Abstract
Topoisomerase IIβ binding protein 1 (TopBP1) is a major player in the DNA damage response and interacts with a number of protein partners via its eight BRCA1 carboxy-terminal (BRCT) domains. In particular, the sixth BRCT domain of TopBP1 has been implicated in binding to the phosphorylated transcription factor, E2F1, and poly(ADP-ribose) polymerase 1 (PARP-1), where the latter interaction is responsible for the poly(ADP-ribosyl)ation of TopBP1. To gain a better understanding of the nature of TopBP1 BRCT6 interactions, we solved the crystal structure of BRCT6 to 1.34 Å. The crystal structure reveals a degenerate phospho-peptide binding pocket and lacks conserved hydrophobic residues involved in packing of tandem BRCT repeats, which, together with results from phospho-peptide binding studies, strongly suggest that TopBP1 BRCT6 independently does not function as a phospho-peptide binding domain. We further provide insight into poly(ADP-ribose) binding and sites of potential modification by PARP-1.
Keywords: X-ray crystal structure, DNA damage response, BRCT domain, peptide interaction, ADP-riboyslation
Introduction
DNA damage is an ongoing event that cells must counteract in order to maintain genomic integrity. As a result, cells have developed elaborate mechanisms to detect the damage and transduce signals to ultimately repair the specific lesion encountered. Topoisomerase IIβ binding protein 1 (TopBP1) has been implicated in a number of processes in response to DNA damage, including DNA replication control, DNA repair, and checkpoint control.1 Perhaps the most characterized function of TopBP1 is the activation of the ATR (ATM and Rad3-related) protein kinase, which plays a crucial role in phosphorylating a number of key target proteins in response to replication stress.2
A unique feature of TopBP1 is the eight BRCA1 carboxy-terminal (BRCT) domains spanning the protein, the most of any BRCT domain-containing protein. Studies of the BRCT domain in Breast cancer associated 1 (BRCA1) and other BRCT-containing proteins indentified its role as a protein–protein binding module.3,4 For example, the role of the tandem BRCT repeats of BRCA1 is to specifically recognize the phosphorylated peptide motif pSer-Pro-Thr-Phe that is present in various protein partners involved in the DNA damage response.3–6 Because TopBP1 contains a number of BRCT domains, various protein partners identified through TopBP1 BRCT-dependent interactions have emerged. For instance, the activation of ATR by TopBP1 is dependent on the interaction between TopBP1 BRCT1/2 and phosphorylated Rad9.7,8 The molecular basis of BRCT interaction has also been elucidated from crystal structures of complexes involving BRCA1 and Mediator of DNA damage checkpoint 1 (MDC1) tandem BRCT repeats with their cognate phospho-peptides (BACH1 and γ-H2AX, respectively).9–11 The conserved mode of recognition revealed by these studies spans both BRCT domains and involves two key regions: a phospho-serine (pSer) recognition pocket in the N-terminal BRCT and a hydrophobic specificity pocket formed at the interface of both BRCT repeats.
Interestingly, single BRCT domains have also been implicated in phospho-peptide binding.4 In particular, there is evidence that the sixth BRCT of TopBP1 interacts with the phosphorylated transcription factor, E2F1, to repress E2F1 activity in response to DNA damage.4,12 However, the molecular mechanism of interaction initiated by a single BRCT domain remains unclear. Besides phospho-peptide binding activity, TopBP1 BRCT6 is also a target for poly(ADP-ribosyl)ation by poly(ADP-ribose) polymerase 1 (PARP-1)13 and it also contains a consensus PAR binding sequence that has been shown in other proteins to bind poly(ADP-ribose) (PAR) chains.14
To gain a better understanding of TopBP1 BRCT6 in phospho-peptide and PAR binding, we solved the crystal structure of TopBP1 BRCT6 to 1.34 Å resolution. The structure reveals a canonical BRCT fold containing a degenerate pSer binding pocket. TopBP1 BRCT6 also lacks the hydrophobic surface that is required for packing of two tandem repeats, suggesting TopBP1 BRCT6 independently does not interact with a phospho-peptide. The structure also provides insight into a consensus PAR binding motif in TopBP1 BRCT6 that likely does not bind PAR, as well as common Glu residues that may be poly(ADP-ribosyl)ated by PARP-1.
Results and Discussion
Overall structure of TopBP1 BRCT6
The crystal structure of TopBP1 BRCT6 was determined and refined to 1.34 Å resolution. TopBP1 BRCT6 both purified and crystallized as a monomer, with one BRCT6 molecule present in the asymmetric unit. Like other BRCT domains, the overall fold of TopBP1 BRCT6 consists of a central four-stranded parallel β-sheet flanked on one side by a single helix (α2) and on the opposite side by a pair of helices (α1 and α3) [Fig. 1(A)]. Structural alignment of the BRCT domain with the N-terminal BRCT domains of BRCA1 and MDC1 also confirm a conserved core involving the β-sheet packed against the α-helices [Fig. 1(B)]. In contrast, both the length and orientation of the connecting loops, particularly the β1-α1 and β3-α2 loops, show less conservation. Unique to the TopBP1 BRCT6 structure is the incorporation of a 310-helix, which replaces the majority of the β1-α1 loop found in other BRCT domains.
Figure 1.
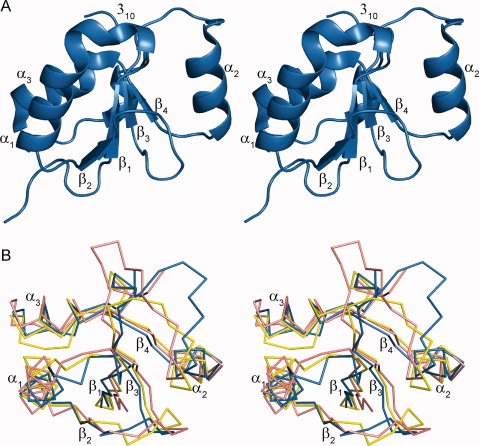
Conserved BRCT fold of TopBP1 BRCT6. (A) Ribbon model of TopBP1 BRCT6 in stereoview. Secondary structure elements are labeled. (B) Superposition of TopBP1 BRCT6 (blue) with the N-terminal BRCT domains of BRCA1 (pink) and MDC1 (yellow) in stereoview.
TopBP1 BRCT6 contains a degenerate phospho-peptide binding pocket
Contrary to previous reports,4 we were unable to observe binding of an E2F1 phospho-peptide (RLLDSpSQIVI) to TopBP1 BRCT6. To further investigate the interaction between TopBP1 BRCT6 and E2F1, we performed binding experiments using isothermal titration calorimetry and fluorescence polarization.15 Although we were able to detect robust binding between the BRCA1 BRCT repeats and its phopho-peptide target derived from BACH1, we were unable to detect interactions between TopBP1 BRCT6 and E2F1 in either of these assays (Supporting Information Fig. S1).
The crystal structures of BRCA1 and MDC1 tandem BRCT repeats in complex with their phospho-peptides (BACH1 and γ-H2AX, respectively) have provided key insights into the mechanism by which the BRCT repeat is selective for a phospho-peptide.9,10 In both BRCA1 and MDC1 structures, the pSer of the bound phospho-peptide is recognized by three structurally conserved residues (Ser1655/Thr1898, Gly1656/Gly1899, and Lys1702/Lys1936 in BRCA1/MDC1, respectively) in the N-terminal BRCT domain [Fig. 2(A)]. These residues make three essential hydrogen bonds with the phosphate moiety and form the phosphate recognition pocket in the BRCT domain. In addition, a conserved Thr1700/Thr1934 residue on the α2 helix makes a hydrogen bond with Ser1655/Thr1898 of BRCA1/MDC1 in order to provide the correct orientation for interaction with the phosphate oxygen. Surprisingly, only Ser1655/Thr1898 is conserved in TopBP1 BRCT6 (Ser913) [Fig. 2(A.B)]. Structural alignment of the phosphate binding pocket of BRCA1 with TopBP1 BRCT6 also supports the lack of conservation. The conserved Ser913 of TopBP1 BRCT6 points away from the solvent as a result of the additional 310-helix between the β1 strand and α1 helix. In addition, the substitution of the conserved Gly1656/Gly1899 of BRCA1/MDC1 to Lys914 on the 310-helix in TopBP1 BRCT6 alters the orientation of the main chain NH such that it is unlikely to form a hydrogen bond with a bound phosphate oxygen. Instead, both Lys914 and Lys915 side chains protrude from the putative pSer binding pocket and are predicted to clash with a putative phospho-peptide. Substitution of the conserved Lys to Glu (Glu957) on α2 helix would also repel the negative charge of the phosphate group. Overall, the degenerate phospho-peptide binding pocket in TopBP1 BRCT6 would likely perturb any phospho-peptide interaction.
Figure 2.
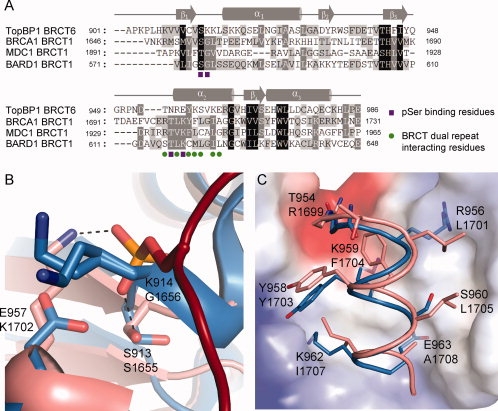
Comparison of TopBP1 BRCT6 to phospho-peptide binding tandem BRCT repeats. (A) Sequence alignment of TopBP1 BRCT6 with N-terminal BRCT domains of BRCA1, MDC1, and BARD1. Residues are shaded based on levels of conservation. Secondary structure elements are plotted above the sequences. Conserved residues involved in pSer binding are shown as purple squares. Conserved residues involved in BRCT repeat packing are shown as green circles. (B) Structural alignment of the pSer binding pocket of BRCA1 (pink) with TopBP1 BRCT6 (blue). The BACH1 phospho-peptide is shown in red. Hydrogen bonding is designated as dotted lines. Hydrogen bonding residues in BRCA1 (below) aligned with TopBP1 (above) are displayed. (C) Structural alignment of the α2 helix of BRCA1 (pink) and TopBP1 BRCT6 (blue) packed against the C-terminal BRCT of BRCA1 (represented as electrostatic surface). Equivalent residues of TopBP1 BRCT6 (above) and BRCA1 (below) are shown.
TopBP1 BRCT6 likely does not form a tandem BRCT repeat
Another requirement of phospho-peptide binding by the tandem BRCT repeats of BRCA1 and MDC1 is the hydrophobic packing of the two BRCT repeats in order to form the specificity pocket for the +3 position of the phospho-peptide. This is facilitated by conserved hydrophobic residues on α2 helix of the N-terminal BRCT and α1 and α3 helices of the C-terminal BRCT. Although TopBP1 BRCT6 purified and crystallized as a monomer, we investigated whether TopBP1 BRCT6 might pack with another BRCT repeat in a similar manner. Interestingly, many of the conserved hydrophobic residues that are involved in packing of the tandem BRCTs in BRCA1 and MDC1 are not present in TopBP1 BRCT6 and are replaced by charged residues [Fig. 2(A,C)]. Crystal packing of TopBP1 BRCT6 also did not provide evidence of a related packing between symmetry molecules. Therefore, it is unlikely that TopBP1 BRCT6 can pack in a dimer analogous to the tandem BRCT repeats in BRCA1 and MDC1. Liu et al. previously suggested that TopBP1 oligomerization could potentially bring two TopBP1 BRCT6 molecules together in a fashion similar to tandem BRCT repeats in order to foster stable phospho-E2F1 recognition.16 Our structural data supports the need for other regions of TopBP1 to interact in order for BRCT6 to form a putative higher order functional unit.
Implications for TopBP1 BRCT6 PAR binding and PARP-1 modification
Pleschke et al. previously identified a consensus PAR binding motif (hxbxhhxbhhb, where h is hydrophobic, b is basic and x is any residue) present in a number of proteins.14 As TopBP1 BRCT6 contains a similar sequence from residues Val908 to Lys918, we wondered if TopBP1 BRCT6 could bind to PAR chains. We were unable to detect any binding to PAR chains in vitro using a PAR binding assay as described by Panzeter et al.17 (data not shown). Mapping of the consensus PAR-binding motif on the BRCT6 structure reveals a region that is partially buried in the core of the BRCT fold [Fig. 3(A)]. We hypothesize that TopBP1 BRCT6 is unlikely to bind to PAR chains. We have also previously shown that TopBP1 BRCT6 can interact with PARP-1 and can be poly-ADP(ribosyl)ated by PARP-1 in vitro.13 TopBP1 BRCT6 interacts with the N-terminal region of PARP-1 that is comprised of a zinc-finger DNA binding domain and an auto-modification domain. Intriguingly, this auto-modification domain consists largely of a BRCT domain that shows a relatively high sequence identity to TopBP1 BRCT6 (25% sequence identity).18 In particular, some putative auto-modification Glu residues in PARP-1 are conserved in TopBP1 BRCT6 and are surface exposed (Fig. 3). Because TopBP1 binds to a number of proteins in the DNA damage response, Glu922, Glu963, Glu971, and Glu986 of TopBP1 may serve as potential sites of PARP-1 poly(ADP-ribosyl)ation as an important way of regulating the activities of both TopBP1 and its protein partners. Further studies will be needed to determine if these specific Glu residues in TopBP1 are indeed modified by PARP-1. Interestingly, TopBP1 BRCT6 also displays high sequence identity to the first BRCT domain of XRCC1 (30% sequence identity), which also exists as a single BRCT domain and has been shown to interact with the N-terminal region of PARP-1.19 This may suggest a conserved regulatory interaction between PARP-1 and targeting proteins containing single BRCT domains in the DNA damage response.
Figure 3.
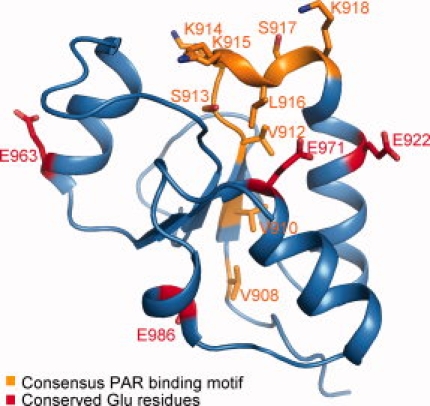
Mapping of putative PAR binding and PARP-1 modification residues in TopBP1 BRCT6. Consensus PAR-binding motif residues are shown in orange. Putative PARP-1 auto-modification Glu residues common in TopBP1 BRCT6 are indicated in red.
Materials and Methods
Expression and purification
TopBP1 BRCT6 (893–996) was cloned into the pGEX-6P-1 vector (GE Healthcare) encoding an N-terminal GST tag. The resultant GST-tag fusion protein was expressed in E. coli BL21-Gold cells and purified using glutathione affinity chromatography. TopBP1 BRCT6 was then cleaved from GST with Prescission protease, and the C-terminal TopBP1 BRCT6 polypeptide was purified from GST by gel filtration chromatography on a Superdex 75 column.
Crystallization
Purified TopBP1 BRCT6 was concentrated to 10 mg/mL in protein buffer (150 mM NaCl, 1 mM DTT and 10 mM Tris-HCl, pH 8) for crystallization. Crystals were grown at 4°C using hanging drop vapor diffusion by mixing 1 μL of protein with 2 μL of reservoir containing 0.1M Tris-HCl, pH 6.8 and 16% PEG 2000 MME. After 2 weeks, crystals were flash-cooled in a cryo-protectant supplemented with 20% glycerol.
Data collection and structure determination
Data was collected at the CMCF-1 beamline at the Canadian Light Source (CLS, Saskatoon). Intensity data were scaled and reduced using the HKL-2000 package.20 Crystals of a TopBP1 BRCT6 variant (W936R), which was identified in the original construct, belonged to the space group C2 with unit cell parameters a = 120.59, b = 88.88, c = 43.31 Å. The Rosetta comparative modeling methodology21 was used to generate models for TopBP1 using 2d8m as a starting template. The crystal structure TopBP1 BRCT6 variant was solved using molecular replacement by the program Phaser22 with the Rosetta-built model, placing three molecules in the asymmetric unit. Automated model building with flex-warp23 was used to build 263 of 270 residues, resulting in an initial Rwork and Rfree of 18.7% and 23.7%, respectively.
Wildtype TopBP1 BRCT6 crystals were obtained in similar crystallization conditions and in the space group P212121 with unit cell dimensions a = 35.77, b = 51.82, c = 62.09 Å (Table I). The partially refined BRCT6 variant was used as a search model for molecular replacement with Phaser to find a solution. The final structure was refined at 1.34 Å resolution to an Rwork and Rfree of 17.2% and 19.4%, respectively, using TLS refinement in REFMAC.24,25 Further TLS and anisotropic B-factor refinement yielded an Rwork of 15.8% and Rfree of 17.3%. The final model contains 818 protein atoms and 153 waters. We were unable to model residues 893–900 from the N-terminus and residue 996 of the C-terminus, which are presumed to be disordered in the crystals. All model building was carried in COOT.26 The Ramachandran plot contained 90.5% of all residues in most favored regions, and 9.5% of residues in additionally allowed regions. The model was validated with PROCHECK.27
Table 1.
Data Collection and Refinement Statistics
| TopBP1 BRCT6 | |
|---|---|
| Data Collection | |
| Space group | P212121 |
| Unit-cell parameters (Å) | a = 35.77, b = 51.82, c = 62.09 |
| Resolution range (Å) | 50–1.34 (1.39–1.34) |
| No. of reflections | 173,010 |
| Unique reflections | 26,433 (2396) |
| Rsyma (%) | 4.3 (32.6) |
| I/σ(I) | 30.2 (2.1) |
| Completeness (%) | 99.0 (91.6) |
| Redundancy | 6.5 (3.3) |
| Refinement | |
| Resolution (Å) | 39.78–1.34 |
| No. of reflections | 25,049 (1644) |
| Rwork/Rfree (%)b | 15.8/17.3 |
| No. of atoms | |
| Overall | 971 |
| Protein | 818 |
| Waters | 153 |
| Average B factor (Å2) | |
| Overall | 12.11 |
| Protein | 9.97 |
| Waters | 23.57 |
| R.m.s. deviations | |
| Bonds (Å) | 0.009 |
| Angles (°) | 1.208 |
| Ramachandran | |
| Most favored (%) | 90.5 |
| Additionally allowed (%) | 9.5 |
| Generously allowed (%) | 0 |
Rsym = S|I−〈I〉|2/S|I2|
R = S||Fo|−|Fc||/S|Fo|, Rfree was calculated from 5% of the data excluded from refinement.
Coordinates
Coordinates for TopBP1 BRCT6 have been deposited in Protein Data Bank with the accession code 3JVE.
Fluorescence polarization
FP measurements were carried out using an Envision multilabel plate reader (Perkin Elmer). Each well consisted of 100 nM FITC-labeled phospho-peptide and various protein concentrations in assay buffer (10 mM Tris-HCl pH 7.5, 400 mM NaCl, 1 mM DTT, 0.5% Tween-20). Binding assays were incubated for 15 min at room temperature before taking FP measurements. FITC-labeled phopsho-peptide for E2F1 (FITC-RLLDSpSQIVIKK-NH2) and BACH1 (FITC-GGSRSTpSPTFNK-NH2) were used for FP (Biomatik).
Isothermal titration calorimetry
The Micro Calorimetry System (Microcal) was used to perform ITC measurements. GST-BRCT6 and E2F1 phospho-peptide (Ac-RLLDSpSQIVI-NH2) were prepared in 10 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.05% BME and titration data was measured at 22°C. Data was analyzed using ORIGIN software (Microcal).
Acknowledgments
The authors thank Dr. Junjie Chen for providing the original TopBP1 constructs and Pawel Grochulski and the Canadian Light Source staff for assistance with synchrotron data collection. They also thank Dr. Ross Edwards for his technical assistance.
References
- 1.Garcia V, Furuya K, Carr AM. Identification and functional analysis of TopBP1 and its homologs. DNA Repair (Amst) 2005;4:1227–1239. doi: 10.1016/j.dnarep.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 2.Kumagai A, Lee J, Yoo HY, Dunphy WG. TopBP1 activates the ATR-ATRIP complex. Cell. 2006;124:943–955. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- 3.Manke IA, Lowery DM, Nguyen A, Yaffe MB. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science. 2003;302:636–639. doi: 10.1126/science.1088877. [DOI] [PubMed] [Google Scholar]
- 4.Yu X, Chini CC, He M, Mer G, Chen J. The BRCT domain is a phospho-protein binding domain. Science. 2003;302:639–632. doi: 10.1126/science.1088753. [DOI] [PubMed] [Google Scholar]
- 5.Wang B, Matsuoka S, Ballif BA, Zhang D, Smogorzewska A, Gygi SP, Elledge SJ. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science. 2007;316:1194–1198. doi: 10.1126/science.1139476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu X, Chen J. DNA damage-induced cell cycle checkpoint control requires CtIP, a phosphorylation-dependent binding partner of BRCA1 C-terminal domains. Mol Cell Biol. 2004;24:9478–9486. doi: 10.1128/MCB.24.21.9478-9486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delacroix S, Wagner JM, Kobayashi M, Yamamoto K, Karnitz LM. The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev. 2007;21:1472–1477. doi: 10.1101/gad.1547007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee J, Kumagai A, Dunphy WG. The Rad9-Hus1-Rad1 checkpoint clamp regulates interaction of TopBP1 with ATR. J Biol Chem. 2007;282:28036–28044. doi: 10.1074/jbc.M704635200. [DOI] [PubMed] [Google Scholar]
- 9.Clapperton JA, Manke IA, Lowery DM, Ho T, Haire LF, Yaffe MB, Smerdon SJ. Structure and mechanism of BRCA1 BRCT domain recognition of phosphorylated BACH1 with implications for cancer. Nat Struct Mol Biol. 2004;11:512–518. doi: 10.1038/nsmb775. [DOI] [PubMed] [Google Scholar]
- 10.Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 11.Williams RS, Lee MS, Hau DD, Glover JN. Structural basis of phosphopeptide recognition by the BRCT domain of BRCA1. Nat Struct Mol Biol. 2004;11:519–525. doi: 10.1038/nsmb776. [DOI] [PubMed] [Google Scholar]
- 12.Liu K, Lin FT, Ruppert JM, Lin WC. Regulation of E2F1 by BRCT domain-containing protein TopBP1. Mol Cell Biol. 2003;23:3287–3304. doi: 10.1128/MCB.23.9.3287-3304.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wollmann Y, Schmidt U, Wieland GD, Zipfel PF, Saluz HP, Hanel F. The DNA topoisomerase IIbeta binding protein 1 (TopBP1) interacts with poly (ADP-ribose) polymerase (PARP-1) J Cell Biochem. 2007;102:171–182. doi: 10.1002/jcb.21292. [DOI] [PubMed] [Google Scholar]
- 14.Pleschke JM, Kleczkowska HE, Strohm M, Althaus FR. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J Biol Chem. 2000;275:40974–40980. doi: 10.1074/jbc.M006520200. [DOI] [PubMed] [Google Scholar]
- 15.Lokesh GL, Rachamallu A, Kumar GD, Natarajan A. High-throughput fluorescence polarization assay to identify small molecule inhibitors of BRCT domains of breast cancer gene 1. Anal Biochem. 2006;352:135–141. doi: 10.1016/j.ab.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 16.Liu K, Paik JC, Wang B, Lin FT, Lin WC. Regulation of TopBP1 oligomerization by Akt/PKB for cell survival. EMBO J. 2006;25:4795–4807. doi: 10.1038/sj.emboj.7601355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Panzeter PL, Zweifel B, Malanga M, Waser SH, Richard M, Althaus FR. Targeting of histone tails by poly(ADP-ribose) J Biol Chem. 1993;268:17662–17664. [PubMed] [Google Scholar]
- 18.Yamane K, Kawabata M, Tsuruo T. A DNA-topoisomerase-II-binding protein with eight repeating regions similar to DNA-repair enzymes and to a cell-cycle regulator. Eur J Biochem. 1997;250:794–799. doi: 10.1111/j.1432-1033.1997.00794.x. [DOI] [PubMed] [Google Scholar]
- 19.Masson M, Niedergang C, Schreiber V, Muller S, Menissier-De Murcia J, De Murcia G. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol Cell Biol. 1998;18:3563–3571. doi: 10.1128/mcb.18.6.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 21.Qian B, Raman S, Das R, Bradley P, Mccoy AJ, Read RJ, Baker D. High-resolution structure prediction and the crystallographic phase problem. Nature. 2007;450:259–264. doi: 10.1038/nature06249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mccoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cohen SX, Ben Jelloul M, Long F, Vagin A, Knipscheer P, Lebbink J, Sixma TK, Lamzin VS, Murshudov GN, Perrakis A. ARP/wARP and molecular replacement: the next generation. Acta Crystallogr D Biol Crystallogr. 2008;64:49–60. doi: 10.1107/S0907444907047580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–245. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 25.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 26.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 27.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]


