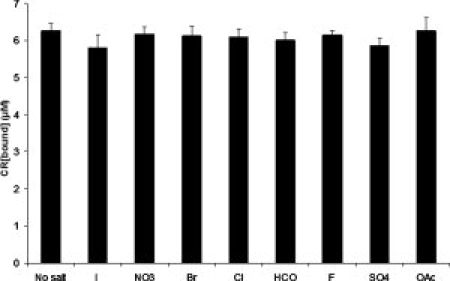
Congo Red binding to an amyloid in the presence of Hofmeister salts. The amyloids were formed in the absence of salts, but CR assay was conducted in the presence of salt as described in “Material and Methods” section. No inhibitory effect of any salt on CR binding is detected.
Sup35NM polymerization rates in sodium salt solutions
The highly concentrated unaggregated stock was diluted at least 50-fold into 10 μM with various sodium salt solutions, NaI, NaNO3, NaBr, NaCl, NaHCO2, NaF, Na2SO4, and NaCH3CO2 (pH 7.4, 0.97 water activity, and 5 mM potassium phosphate) and allowed to aggregate at 23°C. Salt concentrations were chosen to maintain constant thermodynamic water activity between salt solutions and to allow direct comparison with a previous study.19 A control was also established using Congo Red Binding Buffer (CRBB; 150 mM NaCl, pH 7.4, 5 mM potassium phosphate) as a reference to previous studies in literature. Polymerization was then measured by Congo Red binding over the course of 3 days. For seeded aggregation experiments, sonicated preformed fibrils were added to the salt solutions at 2% (1:50 by mass) of the total volume. For the agitated experiments, the sample solutions were placed on a 12 rpm rotator (Fisher Scientific, Chemistry Mixer Model 346) and polymerization was followed by Congo Red assay for the next 4–5 hours, or until polymerization is complete. The polymerization rate was then obtained from the data by measuring the maximum slope. Although protein batch variations were observed between each experiment, the general trend remains consistent.
Acknowledgments
V. Y. gratefully acknowledges assistantships from the Presidential Undergraduate Research Award (Georgia Institute of Technology) and the Undergraduate Research Scholar Program (Parker H. Petit Institute for Bioengineering and Bioscience). J. M. B. gratefully acknowledges support from a National Science Foundation Graduate Research Fellowship and a GAANN Fellowship.
References
- 1.Sipe JD, Cohen AS. Review: history of the amyloid fibril. J Struct Biol. 2000;130:88–98. doi: 10.1006/jsbi.2000.4221. [DOI] [PubMed] [Google Scholar]
- 2.Kyle RA. Amyloidosis: a convoluted story. Br J Haematol. 2001;114:529–538. doi: 10.1046/j.1365-2141.2001.02999.x. [DOI] [PubMed] [Google Scholar]
- 3.Lai Z, Colon W, Kelly JW. The acid-mediated denaturation pathway of transthyretin yields a conformational intermediate that can self-assemble into amyloid. Biochemistry. 1996;35:6470–6482. doi: 10.1021/bi952501g. [DOI] [PubMed] [Google Scholar]
- 4.Prusiner SB. Prions. Proc Natl Acad Sci USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dobson CM. The structural basis of protein folding and its links with human disease. Philos Trans R Soc Lond B Biol Sci. 2001;356:133–145. doi: 10.1098/rstb.2000.0758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prusiner SB, Scott MR, Dearmond SJ, Cohen FE. Prion protein biology. Cell. 1998;93:337–348. doi: 10.1016/s0092-8674(00)81163-0. [DOI] [PubMed] [Google Scholar]
- 7.Wickner RB, Edskes HK, Shewmaker F, Nakayashiki T. Prions of fungi: inherited structures and biological roles. Nat Rev Microbiol. 2007;5:611–618. doi: 10.1038/nrmicro1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tuite MF, Cox BS. Propagation of yeast prions. Nat Rev Mol Cell Biol. 2003;4:878–890. doi: 10.1038/nrm1247. [DOI] [PubMed] [Google Scholar]
- 9.Chernoff YO. Amyloidogenic domains, prions and structural inheritance: rudiments of early life or recent acquisition? Curr Opin Chem Biol. 2004;8:665–671. doi: 10.1016/j.cbpa.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 10.Glover JR, Kowal AS, Schirmer EC, Patino MM, Liu JJ, Lindquist S. Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell. 1997;89:811–819. doi: 10.1016/s0092-8674(00)80264-0. [DOI] [PubMed] [Google Scholar]
- 11.Chernoff YO, Uptain SM, Lindquist SL. Analysis of prion factors in yeast. Methods Enzymol. 2002;351:499–538. doi: 10.1016/s0076-6879(02)51867-x. [DOI] [PubMed] [Google Scholar]
- 12.King CY, Diaz-Avalos R. Protein-only transmission of three yeast prion strains. Nature. 2004;428:319–323. doi: 10.1038/nature02391. [DOI] [PubMed] [Google Scholar]
- 13.Tanaka M, Chien P, Naber N, Cooke R, Weissman JS. Conformational variations in an infectious protein determine prion strain differences. Nature. 2004;428:323–328. doi: 10.1038/nature02392. [DOI] [PubMed] [Google Scholar]
- 14.Klunk WE, Pettegrew JW, Abraham DJ. Two simple methods for quantifying low-affinity dye-substrate binding. J Histochem Cytochem. 1989;37:1293–1297. doi: 10.1177/37.8.2666512. [DOI] [PubMed] [Google Scholar]
- 15.Klunk WE, Jacob RF, Mason RP. Quantifying amyloid by congo red spectral shift assay. Methods Enzymol. 1999;309:285–305. doi: 10.1016/s0076-6879(99)09021-7. [DOI] [PubMed] [Google Scholar]
- 16.Collins KD, Washabaugh MW. The Hofmeister effect and the behavior of water at interfaces. Q Rev Biophys. 1985;18:323–422. doi: 10.1017/s0033583500005369. [DOI] [PubMed] [Google Scholar]
- 17.Lavelle L, Fresco JR. Stabilization of nucleic acid triplexes by high concentrations of sodium and ammonium salts follows the Hofmeister series. Biophys Chem. 2003;105:681–699. doi: 10.1016/s0301-4622(03)00097-8. [DOI] [PubMed] [Google Scholar]
- 18.Jones GD. The viscosity of aqueous solutions of strong electrolytes with special reference to barium chloride. J Am Chem Soc. 1929;51:2950. [Google Scholar]
- 19.Broering JM, Bommarius AS. Evaluation of Hofmeister effects on the kinetic stability of proteins. J Phys Chem B. 2005;109:20612–20619. doi: 10.1021/jp053618+. [DOI] [PubMed] [Google Scholar]
- 20.Munishkina LA, Henriques J, Uversky VN, Fink AL. Role of protein-water interactions and electrostatics in alpha-synuclein fibril formation. Biochemistry. 2004;43:3289–3300. doi: 10.1021/bi034938r. [DOI] [PubMed] [Google Scholar]
- 21.Kalhor HR, Kamizi M, Akbari J, Heydari A. Inhibition of amyloid formation by ionic liquids: ionic liquids affecting intermediate oligomers. Biomacromolecules. 2009;10:2468–2475. doi: 10.1021/bm900428q. [DOI] [PubMed] [Google Scholar]
- 22.Scheibel T, Lindquist SL. The role of conformational flexibility in prion propagation and maintenance for Sup35p. Nat Struct Biol. 2001;8:958–962. doi: 10.1038/nsb1101-958. [DOI] [PubMed] [Google Scholar]
- 23.Ono B, Kawaminami H, Kobayashi H, Kubota M, Murakami Y. Effects of mutations in yeast prion [PSI+] on amyloid toxicity manifested in Escherichia coli strain BL21. Prion. 2008;2:37–41. doi: 10.4161/pri.2.1.6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andrews JM, Roberts CJ. A Lumry-Eyring nucleated polymerization model of protein aggregation kinetics: 1. Aggregation with pre-equilibrated unfolding. J Phys Chem B. 2007;111:7897–7913. doi: 10.1021/jp070212j. [DOI] [PubMed] [Google Scholar]
- 25.Li Y, Roberts CJ. Lumry-eyring nucleated-polymerization model of protein aggregation kinetics. 2. Competing growth via condensation and chain polymerization. J Phys Chem B. 2009;113:7020–7032. doi: 10.1021/jp8083088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Collins SR, Douglass A, Vale RD, Weissman JS. Mechanism of prion propagation: amyloid growth occurs by monomer addition. PLoS Biol. 2004;2:e321. doi: 10.1371/journal.pbio.0020321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Serio TR, Cashikar AG, Kowal AS, Sawicki GJ, Moslehi JJ, Serpell L, Arnsdorf MF, Lindquist SL. Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science. 2000;289:1317–1321. doi: 10.1126/science.289.5483.1317. [DOI] [PubMed] [Google Scholar]
- 28.Xu S, Bevis B, Arnsdorf MF. The assembly of amyloidogenic yeast sup35 as assessed by scanning (atomic) force microscopy: an analogy to linear colloidal aggregation? Biophys J. 2001;81:446–454. doi: 10.1016/S0006-3495(01)75712-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Broering JM, Bommarius AS. Kinetic model for salt-induced protein deactivation. J Phys Chem B. 2008;112:12768–12775. doi: 10.1021/jp7120164. [DOI] [PubMed] [Google Scholar]
- 30.Der A, Kelemen L, Fabian L, Taneva SG, Fodor E, Pali T, Cupane A, Cacace MG, Ramsden JJ. Interfacial water structure controls protein conformation. J Phys Chem B. 2007;111:5344–5350. doi: 10.1021/jp066206p. [DOI] [PubMed] [Google Scholar]
- 31.Yang DS, Yip CM, Huang TH, Chakrabartty A, Fraser PE. Manipulating the amyloid-beta aggregation pathway with chemical chaperones. J Biol Chem. 1999;274:32970–32974. doi: 10.1074/jbc.274.46.32970. [DOI] [PubMed] [Google Scholar]
- 32.Sikkink LA, Ramirez-Alvarado M. Salts enhance both protein stability and amyloid formation of an immunoglobulin light chain. Biophys Chem. 2008;135:25–31. doi: 10.1016/j.bpc.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lodderstedt G, Sachs R, Faust J, Bordusa F, Kuhn U, Golbik R, Kerth A, Wahle E, Balbach J, Schwarz E. Hofmeister salts and potential therapeutic compounds accelerate in vitro fibril formation of the N-terminal domain of PABPN1 containing a disease-causing alanine extension. Biochemistry. 2008;47:2181–2189. doi: 10.1021/bi701322g. [DOI] [PubMed] [Google Scholar]
- 34.Allen KD, Wegrzyn RD, Chernova TA, Muller S, Newnam GP, Winslett PA, Wittich KB, Wilkinson KD, Chernoff YO. Hsp70 chaperones as modulators of prion life cycle: novel effects of Ssa and Ssb on the Saccharomyces cerevisiae prion [PSI+] Genetics. 2005;169:1227–1242. doi: 10.1534/genetics.104.037168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mach H, Middaugh CR, Lewis RV. Statistical determination of the average values of the extinction coefficients of tryptophan and tyrosine in native proteins. Anal Biochem. 1992;200:74–80. doi: 10.1016/0003-2697(92)90279-g. [DOI] [PubMed] [Google Scholar]


