Abstract
Detergents such as Triton X-100 are often used in drug discovery research to weed out small molecule promiscuous and non-specific inhibitors which act by aggregation in solution and undesirable precipitation in aqueous assay buffers. We evaluated the effects of commonly used detergents, Triton X-100, Tween-20, Nonidet-40 (NP-40), Brij-35, and CHAPS, on the enzymatic activity of West Nile virus (WNV) protease. Unexpectedly, Triton X-100, Tween-20, and NP-40 showed an enhancement of in vitro WNV protease activity from 2 to 2.5-fold depending on the detergent and its concentration. On the other hand, Brij-35, at ≥ ).001% enhanced the protease activity by 1.5-fold at and CHAPS had the least enhancing effect. The kinetic analysis showed that the increase in protease activity by Triton X-100 was dose-dependent. Furthermore, at Triton X-100 and Tween-20 concentrations higher than 0.001%, the inhibition of compound B, one of the lead compounds against WNV protease identified in a high throughput screen (IC50 value of 5.7 ± 2.5 μM), was reversed. However, in the presence of CHAPS, compound B still showed good inhibition of WNV protease. Our results, taken together, indicate that nonionic detergents, Triton X-100, Tween, and NP-40 are unsuitable for the purpose of discrimination of true versus promiscuous inhibitors of WNV protease in high throughput assays.
Introduction
The family of Flaviviridae contains more than 70 viruses including Yellow fever virus (YFV), Dengue virus (subtypes 1-4) (DENV1-4), West Nile virus (WNV), Kunjin, and Japanese encephalitis virus primarily transmitted by arthropods1. The mosquito-borne flaviviruses such as Dengue and West Nile viruses have recently emerged in major epidemics causing severe and lethal diseases such as dengue hemorrhagic fever and WNV encephalitis with significant morbidity 2,3. Currently, there is no antiviral therapy or vaccines available for treatment or prevention of DENV and WNV infections.
Flaviviruses are composed of a positive-sense RNA which is translated into a single polyprotein comprising three structural (C, prM, E) and seven non-structural (NS) proteins arranged in the order NH2-C-prM-E-NS1-NS2A-NS2B-NS3-NS4A-NS4B-NS5-COOH. The polyprotein is processed by the host proteases in the endoplasmic reticulum at the C-prM, prM-E, and E-NS1 as well as NS4A-NS4B sites and by the viral serine protease at specific sites containing two basic amino acid residues at P1 and P2 positions of the substrate followed by Ala, Ser, or Gly at the P1’ position (according to the nomenclature of Schechterand Berger) within the nonstructural regions1.
The NS3 protein was identified as a multifunctional protein exhibiting enzymatic activities of a serine protease within the N-terminal region as well as an NTPase, 5′-RNA triphosphatase, and RNA helicase within the C-terminal region 4-12. The serine protease domain (NS3-pro; ~184 aa) consists of three highly conserved amino acids, His51, Asp75, and Ser135, in the catalytic triad 13,14. In previous studies, it was shown that NS3-pro requires the NS2B (130 aa) cofactor for protease activity in processing of the polyprotein at NS2A-NS2B, NS2B-NS3, NS3-NS4A, and NS4B-NS5 sites 15-21. NS2B consists of three hydrophobic regions flanking a conserved ~40 residue hydrophilic domain (NS2BH) which is sufficient for protease activity in vitro 20,22,23. The role of flavivirus protease in polyprotein processing, which is a prerequisite for viral RNA replication, renders the protease as an attractive target for development of inhibitors as potential therapeutics.
One of the frequently used experimental approaches in identification of small molecule lead compounds is high throughput screening (HTS) based on robust in vitro as well as cell-based assays. Using the HTS, hundreds of thousands of compounds can be screened within a short time. However, potential false-positive “hits” could arise due to compounds having undesirable properties such as aggregation leading to nonspecific inhibition. Compounds which exhibit these properties are known as “promiscuous” inhibitors, which could inhibit a variety of targets in a nonspecific manner 24,25. The aggregates of compounds may envelop the protein and block the binding site for the substrate inhibiting the protein’s function 24,26. To identify such compounds and eliminate them from the list of true inhibitors, previous studies have reported the use of detergents that modulate surface properties and cause disaggregation of the compounds 24,27. Thus, if a compound shows inhibition in the presence of a detergent, it is then considered as a true lead compound that warrants further analysis. Triton X-100 is commonly used for distinguishing “false positive hits” from the true inhibitors 25. In this study, we demonstrate that Triton X-100 and Tween-20, and NP-40, the three nonionic detergents are not suitable for discrimination of true from “promiscuous” inhibitors in HTS because they enhance the WNV protease activity by 2- to 2.5-fold. However, a zwitterionic detergent, CHAPS (3-[(3-Cholamidopropyl)dimethylammonio]-1-propanesulfonate), has no significant effect on protease activity per se and hence could be used to distinguish false positive hits from true inhibitors. A lead inhibitor of WNV protease previously identified by HTS shows good inhibition in the presence of CHAPS but not in the presence of Triton X-100.
Methods and Materials
Materials
Triton X-100 was purchased from ICN Biomedicals. The purified WNV protease (NS2B(H)/NS3-pro) was obtained as reported previously 28. The fluorogenic WNV substrate, t-butyl-oxycarbonyl (Boc)-Gly-Lys-Arg-7-amino-4-methylcoumarin (AMC), was purchased from Bachern. Compound B was purchased from I.F. Lab (vendor ID, F0842-0004) (Life Chemicals Inc., Burlington, ON, Canada). CHAPS was purchased from Pierce, Rockford, IL.
In vitro protease assay
All assays were done in triplicate in black 96-well plates. The in vitro protease assays were performed as described before 28,29. The reaction mixture of 100 μl/assay contained 200 mM Tris pH 9.5, 30% glycerol, 27 nM WNV protease, 2% DMSO, and fluorogenic WNV substrate Boc-Gly-Lys-Arg-AMC at concentrations that varied from 15.5 μM to 2 mM or at 100 μM for fixed substrate concentrations. In some assays, the effect of Triton X-100 or CHAPS on the WNV protease activity was verified using a tetrapeptide substrate, N-Carbobenzyloxy-Val-Lys-Lys-Arg-4-Methoxy-β-naphthylamide. Aprotinin (bovine pancreatic trypsin inhibitor, BPTI) was used as a positive control for inhibition of WNV protease (Ki=~162 nM) 28 at a final concentration of 10 μM, and dimethylsulfoxide (DMSO) for the no-inhibitor control at a concentration of 2%. In vitro protease assays in presence of Triton X-100 were performed by adding the detergent to the protease buffer (31 nM WNV protease/200 mM Tris-HCI, pH 9.0/30% glycerol). The fluorescence of the AMC, the product of the protease activity on the tripeptide substrate was measured at excitation and emission wavelengths of 385 and 465 nm, respectively. The hydrolysis of the tetrapeptide substrate to the product, 4-methoxy-b-naphthylamine, was monitored at excitation and emission wavelengths of 290 and 420 nm, respectively. Fluorescence was measured using the SpectraMax Gemini EM spctrofluorometer (Molecular Devices). The relative fluorescence units (RFU) were converted to micromolar (μM) concentrations of AMC using a standard curve generated from the fluorescence values versus micromolar concentrations of free AMC. Kinetic values were determined using Microsoft Excel and GraphPad Prism 5 (La Jolla, CA). RFUs, obtained in a standard assay in the absence of detergents, were taken as 100% (control).
To assess the effects of inhibitors in the presence and absence of detergents, protease assays were performed as above except in the presence of various concentrations of an inhibitor (in the range, 0.3125-100μM), serially diluted in dimethylsulfoxide (DMSO; final concentration, 2%). In these assays, the inhibitor-enzyme complex was allowed to form by preincubation for 15 min at room temperature before addition of the substrate (100 μM final concentration). After five minutes incubation, fluorescence was measured as described above. From the initial velocities of the reactions, the IC50 values were determined using GraphPad Prism 5. The lowest and highest fluorescence values were set as 100% and 0% inhibition, respectively.
Effect of Triton X-IOO on kinetic constants of WNV protease
In the standard assay, the WNV substrate concentrations were varied from 0 to 2 mM to determine the kinetic parameters, Km and Vmax, of the WNV protease in the presence of DMSO (2%). Time course of WNV protease reaction, monitored by Spectra Max Gemini EM, were analyzed by linear regression to obtain the slopes in RFU/min. This value was then converted to μM/min using a standard curve of free AMC as described above. Using GraphPad Prism 5 software, initial rates (v) of enzyme activity were fit by non-linear regression analysis into Michaelis-Menten equation where the Vmax and Km are the maximum velocity and Michaelis-Menten constant, respectively, and [S] is the substrate concentration.
Results
(i) Triton X-IOO enhances the WNV protease activity in a dose-dependent manner
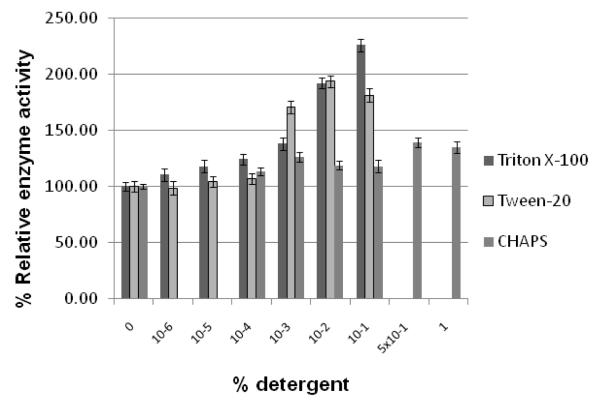
Since Triton X-100 is often used to discriminate between the “false positives” hits and the true inhibitors in the in vitro assays, we first tested the effect of Triton X-100 on the activity of WNV protease. The non-ionic detergents, Tween-20, NP-40, and Brij-35 as well as the zwitterionic detergent, CHAPS, were also used for comparison. As shown in Fig. 1, Triton X-100, Tween-20, and NP-40 enhanced the relative activity of the protease at concentrations ≥ 0.001%. The level of enhancement reached about 2- to 2.3-fold when these detergents were present in the range of 0.01 -0.1%. This stimulatory effect of Triton X-100 is not unique to the tripeptide substrate (Boc-Gly-Lys-Arg-AMC) used in the protease assay. The same enhancement effect of Triton X-100 was also seen when the tetrapeptide substrate, 4-methoxy-naphthalamide, was used (Fig. 1C). Another non-ionic detergent, Brij-35, enhanced the protease activity only about 1.5-fold in the same range of detergent concentration. On the other hand, the protease activity was not significantly affected by the zwitterionic detergent, CHAPS (Fig. 1 ).
Figure 1. WNV protease activity in presence of different detergents.
Panel A. Standard protease assays were carried out using the tripeptide substrate, t-butyl-oxycarbonyl (Boc)-Gly-Lys-Arg-7-amino-4-methylcoumarin (AMC) as described under Materials and Methods in the absence of any detergent or in the presence of Triton-X100, Tween-20, or CHAPS at the indicated concentrations of 10−6, 10−5, 10−4, 10−3, 10−2, 10−1, or 1 % in the assay mixture.
Panel B. The experimental conditions for the assay were the same as in Panel A except that NP-40, Brij-35, or CHAPS was used at the indicated concentrations.
Panel C. The conditions of the assay were the same except that the effects of Triton X-100 and CHAPS at indicated concentrations on the WNV protease activity using a tetrapeptide substrate, N-Carbobenzyloxy-Val-Lys-Lys-Arg-4-Methoxy-b-naphthylamide as described under Materials and Methods. The percent protease activity was plotted compared to that of the no-detergent control set at 100%.
(ii) Effect of Triton X-IOO on the Km and the Vmax of the WNV protease
The Km and Vmax values were determined for the WNV protease in presence and absence of Triton X-100 to examine whether the detergent affects the affinity of the enzyme for the substrate. As shown in Fig. 2, the Km values do not differ significantly in the presence and absence of Triton X-100 whereas the Vmax values are increased with increasing concentrations of Triton X-100 (Table 1 ) in a dose-dependent manner. This experiment was repeated three times with similar values for Km and Vmax supporting our conclusion.
Figure 2. Effect of Triton X-100 on Km and Vmax values of WNV protease.
Different WNV substrate concentrations (0 – 2000 μM) were used to determine WNV protease activity in the presence and absence of Triton X-100 in standard protease assays as described under Materials and Methods. The experiments were repeated three times and the error bars represent the standard deviation of the mean.
Table 1.
Effect of Triton X-100 on the Km and vmax values of WNV protease
| Triton X-100 cone (%) |
Km [μM] |
Vmax [μM/min] |
kcat* [sec−1] |
kcat/Km 10−3 [μM−1 sec− 1] |
|---|---|---|---|---|
| 0 | 172 ± 60 | 0.97 ± 0.18 | 0.60 ± 0.11 | 3.5 ± 0.6 |
| 0.0001 | 182.6 ± 45 | 0.84 ± 0.11 | 0.52 ± 0.07 | 2.8 ± 0.4 |
| 0.001 | 187.1 ± 15 | 1.25 ± 0.25 | 0.77 ± 0.15 | 4.1 ± 0.5 |
| 0.01 | 240 ± 30 | 1.65 ± 0.20 | 1.02 ± 0.12 | 4.3 ± 0.1 |
| 0.1 | 220 ± 20 | 1.70 ± 0.01 | 1.05 ± 0.01 | 4.8 ± 0.4 |
assuming a total enzyme concentration of 27 nM
The protease assays were performed as described under Materials and Methods in the presence of indicated concentrations of Triton X-100. The Km and Vmax values were calculated using the GraphPad Prism 5.0 software.
(iii) Effect of Triton X-IOO on the inhibition of WNV protease by compound B
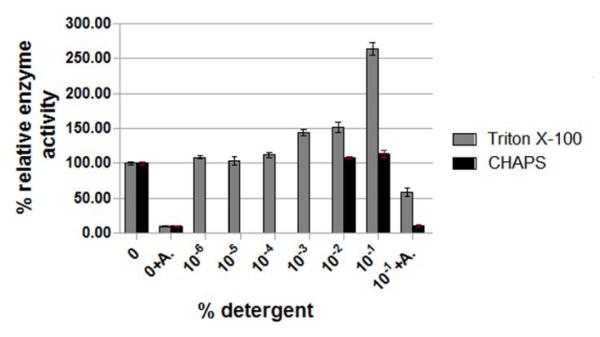
Compound B was identified as a lead inhibitor against WNV protease with a Ki of 3.4 μM ± 0.6 μM in the in vitro assay and selectivity index of 100 in a replicon-based assay in mammalian (Vero) cells 29. Therefore, we sought to examine the inhibitory effect of the lead compound in the presence of Triton X-100. As shown in Fig. 3, when Triton X-100 was included at concentrations well below its critical micelle concentration (up to 0.001%) (CMC= ~0.02%), compound B still retained its inhibitory activity. However, at 0.01% Triton X-100 (~0.5 CMC) in the assay, the percent inhibition of the WNV protease activity by compound B was essentially abolished (Fig. 3A and 3B). However, the inhibitory activity of bovine pancreatic trypsin inhibitor (BPTI), also known as aprotinin, which is a potent inhibitor of DENV and WNV proteases with Ki values of 26 and 162 nM, respectively 28, was reduced only by about 20% in the presence of Triton X-100 (Fig. 1C and Fig. 3A).
Figure 3. Effect of Triton X-100 on the inhibition of WNV protease by compound B.
A. The assays were performed as described under Materials and Methods in the presence and absence of Triton X-100 and 2 μM aprotinin (light grey bars) or 20 μM compound B (dark grey bars). At Triton X-100 concentrations at 0.5 CMC (CMC=0.02%), the inhibition of compound B is reduced ~85%. B. The concentrations of compound B were varied as indicated. The protease assays were performed at two different Triton X-100 concentrations (0.001 %-grey bars and 0.01%-white bars) along with no-detergent control (black bars). The experiments in panels A and B were repeated at least three times with similar results. The error bars represent standard deviation of the mean.
Discussion
Triton X-100, polyethylglycol tert-octylphenyl ether, is a nonionic detergent, and is used extensively for solubilization of membrane proteins and their biochemical characterization. In drug discovery field, this detergent is used to discriminate the “promiscuous” inhibitors from true inhibitors of a target protein or enzyme in the in vitro assays 24,25,30,31. Compounds that inhibit unrelated enzyme targets in the in vitro assays often do so by aggregation and/or formation of colloidal particles which could bind to the targets nonspecifically and interfere with substrate or inhibitor binding leading to false “hits”. The presence of a detergent below its CMC, provided it does not interfere with the assay, prevents aggregation of the inhibitor compound and allows interaction between the inhibitor and the target.
However, the findings of the present study using the WNV protease as the enzyme target indicate that Triton X-100 interferes with the inhibitory activity of compound B (Fig. 3) and other compounds tested (data not shown). However, inclusion of a zwitterionic detergent such as CHAPS did not reverse the inhibitory activity of compound B (Fig. 1A). There are other examples of Triton X-100, Nonidet P-40 (NP-40) and/or Tween-20 interfering with target assays and those that describe that CHAPS or Brij-35 could also be used as a detergent to distinguish between true inhibitors from false-positive ones 32-34 35. For example, it was previously reported that Triton X-100 may act as a nonspecific activator of chymotrypsin 36. In the investigation of small molecule inhibitors of Rcel p protease, which is required for Ras GTPase maturation, Triton X-100, NP-40, and CHAPS at their CMC inactivated the protease. Tween-20 at its CMC, on the other hand, was also ineffective for this purpose. Therefore, these detergents could not be used for distinguishing promiscuous versus true inhibition by the compounds 37. Moreover, Triton X-100 and NP-40 decreased the inhibition of reverse transcriptase (RT) activity by inophyllum B whereas CHAPS had no effect 38. In this regard, it was noted that Triton X-100 is an activator of HIV-1 RT activity and reverses the inhibition of TIBO R82150 39.
The possibility that compound B is a promiscuous inhibitor rather than a specific inhibitor of WNV protease is unlikely because of the following properties of compound B as an inhibitor of WNV protease. (1) Compound B shows some specificity as it did not have any inhibitory effect on cellular serine proteases such as trypsin and factor Xa 29. (2) Other derivatives of compound B also exhibit potent inhibition of WNV protease activity in vitro (P. Viswanathan et al. unpublished results; data not shown). (3) Compound B inhibited WNV RNA replication in a cell-based assay with an EC50 value of 1.4 ± 0.4 μM29. Moreover, in the presence of Triton X-100 at 0.001% compound B showed good inhibition of WNV protease with no change in the IC50 value compared to no-detergent control (Fig. 3B). In the presence of Triton X-100 greater than 0.001% and Tween-20, the inhibition of WNV protease by compound B is reduced by up to 70% whereas the ionic detergent, CHAPS, did not have any affect. This study is the first to report on the enhancement of trypsin-like WNV serine protease activity by Triton X-100, Tween-20, and NP-40 whereas CHAPS shows no significant effect on the activity. In fact, the lack of appreciable stimulation by CHAPS up to 0.5% (CMC = 0.48% or 7.4 mM)40 is not due to high pH of the assay buffer because it has been reported that CHAPS maintains its zwitter ionic property in a wide pH range of 2 to 12 41. CHAPS was a component of the purification/assay buffer used in the characterization of the WNV protease activity in an earlier study23; however, its property in the context of enhancing effects of detergents on the WNV protease activity was not investigated earlier to this study.
The mechanism by which Triton X-100 enhances the WNV protease activity is unknown at present. However, our kinetic analysis suggests that the detergent increases the Vmax values of the enzyme in a dose-dependent manner without affecting the affinity of the enzyme (Km values) for the substrate. One possibility is that Triton X-100, Tween-20, and NP-40 prevent the protease sticking to the wall of the 96-well plate thereby increasing the local concentration of the enzyme compared to the well without the detergent. Alternately, Triton X-100 may prevent aggregation of the enzyme thereby increasing the concentration of the active enzyme which would only influence the Vmax but not the Km. This possibility is supported by our data that Km values for WNV protease remain almost the same, whereas Vmax slightly increases with higher Triton X-100 concentrations (Table 1 ). However, the result that increasing the Triton X-100 concentration from 0.001 to 0.01 reduced the inhibition of the compound B by 70% (Fig. 3B), is not consistent with this interpretation because the total amount of the enzyme in each assay was only 2.7 pmol, the substrate 10 nmol and the inhibitor 2 nmol and therefore, increasing the concentration of the detergent is not expected to have any effect on the inhibition of compound B. Moreover, Triton X-100 does not have any effect on the substrate in the absence of WNV protease (data not shown). Another possibility should be considered that Triton X-100 at higher concentrations not only enhances the Vmax of the hydrolysis of the substrate without significant change of Km, but may also cause a conformational change at a different site that affects the binding of the compound B to the enzyme thereby attenuating its inhibition.
Therefore, our study emphasizes the need to test the effect of the chosen detergent on the enzyme (or any molecular target) prior to using it in the HTS for identification and elimination of false positive hits.
Acknowledgement
This work was supported by a grant from NIAID/NIH (AI-070791). We thank Dr. Kurt E. Ebner for helpful discussion.
Abbreviations
- AMC
7-amino-4-methylcoumarin
- CHAPS
(3-[(3-Cholamidopropyl)dimethylammonio]-1-propanesulfonate)
- CMC
critical micelle concentration
- DENV
Dengue virus
- DMSO
dimethylsulfoxide
- IC50
50% inhibitory concentration
- Km
Michaelis-Menten constant
- NS2BH
hydrophilic domain of non-structural protein 2B
- NS3-pro
protein domain of non-structural protein 3
- RFU
relative fluorescence unit
- vmax
maximum enzyme velocity
- WNV
West Nile virus
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lindenbach BD, Rice CM. Adv Virus Res. 2003;59:23. doi: 10.1016/s0065-3527(03)59002-9. [DOI] [PubMed] [Google Scholar]
- 2.Gubler DJ. Novartis Found Symp. 2006;277:3. doi: 10.1002/0470058005.ch2. [DOI] [PubMed] [Google Scholar]
- 3.Roehrig JT, Layton M, Smith P, Campbell GL, Nasci R, Lanciotti RS. Curr Top Microbiol Immunol. 2002;267:223. doi: 10.1007/978-3-642-59403-8_11. [DOI] [PubMed] [Google Scholar]
- 4.Wengler G. Virology. 1991;184:707. doi: 10.1016/0042-6822(91)90440-m. [DOI] [PubMed] [Google Scholar]
- 5.Warrener P, Tamura JK, Collett MS. J Virol. 1993;67:989. doi: 10.1128/jvi.67.2.989-996.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuo MD, Chin C, Hsu SL, Shiao JY, Wang TM, Lin JH. J Gen Virol. 1996;77:2077. doi: 10.1099/0022-1317-77-9-2077. [DOI] [PubMed] [Google Scholar]
- 7.Cui T, Sugrue RJ, Xu Q, Lee AK, Chan YC, Fu J. Virology. 1998;246:409. doi: 10.1006/viro.1998.9213. [DOI] [PubMed] [Google Scholar]
- 8.Li H, Clum S, You S, Ebner KE, Padmanabhan R. J. Virol. 1999;73:3108. doi: 10.1128/jvi.73.4.3108-3116.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matusan AE, Pryor MJ, Davidson AD, Wright PJ. J Virol. 2001;75:9633. doi: 10.1128/JVI.75.20.9633-9643.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borowski P, Niebuhr A, Mueller O, Bretner M, Felczak K, Kulikowski T, Schmitz H. J Virol. 2001;75:3220. doi: 10.1128/JVI.75.7.3220-3229.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartelma G, Padmanabhan R. Virology. 2002;299:122. doi: 10.1006/viro.2002.1504. [DOI] [PubMed] [Google Scholar]
- 12.Benarroch D, Selisko B, Locatelli GA, Maga G, Romette JL, Canard B. Virology. 2004;328:208. doi: 10.1016/j.virol.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 13.Bazan JF, Fletterick RJ. Virology. 1989;171:637. doi: 10.1016/0042-6822(89)90639-9. [DOI] [PubMed] [Google Scholar]
- 14.Gorbalenya AE, Donchenko AP, Koonin E,V, Blinov VM. Nucl. Acid Res. 1989;17:3889. doi: 10.1093/nar/17.10.3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chambers TJ, Grakoui A, Rice CM. J Virol. 1991;65:6042. doi: 10.1128/jvi.65.11.6042-6050.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Falgout B, Pethel M, Zhang YM, Lai CJ. J Virol. 1991;65:2467. doi: 10.1128/jvi.65.5.2467-2475.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang L, Mohan PM, Padmanabhan R. J Virol. 1992;66:7549. doi: 10.1128/jvi.66.12.7549-7554.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Falgout B, Miller RH, Lai C-J. Journal of Virology. 1993;67:2034. doi: 10.1128/jvi.67.4.2034-2042.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chambers TJ, Nestorowicz A, Amberg SM, Rice CM. J Virol. 1993;67:6797. doi: 10.1128/jvi.67.11.6797-6807.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clum S, Ebner KE, Padmanabhan R. J Biol Chem. 1997;272:30715. doi: 10.1074/jbc.272.49.30715. [DOI] [PubMed] [Google Scholar]
- 21.Bera AK, Kuhn RJ, Smith JL. J Biol Chem. 2007;282:12883. doi: 10.1074/jbc.M611318200. [DOI] [PubMed] [Google Scholar]
- 22.Yusof R, Clum S, Wetzel M, Murthy HM, Padmanabhan R. J Biol Chem. 2000;275:9963. doi: 10.1074/jbc.275.14.9963. [DOI] [PubMed] [Google Scholar]
- 23.Leung D, Schroder K, White H, Fang NX, Stoermer MJ, Abbenante G, Martin JL, Young PR, Fairlie DP. J Biol Chem. 2001;276:45762. doi: 10.1074/jbc.M107360200. [DOI] [PubMed] [Google Scholar]
- 24.McGovern SL, Helfand BT, Feng B, Shoichet BK. J Med Chem. 2003;46:4265. doi: 10.1021/jm030266r. [DOI] [PubMed] [Google Scholar]
- 25.Feng BY, Shelat A, Doman TN, Guy RK, Shoichet BK. Nat Chem Biol. 2005;1:146. doi: 10.1038/nchembio718. [DOI] [PubMed] [Google Scholar]
- 26.McGovern SL, Caselli E, Grigorieff N, Shoichet BK. J Med Chem. 2002;45:1712. doi: 10.1021/jm010533y. [DOI] [PubMed] [Google Scholar]
- 27.Ryan AJ, Gray NM, Lowe PN, Chung CW. J Med Chem. 2003;46:3448. doi: 10.1021/jm0340896. [DOI] [PubMed] [Google Scholar]
- 28.Mueller NH, Yon C, Ganesh VK, Padmanabhan R. Int J Biochem Cell Biol. 2007;39:606. doi: 10.1016/j.biocel.2006.10.025. [DOI] [PubMed] [Google Scholar]
- 29.Mueller NH, Pattabiraman N, Ansarah-Sobrinho C, Viswanathan P, Pierson TC, Padmanabhan R. Antimicrob Agents Chemother. 2008;52:3385. doi: 10.1128/AAC.01508-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feng BY, Shoichet BK. Nat Protoc. 2006;1:550. doi: 10.1038/nprot.2006.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feng BY, Shoichet BK. J Med Chem. 2006;49:2151. doi: 10.1021/jm060029z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taylor PB, Culp JS, Debouck C, Johnson RK, Patil AD, Woolf DJ, Brooks I, Hertzberg RP. J Biol Chem. 1994;269:6325. [PubMed] [Google Scholar]
- 33.von Ahsen O, Bomer U. Chembiochem. 2005;6:481. doi: 10.1002/cbic.200400211. [DOI] [PubMed] [Google Scholar]
- 34.Teklu S, Gundersen LL, Larsen T, Malterud KE, Rise F. Bioorg Med Chem. 2005;13:3127. doi: 10.1016/j.bmc.2005.02.056. [DOI] [PubMed] [Google Scholar]
- 35.Johnston PA, Phillips J, Shun TY, Shinde S, Lazo JS, Huryn DM, Myers MC, Ratnikov BI, Smith JW, Su Y, Dahl R, Cosford ND, Shiryaev SA, Strongin AY. Assay Drug Dev Technol. 2007;5:737. doi: 10.1089/adt.2007.101. [DOI] [PubMed] [Google Scholar]
- 36.Goode DR, Totten RK, Heeres JT, Hergenrother PJ. J. Med. Chem. 2008;51:2346. doi: 10.1021/jm701583b. [DOI] [PubMed] [Google Scholar]
- 37.Manandhar SP, Hildebrandt ER, Schmidt WK. J Blomol Screen. 2007;12:983. doi: 10.1177/1087057107307226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Debyser Z, Vandamme AM, Pauwels R, Baba M, Desmyter J, De Clercq E. J Biol Chem. 1992;267:11769. [PubMed] [Google Scholar]
- 39.Debyser Z, Pauwels R, Andries K, Desmyter J, Engelborghs Y, Janssen PA, De Clercq E. Mol Pharmacol. 1992;41:203. [PubMed] [Google Scholar]
- 40.Chattopadhyay A, London E. Anal Blochem. 1984;139:408. doi: 10.1016/0003-2697(84)90026-5. [DOI] [PubMed] [Google Scholar]
- 41.Perdew GH, Schaup HW, Selivonchick DP. Anal Blochem. 1983;135:453. doi: 10.1016/0003-2697(83)90711-x. [DOI] [PubMed] [Google Scholar]