Abstract
DNA transfections are widely performed in research laboratories and in vivo gene delivery holds the promise for curing many diseases. The synthetic carriers or vectors for DNA are typically cationic lipids. However, in biology, the recognition of nucleic acids by proteins involves both electrostatic and stacking contributions. As such we have prepared a series of new lipophilic peptide vectors that possess lysine and tryptophan amino acids for evaluation. These lipophilic peptides show minimal cytotoxicity and enhanced in vitro gene transfection activity.
The complexation of cationic amphiphiles with DNA or RNA and the resulting supramolecular structures are of interest for the delivery of nucleic acids to cells. As such, a means to correct a defective gene, introduce a new gene, or knock-down a gene is provided for a specific in vitro or in vivo application (1-7). This complexation between cationic amphiphiles and nucleic acids is governed by electrostatic interactions between the positively charged amphiphiles and the negatively charged phosphate backbone of the nucleic acid and the hydrophobic chain-chain packing forces between the individual assembled amphiphiles. In biology, the recognition of nucleic acids by proteins involves more than electrostatic interactions (8-10). Examination of these protein nucleic acid recognition motifs typically reveals structures rich in basic and aromatic amino acids that provide important electrostatic and stacking contributions to binding. With this inspiration in mind, we are designing peptide-based amphiphiles for gene delivery. Herein we describe a series of lipophilic peptides including the synthesis, physiochemical properties, cytotoxicity, and in vitro gene transfection activity.
To mimic structural characteristics present in nucleic acid binding proteins, we selected the tripeptide Lys-Trp-Lys, KWK, to be the headgroup for the amphiphile because it possesses cationic charges and an aromatic side chain and is relatively small in size. Moreover, this peptide is known to bind DNA with a binding constant on the order of 104 M and is a model peptide used to study protein-nucleic acid interactions, and lysine and tryptophan provide important interactions in a number of structurally characterized DNA binding domains of proteins (11-14). In addition, lysine-based small molecule amphiphiles and macromolecules (e.g., polylysine) are being explored and used for gene transfection (15-24). In order to perform a systematic study to identify the key molecular components responsible for transfection activity with these new lipophilic peptides, we prepared a series of amphiphiles where the headgroup (charge, aliphatic content, aromatic content) and fatty acid chain length (C12:0, C14:0, C16:0, and C18:0) were varied. As shown in Figure 1, fourteen different lipophilic peptides were synthesized and investigated. The lipophilic peptides were synthesized in good yield. First, the fatty acids were coupled to 3-t-butyldiphenylsilylglycerol using EDCI and DMAP in CH2Cl2. The silyl protecting group was removed with TBAF and then the corresponding Boc-protected amino acids were coupled to the free primary hydroxyl. The complete synthetic and characterization details as well as the reaction scheme can be found in the Supporting Information document.
Figure 1.
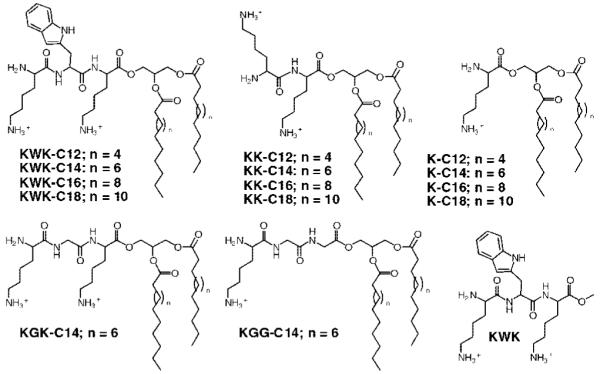
Lipophilic peptides under investigation for gene delivery.
To determine whether the lipophilic peptides bind DNA, we performed a standard ethidium bromide-DNA displacement fluorescence assay. All of the lipophilic peptides bind DNA with approximately the same affinity 106 M as measured by displacement of ethidium bromide. Most of the lipophilic peptides form DNA complexes with a cation to anion ratio between 1.5 and 2, except for the C18:0 analogues which have a cation to anion ratio of 3. Binding curves for all of the lipophilic peptides can be found in the SI.
These lipophilic peptides are likely to form bilayer vesicles in aqueous solution, given the polar headgroup and long hydrophobic acyl chains present in the structure. We prepared liposome solutions of all the lipophilic peptides with and without DNA by sonicating a hydrated film of the lipid in an aqueous solution (see SI for details). Dynamic light scattering measurements revealed the average diameter to range from 170 to 850 nm for the various lipophilic peptides. As expected, vesicles were not observed with the tripeptide alone, KWK. Next, X-ray diffraction experiments were performed at 25 °C on samples in the absence and presence of DNA to characterize the supramolecular structure (see SI). A hydrated vesicle pellet was obtained for all samples except K-C14, KK-C16, K-C16, and KGG-C14. For the specimens that formed pellets, oriented multilayers were prepared for X-ray analysis by placing the pellets on a curved glass substrate and incubating at 66% relative humidity. All of the samples that did diffract showed multiple reflections indexing as orders of lamellar repeat periods, characteristic of lipid bilayers. For a specific headgroup the lamellar repeat period systematically increased with increasing fatty acid chain length. For example, the repeat periods for KK-C12, KK-C14, and KK-C18 were 3.7 nm, 4.1 nm, and 5.1 nm, respectively. For these KK samples the repeat period did not increase appreciably upon the addition of DNA, indicating that DNA was not present within the multilayers. The lipophilic peptides KWK-C14 and KWK-C16, possessing the tripeptide headgroup and either the C14:0 or C16:0 acyl chains, gave larger lamellar repeat periods in the presence of DNA. The addition of DNA increased the repeat periods from 4.4 to 7.3 nm and 6.6 to 7.1 nm for KWK-C14 and KWK-C16, respectively. For KGK-C14 a repeat period of 4.1 nm was obtained without DNA and a satisfactory pellet with DNA was not obtained. For KGG-C14 the repeat period in the presence of DNA was 6.8 nm. These data with the tripeptide headgroups are consistent with a structural model where a smectic phase is formed with the DNA chains located between the adjacent lipid bilayers within the multilamellar liposome. This structural model has been reported for complexes of DNA with DOTAP (25) and cationic triesters of phosphatidylcholine (26).
Transfection experiments using the reporter gene, β-galactosidase (β-gal, pVax-LacZ1, Invitrogen), were performed with chinese hamster ovarian (CHO) and NIH 3T3 cells (see SI for details). Gene transfection results were determined after 48 h as a function of both lipophilic peptides and cation/anion ratio. Only those lipophilic peptides possessing the tripeptide headgroup afforded significant transfection of the reporter gene. The peptide itself does not transfect. As shown in Figure 2 for the CHO cells, lipophilic peptides KWK-C14 and KWK-C16 were more effective synthetic vectors than the KWK-C12 and KWK-C18 based lipids. To assess the role of the aromatic tryptophan and lysine in the headgroup, we tested peptidic amphiphile KGK-C14 where the W was replaced with a glycine (G) residue and KGG-C14 where the alpha K was also replaced with a G. As shown in Figure 2, good transfection activity was observed with KGK-C14, and higher levels were obtained with the KGG-C14 transfection agent. The lipophilic peptide KGG-C14 was comparable to our positive control. Successful transfection was also observed with the NIH 3T3 cells as shown in Figure 2 with the KWK-C16 being the most active. The lipophilic peptides transfected a large number of cells in the population as confirmed by histochemical staining (Figure 3). Cytotoxity experiments performed with all the tripeptide lipophilic peptides showed no significant cytotoxicity, similar to nontreated cells and significantly better than Lipofectamine 2000 when exposed to chinese hamster ovarian (CHO) or NIH 3T3 cells (see SI).
Figure 2.
Gene transfection results after 48 h in CHO cells (top) and in NIH 3T3 (bottom) as a function of lipophilic peptide and mole ratio. N = 3.
Figure 3.

Photograph of CHO cells transfected with the reporter gene, β-galactosidase (β-gal) using KWK-C14 (right) and Lipofectamine 2000 (left).
In summary, a series of new lipophilic peptides are reported and those possessing a cationic tripeptide headgroup are effective nonviral vectors for gene delivery. A number of observations can be made for these lipophilic peptides including (1) the necessity for lipophilicity in the vector as the tripeptide itself does not form vesicular structures nor afford transfection (KWK-C14 vs KWK); (2) more cationic charges are not the sole contributor to efficient transfection (K-C16 vs KK-C16 vs KWK-C16); (3) a peptide-based headgroup containing an aromatic residue capable of DNA intercalation is not required as substitution by an aliphatic residue gives activity; (4) spacing of the cationic charges within the headgroup affords better transfection performance (KWK-C14 vs KK-C14); and (5) lipoplexes forming a lamellar structure with DNA which possess >6 nm repeat periods perform better compared to the structures that do not (KWK-C14 vs KK-C14). These results highlight the sensitivity of gene transfection to subtle changes in chemical structure and the importance of identifying the optimized noncovalent interactions between the transfection agent and DNA. Importantly, these results with the lipophilic peptides expand the current repertoire of efficient transfection vectors available for use. Continued research with synthetic nonviral vectors will facilitate the design, development, and subsequent in vitro and in vivo evaluation toward the goal of efficient and safe delivery of nucleic acids.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by the National Institutes of Health (EB005658 to M.W.G. and GM27278 to T.J.M.).
LITERATURE CITED
- (1).Saltzman WM, Luo D. Synthetic DNA delivery systems. Nat. Biotechnol. 2000;18:33–37. doi: 10.1038/71889. [DOI] [PubMed] [Google Scholar]
- (2).Davis ME. Curr. Opin. Biotechnol. 2002;13:128–131. doi: 10.1016/s0958-1669(02)00294-x. [DOI] [PubMed] [Google Scholar]
- (3).Kabanov AV, Felgner PL, Seymour LW. From laboratory to clinical trial. John Wiley and Sons; New York: 1998. Self-assembling complexes for gene delivery. [Google Scholar]
- (4).Putnam D. Polymers for gene delivery across length scales. Nat. Mater. 2006;5:439–451. doi: 10.1038/nmat1645. [DOI] [PubMed] [Google Scholar]
- (5).Behr JP. Gene transfer with synthetic cationic amphiphiles: Prospects for gene therapy. Bioconjugate Chem. 1994;5:382–389. doi: 10.1021/bc00029a002. [DOI] [PubMed] [Google Scholar]
- (6).Yan W, Huang L. Recent advances in liposome-based nanoparticles for antigen delivery. Polym. Rev. 2007;47:329–344. [Google Scholar]
- (7).Karmali PP, Chaudhuri A. Cationic liposomes as non-viral carriers of gene medicines: Resolved issues, open questions, and future promises. Med. Res. Rev. 2007;27:696–722. doi: 10.1002/med.20090. [DOI] [PubMed] [Google Scholar]
- (8).Nadassy K, Wodak SJ, Janin J. Structural features of protein-nucleic acid recognition sites. Biochemistry. 1999;38:1999–2017. doi: 10.1021/bi982362d. [DOI] [PubMed] [Google Scholar]
- (9).Jones S, van Heyningen P, Berman HM, Thornton JM. Protein-DNA interactions: A structural analysis. J. Mol. Biol. 1999;287:877–896. doi: 10.1006/jmbi.1999.2659. [DOI] [PubMed] [Google Scholar]
- (10).Ahmad S, Gromiha MM, Sarai A. Analysis and prediction of DNA-binding proteins and their binding residues based on composition, sequence and structural information. Bioinformatics. 2004;20:477–486. doi: 10.1093/bioinformatics/btg432. [DOI] [PubMed] [Google Scholar]
- (11).Brun F, Toulmé JJ, Hélène C. Interactions of aromatic residues of proteins with nucleic acids. Fluorescence studies of the binding of oligopeptides containing tryptophan and tyrosine residues to polynucleotides. Biochemistry. 1975;14:558–563. doi: 10.1021/bi00674a015. [DOI] [PubMed] [Google Scholar]
- (12).Behmoaras T, Toulmé JJ, Hélène C. Specific recognition of apurinic sites in DNA by a tryptophan containing peptide. Proc. Nat. Acad. Sci. 1982;78:926–930. doi: 10.1073/pnas.78.2.926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Masse JE, Wong B, Yen Y-M, Allain FH-T, Johnson RC, Feigon J. The s. Cerevisiae architectural hmgb protein nhp6a complexed with DNA: DNA and protein conformational changes upon binding. J. Mol. Biol. 2002;323:263–284. doi: 10.1016/s0022-2836(02)00938-5. [DOI] [PubMed] [Google Scholar]
- (14).Redinbo MR, Champoux JJ, Hol WGJ. Novel insights into catalytic mechanism from a crystal structure of human topoisomerase i complex with DNA. Biochemistry. 2000;39:6832–6840. doi: 10.1021/bi992690t. [DOI] [PubMed] [Google Scholar]
- (15).Kabanov AV, Kabanov VA. DNA complexes with polycations for the delivery of genetic material into cells. Bioconjugate Chem. 1995;6:7–20. doi: 10.1021/bc00031a002. [DOI] [PubMed] [Google Scholar]
- (16).Karmali PP, Kumar VV, Chaudhuri A. Design, syntheses, and in vitro gene delivery efficacies of novelmono, d-, and trilysinated cationic lipids: A structure-activity investigation. J. Med. Chem. 2004;47:2123–2132. doi: 10.1021/jm030541+. [DOI] [PubMed] [Google Scholar]
- (17).Choi JS, Joo DK, Kim CH, Kim K, Park JS. Synthesis of barbell-like tri-block copolymers, poly(l-lysine)dendrimer-block-poly(ethylene glycol)-block-poly(l-lysine) dendrimers and its self-assembly with plasmid DNA. J. Am. Chem. Soc. 2000;122:474–480. [Google Scholar]
- (18).Zauner W, Ogris M, Wagner E. Polylysine-based transfection systems utilizing receptor-mediated delivery. Adv. Drug. Delivery Rev. 1998;30:97–113. doi: 10.1016/s0169-409x(97)00110-5. [DOI] [PubMed] [Google Scholar]
- (19).Deshmukh HM, Huang L. Liposome and polylysine mediated gene transfer. New. J. Chem. 1997;21:113–124. [Google Scholar]
- (20).Slimani H, Guenin E, Briane D, Coudert R, Charnaux N, Starzec A, Vassy R, Lecouvey M, Perret YG, Cao A. Lipopeptide-based liposomes for DNA delivery into cells expressing neuropilin-1. J. Drug Targeting. 2006;14:694–706. doi: 10.1080/10611860600947607. [DOI] [PubMed] [Google Scholar]
- (21).Murphy EA, Waring AJ, Murphy JC, Willson RC, Longmuir KJ. Development of an effective gene delivery system: A study of complexes composed of a peptide-based amphiphilic DNA compaction agent and phospholipid. Nucleic Acid Res. 2001;29:3694–3704. doi: 10.1093/nar/29.17.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Gopal V, Prasad TK, Rao NM, Takafuji M, Rahman MM, Ihara H. Synthesis and in vitro evaluation of glutamide-containing cationic lipids for gene delivery. Bioconjugate Chem. 2006;17:1530–1536. doi: 10.1021/bc0601020. [DOI] [PubMed] [Google Scholar]
- (23).Chan E, Amon M, Marano RJ, Wimmer N, Kearns PS, Manolios N, Rakoczy PE, Toth I. Novel cationic lipophilic peptides for oligodeoxynucleotide delivery. Bioorg. Med. Chem. 2007;15:4091–4097. doi: 10.1016/j.bmc.2007.03.078. [DOI] [PubMed] [Google Scholar]
- (24).Waterhouse JE, Harbottle RP, Keller M, Kostarelos K, Coutelle C, Jorgensen MR, Miller AD. Synthesis and application of integrin targeting lipopeptides in targeted gene delivery. ChemBioChem. 2005;6:1212–1223. doi: 10.1002/cbic.200400408. [DOI] [PubMed] [Google Scholar]
- (25).Radler JO, Koltover I, Salditt T, Safinya CR. Structure of DNA-cationic liposome complexes: DNA intercalation in multilamellar membranes in distinct interhelical packing regimes. Science. 1997;275:810–814. doi: 10.1126/science.275.5301.810. [DOI] [PubMed] [Google Scholar]
- (26).MacDonald RC, Ashley GW, Shida MM, Rakhmanova VA, Tarahovsky YS, Kennedy MT, Pozharski EV, Baker KA, Jones RD, Rosenzweig HS, Choi KL, Qiu R, McIntosh TJ. Physical and biological properties of cationic triesters of phosphatidylcholine. Biophys. J. 1999;77:2612–2629. doi: 10.1016/S0006-3495(99)77095-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



