Abstract
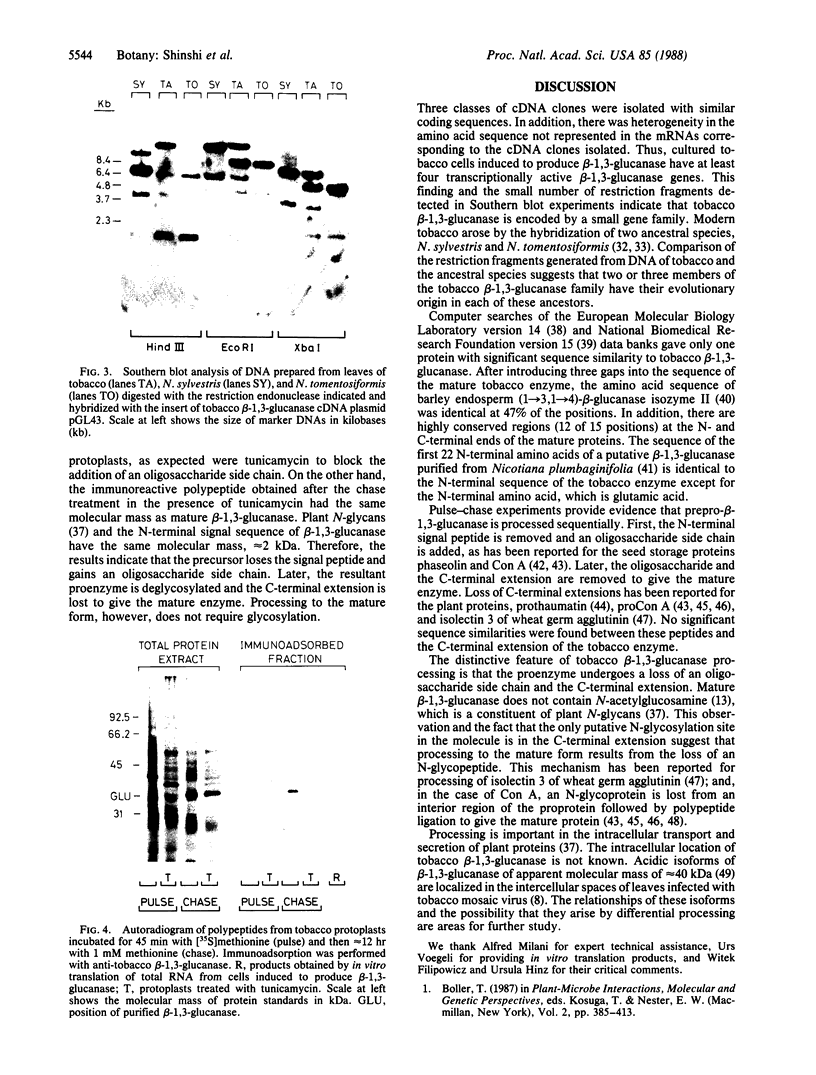
Tobacco glucan endo-1,3-β-glucosidase (β-1,3-glucanase; 1,3-β-D-glucan glucanohydrolase; EC 3.2.1.39) exhibits complex hormonal and developmental regulation and is induced when plants are infected with pathogens. We determined the primary structure of this enzyme from the nucleotide sequence of five partial cDNA clones and the amino acid sequence of five peptides covering a total of 70 residues. β-1,3-Glucanase is produced as a 359-residue preproenzyme with an N-terminal hydrophobic signal peptide of 21 residues and a C-terminal extension of 22 residues containing a putative N-glycosylation site. The results of pulse-chase experiments with tunicamycin provide evidence that the first step in processing is loss of the signal peptide and addition of an oligosaccharide side chain. The glycosylated intermediate is further processed with the loss of the oligosaccharide side chain and C-terminal extension to give the mature enzyme. Heterogeneity in the sequences of cDNA clones and of mature protein and in Southern blot analysis of restriction endonuclease fragments indicates that tobacco β-1,3-glucanase is encoded by a small gene family. Two or three members of this family appear to have their evolutionary origin in each of the progenitors of tobacco, Nicotiana sylvestris and Nicotiana tomentosiformis.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abeles F. B., Bosshart R. P., Forrence L. E., Habig W. H. Preparation and purification of glucanase and chitinase from bean leaves. Plant Physiol. 1971 Jan;47(1):129–134. doi: 10.1104/pp.47.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham G. N., Podell D. N. Pyroglutamic acid. Non-metabolic formation, function in proteins and peptides, and characteristics of the enzymes effecting its removal. Mol Cell Biochem. 1981 Aug 11;38(Spec No)(Pt 1):181–190. doi: 10.1007/BF00235695. [DOI] [PubMed] [Google Scholar]
- Bauw G., De Loose M., Inzé D., Van Montagu M., Vandekerckhove J. Alterations in the phenotype of plant cells studied by NH(2)-terminal amino acid-sequence analysis of proteins electroblotted from two-dimensional gel-separated total extracts. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4806–4810. doi: 10.1073/pnas.84.14.4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollini R., Vitale A., Chrispeels M. J. In vivo and in vitro processing of seed reserve protein in the endoplasmic reticulum: evidence for two glycosylation steps. J Cell Biol. 1983 Apr;96(4):999–1007. doi: 10.1083/jcb.96.4.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowles D. J., Marcus S. E., Pappin D. J., Findlay J. B., Eliopoulos E., Maycox P. R., Burgess J. Posttranslational processing of concanavalin A precursors in jackbean cotyledons. J Cell Biol. 1986 Apr;102(4):1284–1297. doi: 10.1083/jcb.102.4.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington D. M., Auffret A., Hanke D. E. Polypeptide ligation occurs during post-translational modification of concanavalin A. Nature. 1985 Jan 3;313(5997):64–67. doi: 10.1038/313064a0. [DOI] [PubMed] [Google Scholar]
- Chrispeels M. J., Hartl P. M., Sturm A., Faye L. Characterization of the endoplasmic reticulum-associated precursor of concanavalin A. Partial amino acid sequence and lectin activity. J Biol Chem. 1986 Aug 5;261(22):10021–10024. [PubMed] [Google Scholar]
- Dean C., Tamaki S., Dunsmuir P., Favreau M., Katayama C., Dooner H., Bedbrook J. mRNA transcripts of several plant genes are polyadenylated at multiple sites in vivo. Nucleic Acids Res. 1986 Mar 11;14(5):2229–2240. doi: 10.1093/nar/14.5.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edens L., Heslinga L., Klok R., Ledeboer A. M., Maat J., Toonen M. Y., Visser C., Verrips C. T. Cloning of cDNA encoding the sweet-tasting plant protein thaumatin and its expression in Escherichia coli. Gene. 1982 Apr;18(1):1–12. doi: 10.1016/0378-1119(82)90050-6. [DOI] [PubMed] [Google Scholar]
- Fincher G. B., Lock P. A., Morgan M. M., Lingelbach K., Wettenhall R. E., Mercer J. F., Brandt A., Thomsen K. K. Primary structure of the (1-->3,1-->4)-beta-D-glucan 4-glucohydrolase from barley aleurone. Proc Natl Acad Sci U S A. 1986 Apr;83(7):2081–2085. doi: 10.1073/pnas.83.7.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George D. G., Barker W. C., Hunt L. T. The protein identification resource (PIR). Nucleic Acids Res. 1986 Jan 10;14(1):11–15. doi: 10.1093/nar/14.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray J. C., Kung S. D., Wildman S. G. Origin of Nicotiana tabacum L. detected by polypeptide composition of Fraction I protein. Nature. 1974 Nov 15;252(5480):226–227. doi: 10.1038/252226a0. [DOI] [PubMed] [Google Scholar]
- Gubler U., Hoffman B. J. A simple and very efficient method for generating cDNA libraries. Gene. 1983 Nov;25(2-3):263–269. doi: 10.1016/0378-1119(83)90230-5. [DOI] [PubMed] [Google Scholar]
- Hamm G. H., Cameron G. N. The EMBL data library. Nucleic Acids Res. 1986 Jan 10;14(1):5–9. doi: 10.1093/nar/14.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofsteenge J., Stone S. R. The effect of thrombomodulin on the cleavage of fibrinogen and fibrinogen fragments by thrombin. Eur J Biochem. 1987 Oct 1;168(1):49–56. doi: 10.1111/j.1432-1033.1987.tb13385.x. [DOI] [PubMed] [Google Scholar]
- Jamet E., Durr A., Fleck J. Absence of some truncated genes in the amphidiploid Nicotiana tabacum. Gene. 1987;59(2-3):213–221. doi: 10.1016/0378-1119(87)90329-5. [DOI] [PubMed] [Google Scholar]
- Kauffmann S., Legrand M., Geoffroy P., Fritig B. Biological function of ;pathogenesis-related' proteins: four PR proteins of tobacco have 1,3-beta-glucanase activity. EMBO J. 1987 Nov;6(11):3209–3212. doi: 10.1002/j.1460-2075.1987.tb02637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kombrink E., Schröder M., Hahlbrock K. Several "pathogenesis-related" proteins in potato are 1,3-beta-glucanases and chitinases. Proc Natl Acad Sci U S A. 1988 Feb;85(3):782–786. doi: 10.1073/pnas.85.3.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornfeld R., Kornfeld S. Assembly of asparagine-linked oligosaccharides. Annu Rev Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- Lütcke H. A., Chow K. C., Mickel F. S., Moss K. A., Kern H. F., Scheele G. A. Selection of AUG initiation codons differs in plants and animals. EMBO J. 1987 Jan;6(1):43–48. doi: 10.1002/j.1460-2075.1987.tb04716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauch F., Hadwiger L. A., Boller T. Ethylene: Symptom, Not Signal for the Induction of Chitinase and beta-1,3-Glucanase in Pea Pods by Pathogens and Elicitors. Plant Physiol. 1984 Nov;76(3):607–611. doi: 10.1104/pp.76.3.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohnen D., Shinshi H., Felix G., Meins F. Hormonal regulation of beta1,3-glucanase messenger RNA levels in cultured tobacco tissues. EMBO J. 1985 Jul;4(7):1631–1635. doi: 10.1002/j.1460-2075.1985.tb03830.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore A. E., Stone B. A. Effect of infection with TMV and other viruses on the level of a -1,3-glucan hydrolase in leaves of Nicotiana glutinosa. Virology. 1972 Dec;50(3):791–798. doi: 10.1016/0042-6822(72)90433-3. [DOI] [PubMed] [Google Scholar]
- Murray M. G., Thompson W. F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980 Oct 10;8(19):4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszkowski J., Shillito R. D., Saul M., Mandák V., Hohn T., Hohn B., Potrykus I. Direct gene transfer to plants. EMBO J. 1984 Dec 1;3(12):2717–2722. doi: 10.1002/j.1460-2075.1984.tb02201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queen C., Korn L. J. A comprehensive sequence analysis program for the IBM personal computer. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):581–599. doi: 10.1093/nar/12.1part2.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raikhel N. V., Wilkins T. A. Isolation and characterization of a cDNA clone encoding wheat germ agglutinin. Proc Natl Acad Sci U S A. 1987 Oct;84(19):6745–6749. doi: 10.1073/pnas.84.19.6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinshi H., Mohnen D., Meins F. Regulation of a plant pathogenesis-related enzyme: Inhibition of chitinase and chitinase mRNA accumulation in cultured tobacco tissues by auxin and cytokinin. Proc Natl Acad Sci U S A. 1987 Jan;84(1):89–93. doi: 10.1073/pnas.84.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- von Heijne G. Patterns of amino acids near signal-sequence cleavage sites. Eur J Biochem. 1983 Jun 1;133(1):17–21. doi: 10.1111/j.1432-1033.1983.tb07424.x. [DOI] [PubMed] [Google Scholar]