Abstract
Background and aims
Several genes have been shown to individually affect plasma lipoprotein metabolism in humans. Studies on gene-gene interactions could offer more insight into how genes affect lipid metabolism and may be useful in predicting lipid concentrations. We tested for gene-gene interactions between TaqIB SNP in the cholesterol ester transfer protein (CETP) and three novel single nucleotide polymorphisms (SNPs), namely rs11774572, rs7819412 and rs6995374 for their effect on metabolic syndrome (MetS) components and related traits.
Methods and results
The aforementioned SNPs were genotyped in 1002 subjects who participated in the Genetics of Lipid Lowering Drugs and Diet Network (GOLDN) study. Lipids were measured by standard procedures and lipoprotein subfractions, by proton nuclear magnetic resonance spectroscopy. Polymorphism rs11774572 was significantly associated with MetS (P=0.020), mainly driven by the association of the C allele with lower HDL-C (P=0.043) and higher triglycerides (P=0.049) and insulin (P=0.040) concentrations than TT subjects. A significant interaction between SNPs rs11774572 and CETP-TaqIB SNPs was found for HDL-C concentrations (P=0.006) and for HDL (P=0.008) and LDL particle sizes (P=0.009), small LDL (P=0.004), and VLDL concentrations (P=0.021), in which TT homozygotes displayed higher HDL-C concentrations and for HDL and LDL particle sizes, and lower small LDL and VLDL concentrations than C carriers, if they were CETP B2 allele carriers (P values ranging from <0.001 to 0.001).
Conclusions
The rs11774572 polymorphism may play a role in the dyslipidemia that characterizes MetS. The interaction between rs11774572 and CETP-TaqIB SNPs on HDL-C concentrations provides some insights into the underlying mechanisms.
Keywords: HDL-cholesterol, metabolic syndrome, polymorphism-polymorphism interaction, CETP gene, pathway
INTRODUCTION
Dyslipidemia, insulin resistance, abdominal obesity and hypertension form a cluster of risk factors that characterize the metabolic syndrome (MetS) (1). As a multifactorial disease, the etiology of MetS involves a genetic susceptibility, environmental influences, as well as their interaction (2–3). Many single nucleotide polymorphisms (SNPs) in candidate genes have been associated with MetS (2); however, the results have not been consistent, and initial positive findings have usually failed replication (4). Several reasons have been put forward to explain these inconsistencies (5,6) including: 1) the analysis of a very limited number of SNPs at a single locus, 2) the failure to account for gene-environment interactions, and 3) the lack of simultaneous information about multiple loci involved in the same metabolic pathway. Moreover, the study of gene-gene interactions may be useful in providing mechanistic insights into novel loci associated with plasma lipid concentrations.
Recently, a number of genome-wide association (GWA) studies have identified novel polymorphisms associated with lipid phenotypes but located in or near loci with no known effect on lipid metabolism (7). This is the case for SNPs rs117744572, rs7819412, and rs6995374 located on human chromosome 8p23, a region that has been linked repeatedly with lipid abnormalities (8) and with early-onset of coronary heart disease (CHD) in a French Canadian population (9). These novel polymorphisms are located near the B-cell lymphocyte kinase (BLK) (rs11774572), the kell blood group complex subunit-related family member 6 (XKR6) (rs7819412), and the methionine sulfoxide reductase A (MSRA) (rs6995374) genes. Although these genes function in lymphocytes and red blood cells differentiation (BLK and XKR6) (10,11) and oxidative stress and aging (MSRA) (12), the underlying mechanisms and metabolic pathways affected by these SNPs and the links to lipid values and other components of the MetS remain unknown.
Low HDL-cholesterol (HDL-C) and high triglyceride (TG) concentrations are the hallmarks of the dyslipidemia associated with MetS. The cholesteryl ester transfer protein (CETP) facilitates the exchange of cholesteryl esters from HDL or LDL into TG-rich lipoproteins, resulting in lower HDL-C concentrations and LDL-particles of smaller size. This cholesteryl ester transfer process plays a pivotal role in the reverse cholesterol transport (RCT), whereby cholesterol is transported from peripheral cells back to the liver for metabolism and excretion in the bile. Importantly, CETP gene is a major determinant of HDL-C variability in the general population (13). Among several polymorphisms described in the CETP gene, the TaqIB polymorphism is the variant that has been most firmly associated with HDL-C concentrations in several populations. This polymorphism, at nucleotide 279 in the first intron of the CETP gene, is characterized by an altered recognition site for the restriction endonuclease TaqI. The B2 allele (absence of the TaqI restriction site) has been consistently associated with increased HDL-C concentrations, and variations in HDL and LDL lipoprotein subfractions (13–17). Moreover, this TaqIB polymorphism recently has been related to MetS risk, implicating novel functions of the CETP gene (14).
The aims of the present study were first to assess the impact of novel intergenic polymorphisms first identified by GWA (rs11774572, rs7819412 and rs6995374) and the CETP-TaqIB on lipoprotein levels and MetS-related phenotypes. Secondly, we investigated whether there was evidence for an interaction between these novel polymorphisms and the CETP-TaqIB polymorphism in determining lipid concentrations and particle size.
METHODS
Participants
In the present study, 1002 participants (482 men/520 women, age 49±16 years) from the Genetics of Lipid Lowering Drugs and Diet Network (GOLDN) study were enrolled. Participants were recruited from three-generational pedigrees from two National Heart, Lung, and Blood Institute Family Heart Study field centers (Minneapolis, MN and Salt Lake City, UT) (18). The study population was homogeneous regarding ethnic background, all being of Caucasian origin. The detailed design and methodology of the study have been described (19). The protocol was approved by the Institutional Review Boards at the University of Alabama, the University of Minnesota, the University of Utah, and Tufts University. Written informed consent was obtained from each participant.
Data collection
Clinical examinations at the baseline visit included anthropometrical and blood pressure (BP) measurements. Weight was measured with a beam balance and height with fixed stadiometer. BMI was calculated as weight in kilograms divided by the square of height in meters. Waist circumference was measured at the umbilicus. BP was measured twice with an oscillometric device (Dinamap Pro Series 100, GE Medical Systems) while subjects were seated and had rested for five min. Reported systolic and diastolic BP values were the mean of two measurements. Questionnaires were administered to assess demographic information, and medical and medication history. Physical activity was expressed as metabolic equivalent task (MET) hours based on self-reported types and durations of activities over a period of 24 h.
Laboratory methods
Blood samples were drawn after fasting overnight. Lipid measurements have been previously described (19). Glucose was measured using the method of a hexokinase-mediated reaction and total cholesterol using a cholesterol esterase cholesterol oxidase reaction on a Hitachi 911 autoanalyzer (Roche Diagnostics). Fasting plasma insulin was measured by specific radioimmunoassay kits (Linco Research).
Lipoprotein particle concentrations and size were measured in 996 subjects by proton nuclear magnetic resonance spectroscopy (20). Data were obtained from the measured amplitudes of the spectroscopically distinct lipid methyl group nuclear magnetic resonance signals (15).
Genetic analyses
DNA was extracted from blood samples and purified using commercial Puregene reagents (Gentra Systems) following the manufacturer’s instructions. Three SNPs located on human chromosome 8p23 (rs11774572, rs7819412, rs6995374), and the CETP-TaqIB polymorphism (rs708272, G>A), on human chromosome 16q21 were selected for the study. Genotyping was performed using a TaqMan® assay with allele-specific probes on the ABIPrism 7900 HT Sequence Detection System (Applied Biosystems) according to routine laboratory protocols (21). The ABI assay-on-demand identification for the aforementioned SNPs were C__31455998_10 (rs11774572), C__30011529_10 (rs7819412), C__11589038_10 (rs6995374), and C___9615318_10 (rs708272), respectively. Analysis showed that none of these three SNPs is in linkage disequilibrium with CETP-TaqIB variant.
Statistical analyses
SPSS software (version 16.0) was used for statistical analyses. A logarithmic transformation was applied to insulin, to normalize the distribution of the data. Presence of MetS was defined following the National Cholesterol Education Program-Adult Treatment Panel III (NCEP-ATPIII) guidelines (1). Data were presented as means ± SD for continuous variables and as frequencies or percentages for categorical variables. Differences in mean values were assessed by using analysis of variance. Categorical variables were compared by using the Pearson chi-square or the Fisher’s exact tests. Potential confounding factors included age, sex, BMI, field center (MN vs. UT), physical activity, smoking habit (current vs. never and past smokers), alcohol consumption (current vs. never and past drinkers), prior CHD, medications (treatment for hypertension, hypercholesterolemia, and diabetes), self-reported use of hormone therapy by women, and family relationships. A multivariate adjusted logistic regression model was used to assess the prevalence of risk of MetS across genetic variants. Potential interactions between polymorphisms in determining lipids and lipoprotein subfractions were tested using analysis of variance. Two-sided P values <0.05 were considered statistically significant.
RESULTS
Main characteristics of GOLDN participants are shown in Table 1. Average weight and waist circumference were higher in men than women; however, BMI did not differ by sex. BP, glucose, LDL-cholesterol (LDL-C), and TG values were higher in men whereas HDL-C concentrations were higher in women. Given that subjects from the Minnesota field center were older (51 vs. 47, years; P<0.001), more likely to drink alcohol (82% vs. 17%, P<0.001) and smoke cigarettes (14% vs. 2%, P<0.001), and displayed higher LDL-C (3.27 vs. 3.09, mmol/L; P=0.001), TG (mmol/L) (1.4 vs. 1.1; P<0.001), and HDL-C concentrations (1.3 vs. 1.2, mmol/L; P<0.001) than those in UT, all the analyses were adjusted for field center. Genotype frequencies were consistent with Hardy-Weinberg equilibrium (P>0.05) and no significant differences in genotype frequencies were observed between men and women (P>0.05) (Table 1).
Table 1.
General characteristics of the GOLDN population.
| Men (n=482) | Women (n=520) | |
|---|---|---|
| Age, years (range) | 49 (18–88) | 49 (18–92) |
| Weight, kg* | 90.8±16.0 | 76.1±17.5 |
| BMI, kg/m2 | 28.6±4.8 | 28.2±6.4 |
| Waist circumference, cm* | 100±14 | 93±18 |
| Systolic BP, mmHg* | 119±15 | 113±18 |
| Diastolic BP, mmHg* | 71±8 | 66±9 |
| Glucose, mmol/L* | 5.83±1.17 | 5.44±0.89 |
| Log (Insulin, mU/L) | 1.09±0.21 | 1.07±0.21 |
| Total cholesterol, mmol/L | 4.95±0.98 | 5.00±1.06 |
| LDL cholesterol, mmol/L† | 3.21±0.78 | 3.11±0.85 |
| HDL cholesterol, mmol/L* | 1.06±0.26 | 1.35±0.36 |
| Triglycerides, mmol/L* | 1.35±0.10 | 1.17±0.09 |
| Metabolic syndrome, n (%) | 155 (32) | 151 (29) |
| SNPs, n (%) | ||
| rs11774572 | ||
| CC+CT | 342 (71) | 370 (71) |
| TT | 140 (29) | 150 (29) |
| rs7819412 | ||
| AA+AG | 352 (73) | 381 (73) |
| GG | 129 (27) | 140 (27) |
| rs6995374 | ||
| GG+CG | 218 (45) | 232 (45) |
| CC | 264 (55) | 288 (55) |
| CETP-TaqIB | ||
| B2 carrier | 319 (66) | 357 (69) |
| B1/B1 | 163 (34) | 163 (31) |
BP: blood pressure. All values are means ± SD.
P<0.001,
P<0.05 between genders by unpaired t-test.
C allele carriers at the rs11774572 SNP displayed higher risk of MetS (OR 1.60, 95% CI 1.08–2.37; P=0.020), mainly driven by the association of the minor C allele with lower HDL-C levels (P=0.043), and higher TG values (P=0.049) than TT homozygotes. Likewise insulin values were higher in C allele carriers compared with TT subjects (P=0.040). Although differences were marginally significant (P=0.074), C allele carriers also displayed higher systolic BP than TT subjects. Further adjustment for the CETP-TaqIB polymorphism strengthened the associations between rs11774572 and HDL-C (P=0.021), and insulin (P=0.039) concentrations, but these differences were weakened for TG (P=0.054) and were no longer significant for systolic BP (P=0.067) (Table 2).
Table 2.
Characteristics of participants according to the rs11774572 polymorphism.
| CC+CT (n=712) | TT (n=290) | |||
|---|---|---|---|---|
| Variable | Mean ± SE | Mean ± SE | P* | P† |
| Weight, Kg | 83.4±0.3 | 83.1±0.4 | 0.529 | 0.564 |
| Waist circumference, cm | 97±0.3 | 97±0.5 | 0.902 | 0.846 |
| Systolic BP, mmHg | 117±0.6 | 115±0.9 | 0.074 | 0.067 |
| Diastolic BP, mmHg | 69±0.3 | 68±0.5 | 0.116 | 0.122 |
| Glucose, mmol/L | 5.67±0.03 | 5.61±0.05 | 0.421 | 0.422 |
| Log (Insulin, mU/L) | 1.09±0.01 | 1.07±0.01 | 0.040 | 0.039 |
| Total cholesterol, mmol/L | 4.99±0.03 | 4.99±0.05 | 0.903 | 0.865 |
| LDL cholesterol, mmol/L | 3.19±0.02 | 3.14±0.04 | 0.464 | 0.645 |
| HDL cholesterol, mmol/L | 1.21±0.01 | 1.25±0.01 | 0.043 | 0.021 |
| Triglycerides, mmol/L | 1.29±0.01 | 1.20±0.01 | 0.049 | 0.054 |
BP: Blood pressure; Data are multivariate adjusted means.
Means and P values were adjusted for age, gender, BMI, field center, physical activity, smoking habit, alcohol consumption, CHD, prior medications, hormone treatment, and family relationships.
Further adjustment for CETP-TaqIB SNP.
Minor A allele carriers of SNP rs7819412 displayed lower values of systolic and diastolic BP than GG homozygotes (115 vs 118, mmHg; P=0.015 and 68 vs 70, mmHg; P=0.013, respectively) (Supplemental Table 1). However, these differences did not manifest as lower MetS risk (OR 1.13, 95% CI 0.77–1.67; P>0.05) (data not shown). For the rs6995374 SNP, we did not observe any significant association with lipid concentrations or other components of MetS (Supplemental Table 2). B2 allele carriers at the CETP gene displayed higher total cholesterol, LDL-C and HDL-C concentrations than B1/B1 homozygotes (P values ranging from 0.001 to 0.005) (Table 3). A marginally significant association between the CETP-TaqIB polymorphism and risk of MetS was found, in which B2 allele carriers had lower risk of MetS than B1/B1 homozygotes (OR 0.76, 95% CI 0.53–1.08; P=0.123) (data not shown).
Table 3.
Effect of the CETP-TaqIB polymorphism on lipid concentrations and particle size*.
| B2 carriers (n=676) | B1/B1 (n=326) | ||
|---|---|---|---|
| Variable | Mean ± SE | Mean ± SE | P† |
| Total cholesterol, mmol/L | 5.04±0.03 | 4.86±0.04 | 0.001 |
| LDL cholesterol, mmol/L | 3.22±0.02 | 3.09±0.04 | 0.005 |
| HDL cholesterol, mmol/L | 1.24±0.01 | 1.17±0.01 | 0.001 |
| Triglycerides, mmol/L | 1.26±0.01 | 1.26±0.01 | 0.942 |
| HDL particle size, nm | 8.87±0.01 | 8.81±0.01 | 0.022 |
| LDL particle size, nm | 20.84±0.03 | 20.65±0.51 | 0.003 |
| VLDL particle size, nm | 50.82±0.29 | 51.17±0.49 | 0.491 |
| Large HDL, g/L | 0.17±0.04 | 0.14±0.07 | <0.001 |
| Medium HDL, g/L | 0.03±0.02 | 0.03±0.03 | 0.377 |
| Small HDL, g/L | 0.21±0.02 | 0.20±0.03 | 0.578 |
| IDL, g/L | 0.04±0.02 | 0.05±0.04 | 0.097 |
| Large LDL, g/L | 0.42±0.02 | 0.37±0.03 | 0.040 |
| Small LDL, g/L | 0.47±0.01 | 0.50±0.02 | 0.192 |
| Medium small LDL, g/L | 0.18±0.01 | 0.20±0.01 | 0.185 |
| Very small LDL, g/L | 0.28±0.01 | 0.32±0.01 | 0.032 |
| Large VLDL, g/L | 0.17±0.02 | 0.16±0.03 | 0.493 |
| Medium VLDL, g/L | 0.34±0.02 | 0.32±0.03 | 0.238 |
| Small VLDL, g/L | 0.12±0.03 | 0.12±0.06 | 0.520 |
Sample size of 996 for particle size (n=674 and 322 for B2 carriers and B1/B1, respectively). Values are mean ± SE.
P values were adjusted for age, gender, BMI, field center, physical activity, smoking habit, alcohol consumption, CHD, prior medications, hormone treatment, and family relationships.
Given that rs11774572 and CETP-TaqIB were the only SNPs related to lipid values and risk of MetS, results for these SNPs were examined in more detail. Because of the low genotype frequencies of homozygotes for the minor alleles, we analyzed all SNPs using two genotype categories using the dominant model to maximize the statistical power.
Regarding lipoprotein concentrations and particle size, carriers of the C allele at the rs11774572 polymorphism displayed higher concentrations of small LDL subfractions, and medium VLDL-TG than TT homozygotes (P values ranging from 0.001 to 0.031) whereas increased particle size for HDL and LDL were found in TT subjects compared to C allele carriers (P=0.034, and P=0.019, respectively) (Table 4). Further adjustment for the CETP-TaqIB polymorphism strengthened some of the observed associations (Table 4). When the effect of the CETP-TaqIB SNP was analyzed alone, B2 allele carriers displayed increased particle size for HDL and LDL, higher number of large HDL particles, and lower very small LDL concentrations than B1/B1 homozygotes (P values ranging from <0.001 to 0.040) (Table 3).
Table 4.
Association between the rs11774572 polymorphism and lipoprotein concentrations and particle size.
| CC+CT (n=708) | TT (n=288) | P* | P† | |
|---|---|---|---|---|
| HDL particle size, nm | 8.83±0.01 | 8.89±0.02 | 0.034 | 0.020 |
| LDL particle size, nm | 20.75±0.03 | 20.84±0.04 | 0.019 | 0.009 |
| VLDL particle size, nm | 51.17±0.29 | 50.58±0.49 | 0.226 | 0.204 |
| Large HDL, g/L | 0.16±0.04 | 0.17±0.07 | 0.042 | 0.018 |
| Medium HDL, g/L | 0.03±0.02 | 0.03±0.03 | 0.780 | 0.839 |
| Small HDL, g/L | 0.21±0.02 | 0.20±0.03 | 0.794 | 0.825 |
| IDL, g/L | 0.04±0.02 | 0.04±0.04 | 0.696 | 0.571 |
| Large LDL, g/L | 0.39±0.02 | 0.42±0.02 | 0.208 | 0.149 |
| Small LDL, g/L | 0.50±0.01 | 0.43±0.02 | 0.001 | 0.001 |
| Medium small LDL, g/L | 0.20±0.01 | 0.17±0.02 | 0.002 | 0.001 |
| Very small LDL, g/L | 0.30±0.01 | 0.27±0.01 | 0.031 | 0.020 |
| Large VLDL, g/L | 0.17±0.02 | 0.15±0.03 | 0.192 | 0.211 |
| Medium VLDL, g/L | 0.35±0.02 | 0.30±0.03 | 0.027 | 0.035 |
| Small VLDL, g/L | 0.12±0.03 | 0.12±0.06 | 0.280 | 0.257 |
Values are mean±SE. P values were adjusted for age, gender, BMI, field center, physical activity, smoking habit, alcohol consumption, CHD, prior medications, hormone treatment, and family relationships.
Further adjustment for CETP-TaqIB SNP.
A significant interaction between both polymorphisms was found in determining HDL-C levels (P=0.006), in which TT homozygotes displayed higher HDL-C levels only if they were B2 allele carriers at the CETP gene (P<0.001); whereas no significant differences in both lipid values were obtained within the B1/B1 genotype (P>0.4) (Figure). No significant interactions between both SNPs were found for TG concentrations (P>0.3) and other MetS components (P>0.5) (data not shown). Regarding lipoprotein particle size, a significant interaction between both polymorphisms was observed for HDL and LDL particle sizes (P=0.008, and P=0.009, respectively) in which TT subjects displayed increased size for both particles compared to C allele carriers, only in the B2 allele carriers at the CETP gene (P<0.001 for both). Likewise, a significant association between both polymorphisms was found for small LDL and medium VLDL-TG concentrations (P values 0.004 and 0.021, respectively) in which C allele carriers displayed higher concentrations of those particles compared to TT homozygotes, only in the B2 allele carriers at the CETP gene (data not shown).
DISCUSSION
In the current study, the rs11774572 polymorphism was significantly associated with risk of MetS, mainly driven by the association of the minor C allele with lower HDL-C concentrations and higher TG, and insulin concentrations than TT subjects. At the same time, CETP-TaqIB polymorphism was associated with total cholesterol, LDL-C, and HDL-C concentrations. Interestingly, a significant interaction between polymorphisms rs11774572 and CETP-TaqIB was found for HDL-C levels, and variations in HDL, LDL, and VLDL-TG lipoprotein subfractions, supporting the hypothesis that both polymorphisms may be involved in a related metabolic pathway.
The present study demonstrates for the first time an association between the novel rs11774572 polymorphism and the risk of MetS, in which C allele carriers displayed lower HDL-C and higher TG concentrations compared with TT subjects. Moreover, higher insulin concentrations were also observed in C allele carriers, underlying the association of this polymorphism with insulin resistance. Importantly, further adjustment for the CETP_TaqIB SNP strengthened the association of the rs11774572 SNP with HDL-C concentrations, supporting the hypothesis that the effect of this SNP on HDL-C depends on the CETP polymorphism.
In agreement with prior results (12), we observed a trend towards an increased risk of MetS in B1/B1 homozygotes at the CETP gene, mainly driven by the association of the B1/B1 genotype with lower HDL-C concentrations. In addition to the well-documented association between CETP-TaqIB polymorphism and HDL-C concentrations (13–17), we found a significant association between this polymorphism and LDL-C concentrations, as well as LDL composition and size. In this regard, B2 allele carriers displayed higher LDL-C concentrations, smaller LDL particle sizes, and lower very small LDL concentrations compared to B1/B1 homozygotes. Although no significant associations between CETP genetic variation and LDL-C concentrations have been reported (15), other authors reported a trend towards lower LDL-C concentrations (17) and an increased LDL particle size in men (15) among B2 allele carriers. Likewise, no significant differences in TG concentrations have been reported previously across CETP-TaqIB genotypes (13,15–17).
We have also examined the associations between rs11774572 polymorphism and lipoprotein subfractions. We found lower concentrations of anti-atherogenic particles (large HDL) and higher concentrations of atherogenic cholesterol-rich (small LDL, and medium small LDL), and TG- (medium VLDL-TG) remnant particles in C allele carriers compared with TT subjects. Overall, these findings suggest that the rs11774572 SNP is in linkage disequilibrium with a locus that plays a potential role in the neutral lipid exchange, as well as in the RCT.
A significant interaction between rs11774572 and CETP-TaqIB was observed in which TT homozygous displayed higher HDL-C concentrations, an increased particle size for HDL and LDL, and lower small LDL subfractions and medium VLDL-TG concentrations than C allele carriers, only if they were B2 allele carriers at the CETP gene. To our knowledge, such an interaction has not been reported earlier. Because of this interaction, it is reasonable to hypothesize that the locus associated with the rs11774572 polymorphism may be involved in the same metabolic pathway as CETP. Additionally, the B2 allele carriers in the CETP-TaqIB polymorphism are associated with lower CETP activity, and higher HDL-C levels compared to B1 allele carriers (13–15). Therefore, the low HDL-C concentrations observed in C allele carriers despite carrying the B2 allele at the CETP gene, suggest that the rs11774572 polymorphism may inhibit the effect of the CETP gene in this subgroup of subjects. However, the mechanism by which the intergenic rs11774572 SNP interacts with CETP-TaqIB polymorphism remains unknown. Given its location between two genes, the likelihood that this SNP represents a functional mutation is low. GATA4 and Retinitis Pigmentosa 1 (RP1) are the two genes located 114953 bp and ≈900 kbp from the rs11774572 polymorphism, respectively. Importantly, the pivotal role of both genes in the cholesterol metabolism, may explain our findings. In this regard, GATA4 encodes a transcription factor that regulates expression of the ATP-binding cassette sterol transporters (ABCG5 and ABCG8), which mediate the efflux of cholesterol and plant sterols from enterocytes back into the intestinal lumen and their excretion into the bile, thus limiting their accumulation in the body and promoting the RCT (22). RP1 genetic variation modifies the lipoprotein phenotype of plasma TG and HDL-C concentrations (23). The most plausible explanation is that rs11774572 polymorphism may be in linkage disequilibrium with a yet undiscovered functional mutation in the regulatory region of either GATA4 or RP1 genes.
Despite the evidence, we should be cautious in the interpretation of our data. Moreover, extension of research efforts to other ethnic populations with increased MetS risk is clearly warranted. Importantly, we have to consider the increased levels of population differentiation among specific genetic variations, as a result of the positive selection in humans (24). In this regard, according to the HapMap data for the rs11774572 polymorphism, carriers of the C allele have derived-allele frequencies of 58% in European, 28% in Sub-Saharan African and 1% and 3% in Japanese and Chinese populations, respectively. Moreover, studying populations such as the European, in which the allele is still segregating, may provide clues to its biological significance. These results open multiple avenues for future research, as they may facilitate genetic explorations of medical conditions, such as MetS, in new loci in which disease prevalence depends in part on ethnic background.
In conclusion, the present study carried out in a North American population supports the hypothesis that the rs11774572 polymorphism plays a significant role in the risk of MetS, mainly driven by the association of the C allele with lower HDL-C, higher TG concentrations and insulin resistance. Moreover, the significant interaction between the polymorphisms rs11774572 and CETP-TaqIB for HDL-C concentrations and particle subfractions supports that the idea that the new locus linked to this SNP may be metabolically related to the pathways involving CETP.
Supplementary Material
Figure.
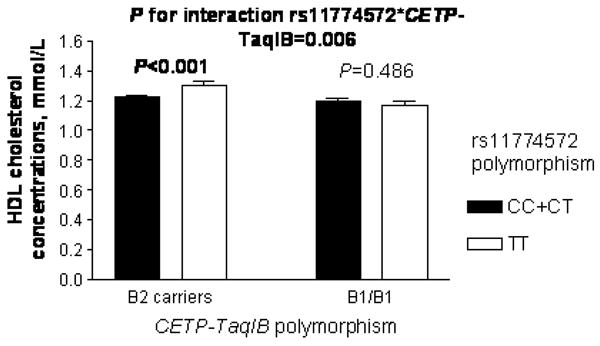
Adjusted HDL-C concentrations depending on rs11774572 and CETP-TaqIB genotypes. Values are mean±SE.
Acknowledgments
This work was supported by NIH Heart, Lung and Blood Institute grant U 01 HL72524, Genetic and Environmental Determinants of Triglycerides, grant HL-54776, by contracts 53-K06-5-10 and 58-1950-9-001 from the US Department of Agriculture Research Service. MJ is supported by a grant from the Fulbright-Spanish Ministry of Education and Science (reference 2007-1086). CES is supported by the grant T32 DK007651-19.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Third Report of the National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults (Adult Treatment Panel III) final report. Circulation. 2002;106:3143–421. [PubMed] [Google Scholar]
- 2.Groop L. Genetics of the metabolic syndrome. Br J Nutr. 2000;83(Suppl 1):S39–48. doi: 10.1017/s0007114500000945. [DOI] [PubMed] [Google Scholar]
- 3.Roche HM, Phillips C, Gibney MJ. The metabolic syndrome: the crossroads of diet and genetics. Proc Nutr Soc. 2005;64:371–7. doi: 10.1079/pns2005445. [DOI] [PubMed] [Google Scholar]
- 4.Pollex RL, Hegele RA. Genetic determinants of the metabolic syndrome. Nat Clin Pract Cardiovasc Med. 2006;3:482–9. doi: 10.1038/ncpcardio0638. [DOI] [PubMed] [Google Scholar]
- 5.Tiret L. Gene-environment interaction: a central concept in multifactorial diseases. Proc Nutr Soc. 2002;61:457–63. doi: 10.1079/pns2002178. [DOI] [PubMed] [Google Scholar]
- 6.Ordovas JM, Corella D. Nutritional genomics. Annu Rev Genomics Hum Genet. 2004;5:71–118. doi: 10.1146/annurev.genom.5.061903.180008. [DOI] [PubMed] [Google Scholar]
- 7.Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40:161–9. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kraja AT, Hunt SC, Pankow JS, Myers RH, Heiss G, Lewis CE, et al. Quantitative trait loci for metabolic syndrome in the hypertension genetic epidemiology network study. Obes Res. 2005;13:1885–90. doi: 10.1038/oby.2005.231. [DOI] [PubMed] [Google Scholar]
- 9.Engert JC, Lemire M, Faith J, Brisson D, Fujiwara TM, Roslin NM, et al. Identification of a chromosome 8p locus for early-onset coronary heart disease in a French Canadian population. Eur J Hum Genet. 2008;16:105–14. doi: 10.1038/sj.ejhg.5201920. [DOI] [PubMed] [Google Scholar]
- 10.Texido G, Su IH, Mecklenbrauker I, Saijo K, Malek SN, Desiderio S, et al. The B-cell-specific src-family kinase Blk is dispensable for B-cell development and activation. Mol Cell Biol. 2000;20:1227–33. doi: 10.1128/mcb.20.4.1227-1233.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee S, Russo D, Redman CM. The Kell blood group system: Kell and XK membrane proteins. Semin Hematol. 2000;37:113–21. doi: 10.1016/s0037-1963(00)90036-2. [DOI] [PubMed] [Google Scholar]
- 12.Cabreiro F, Picot CR, Friguet B, Petropoulos I. Methionine sulfoxide reductases: relevance to aging and protection against oxidative stress. Ann N Y Acad Sci. 2006;1067:37–44. doi: 10.1196/annals.1354.006. [DOI] [PubMed] [Google Scholar]
- 13.Sorli JV, Corella D, Frances F, Ramirez JB, Gonzalez JI, Guillen M, et al. The effect of the APOE polymorphism on HDL-C concentrations depends on the cholesterol ester transfer protein gene variation in a Southern European population. Clin Chim Acta. 2006;366:196–203. doi: 10.1016/j.cca.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 14.Sandhofer A, Tatarczyk T, Laimer M, Ritsch A, Kaser S, Paulweber B, et al. The Taq1B-variant in the cholesteryl ester-transfer protein gene and the risk of metabolic syndrome. Obesity. 2008;16:919–22. doi: 10.1038/oby.2007.130. [DOI] [PubMed] [Google Scholar]
- 15.Ordovas JM, Cupples LA, Corella D, Otvos JD, Osgood D, Martinez A, et al. Association of cholesteryl ester transfer protein-TaqIB polymorphism with variations in lipoprotein subclasses and coronary heart disease risk: the Framingham study. Arterioscler Thromb Vasc Biol. 2000;20:1323–9. doi: 10.1161/01.atv.20.5.1323. [DOI] [PubMed] [Google Scholar]
- 16.Boekholdt SM, Sacks FM, Jukema JW, Shepherd J, Freeman DJ, McMahon AD, et al. Cholesteryl ester transfer protein TaqIB variant, high-density lipoprotein cholesterol levels, cardiovascular risk, and efficacy of pravastatin treatment: individual patient meta-analysis of 13.677 subjects. Circulation. 2005;111:278–87. doi: 10.1161/01.CIR.0000153341.46271.40. [DOI] [PubMed] [Google Scholar]
- 17.Corella D, Saiz C, Guillen M, Portoles O, Mulet F, Gonzalez JI, et al. Association of TaqIB polymorphism in the cholesteryl ester transfer protein gene with plasma lipid levels in a healthy Spanish population. Atherosclerosis. 2000;152:367–76. doi: 10.1016/s0021-9150(99)00477-3. [DOI] [PubMed] [Google Scholar]
- 18.Higgins M, Province M, Heiss G, Eckfeldt J, Ellison RC, Folsom AR, et al. NHLBI Family Heart Study: objectives and design. Am J Epidemiol. 1996;143:1219–28. doi: 10.1093/oxfordjournals.aje.a008709. [DOI] [PubMed] [Google Scholar]
- 19.Lai CQ, Arnett DK, Corella D, Straka RJ, Tsai MY, Peacock JM, et al. Fenofibrate effect on triglyceride and postprandial response of apolipoprotein A5 variants. The GOLDN study Arterioscler Thromb Vasc Biol. 2007;27:1417–25. doi: 10.1161/ATVBAHA.107.140103. [DOI] [PubMed] [Google Scholar]
- 20.Otvos JD. Measurement of lipoprotein subclass profiles by nuclear magnetic resonance spectroscopy. Clin Lab. 2002;48:171–80. [PubMed] [Google Scholar]
- 21.Livak KJ. Allelic discrimination using fluorogenic probes and the 5′ nuclease assay. Genet Anal. 1999;14:143–9. doi: 10.1016/s1050-3862(98)00019-9. [DOI] [PubMed] [Google Scholar]
- 22.Sumi K, Tanaka T, Uchida A, Magoori K, Urashima Y, Ohashi R, et al. Cooperative interaction between hepatocyte nuclear factor 4α and GATA transcription factors regulates ATP-binding cassette sterol transporters ABCG5 and ABCG8. Mol Cell Biol. 2007;27:4248–60. doi: 10.1128/MCB.01894-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fujita Y, Ezura Y, Emi M, Ono S, Takada D, Takahashi K, et al. Hypertriglyceridemia associated with amino acid variation Asn985Tyr of the RP1 gene. J Hum Genet. 2003;48:305–8. doi: 10.1007/s10038-003-0029-z. [DOI] [PubMed] [Google Scholar]
- 24.Barreiro LB, Laval G, Quach H, Patin E, Quintana-Murci L. Natural selection has driven population differentiation in modern humans. Nat Genet. 2008;40:340–5. doi: 10.1038/ng.78. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


