Abstract
Purpose
To implement in vivo detection of lactate in the presence of lipids by proton magnetic resonance spectroscopy at a 3 Tesla (T) field strength for potential applications in human tumors outside of the brain.
Materials and Methods
The BASING J-difference sequence was implemented in the presence of high lipid concentrations in phantoms and in vivo at 3 Tesla.
Results
The effectiveness of the lactate editing scheme is demonstrated in phantoms containing both lactate and lipids and in vivo in ischemic induced human muscle.
Conclusion
The ability of the BASING J-difference technique to detect lactate in the presence of strong lipid signals outside the brain at 3T is feasible. This robust technique should permit noninvasive lactate measurements in human tumors to investigate its potential as a prognostic indicator.
Keywords: lactate, magnetic resonance spectroscopy, BASING, PRESS, 3 Tesla, lipid
While not normally present in well-perfused, healthy tissue, lactate has been detected in solid tumors (1–7). The presence of lactate in tumors is thought to be caused by both aerobic glycolysis, a characteristic unique to tumors, and by anaerobic glycolysis in the hypoxic tumor milieu. In combined bioluminescence and oxygen electrode studies, the level of tumor lactate has been shown to correlate negatively with tumor oxygenation and, additionally, with patient outcome in cervical (7) and head and neck (3) cancers. Thus, in addition to being a potential marker of hypoxia, the in vivo measurement of lactate may provide an a priori prognostic marker of patient outcome.
In the proton NMR spectrum, the J-coupled lactate methyl (CH3) and methine (CH) protons give rise to a doublet at 1.3 ppm and a quartet at 4.1 ppm, respectively. The methyl resonance is preferable for detection because it is more distant from water and the 3 methyl protons give rise to a stronger signal than the single methine proton. However, the lactate CH3 protons are co-resonant with the methylene (CH2) resonance of lipids. Because solid tumors such as head and neck cancers and sarcomas are located in regions of the body where lipids are abundant, detection of the lactate methyl resonance requires an editing scheme. In this respect, the J-coupling of the lactate resonances is advantageous in that it can form the basis of such a scheme.
Numerous investigators have demonstrated lactate editing in the presence of lipids at 1.5 Tesla (T) (5,8). While the expected increase in signal-to-noise ratio at higher field strength should increase the sensitivity for metabolites such as lactate with concentrations in the millimolar range, human in vivo lactate editing in the presence of lipids on the 3T scanner has not been widely reported. To our knowledge, two studies have reported lactate editing in exercised muscle at 3T (9,10). The 3T scanner presents a challenge in that chemical shift effects due to limited radiofrequency (RF) bandwidth in slice selective pulses can result in loss of lactate signal (11–14). The current study extends the use of the dual BASING technique for lactate editing (5,15,16) to 3T and addresses the chemical shift effect as well.
MATERIALS AND METHODS
Theory
The dual band-selective inversion with gradient dephasing (BASING) technique (17) uses two frequency-selective 180° inversion pulses which are flanked by crusher gradients of opposite polarity to suppress chemical species outside of a specified frequency pass-band. Species which are in the inversion band of the pulse are dephased by the crusher gradients while species which are in the passband and are not inverted are refocused by the opposing crushers and are effectively unperturbed. While the BASING sequence was initially implemented for water and lipid suppression in proton spectroscopy (17,18), Star-Lack et al applied the technique to lactate editing in head and neck tumors at 1.5T using a J-difference method (5,15). Briefly, within a PRESS (19) localization scheme, a BASING RF/crusher train was applied after each PRESS 180° pulse as part of a two-cycle experiment. In the first cycle, the frequency of the BASING pulses was offset so that the waveform’s inversion band included the methine quartet moiety of lactate at 4.1 ppm but not metabolites further upfield such as choline, creatine and the lactate methyl doublet at 1.3 ppm. In the second cycle, the inversion band was moved downfield and metabolites in the 4.5- to 0-ppm range were unaffected. Subtraction of the two successive passes provided the coupled resonances such as the methyl doublet of lactate. Addition of the two cycles yielded noncoupled resonances or coupled resonances whose coupling partners were not located in spectral regions which would be effectively edited by this scheme. The current study extends the previous work by implementing the BASING J-difference sequence at 3T in the presence of high lipid concentrations in phantoms and in vivo.
A second issue, which much be addressed when PRESS is used to localize J-coupled metabolites, is anomalous modulation in different regions of the excitation volume due to the chemical shift difference between the coupled species (11–14,20). This spatial variation in lactate modulation can result in net signal loss when the contributions from different regions of the voxel are summed (11,12). This effect is especially severe when the bandwidth of the PRESS RF excitation pulses is on the order of the chemical shift difference of the coupled metabolites. At 3T, this is often the case because relatively low bandwidths are used to keep RF pulse amplitudes within safe limits. One solution is to prescribe a voxel larger than necessary and then eliminate the regions of anomalous modulation by applying saturation pulses to the border regions of the PRESS volume (12). In the present study, we sought to measure the chemical shift effect under our current scanner conditions and incorporate voxel overprescription with spatial saturation into the BASING editing sequence if necessary.
Sequence Implementation
Pulse sequence development was performed using the GE development environment ESE_E2M4 (version 11X). The 3T J-difference BASING sequence was constructed by selecting a BASING waveform from the GE waveform database and inserting it into a PRESS pulse sequence program. The waveform, a 30 ms asymmetrical pulse originally derived from the Shinnar-Le Roux algorithm (21,22) (Fig. 1), was chosen because the pass bandwidth was sufficient for detecting all metabolites of interest resonating upfield from water. The resulting sequence consisted of a traditional PRESS scheme with two additional 180° pulses flanked by bipolar gradients, embedded after each PRESS 180° pulse, that is, 90°–180° PRESS -(+)G-180°BASING-(−)G -180° PRESS -(+)G-180° BASING -(−)G, where G represents the crusher gradient. The time separation between the two BASING pulses is TE/2 where echo time (TE) = 144 ms. The sequence was modified so that the center of the BASING inversion band was alternated between +65 Hz relative to water (cycle 1) or −50 Hz relative to water (cycle 2) on consecutive passes. Therefore, the lactate methine quartet (4.1 ppm) was located in either the passband or the stopband in consecutive cycles and the lactate methyl peak was either inverted or upright at TE = 144ms.
Figure 1.
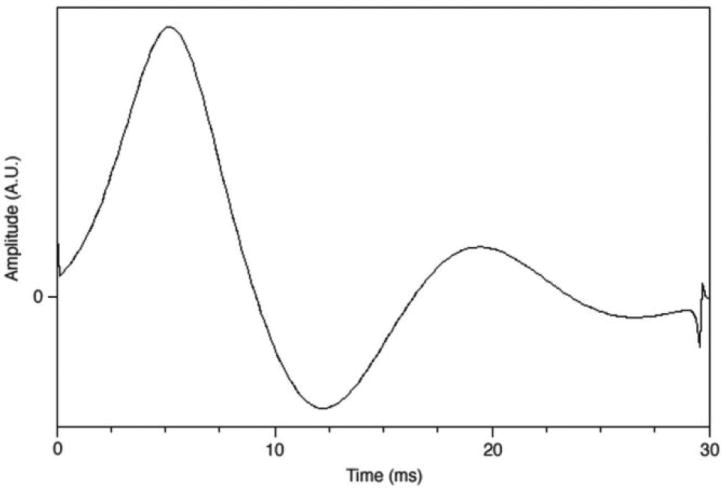
The waveform used for the two BASING (17) 180° pulses in the PRESS sequence. The pulse width was 30 ms. The waveform, obtained from General Electric Medical waveform library, was chosen because its pass-bandwidth was suf-ficient to include resonances from 4 ppm to 0.5 ppm.
J-Difference Pulse Sequence Testing: Phantom Studies
To demonstrate the J-difference editing technique, phantom and in vivo data were collected on a 3T Signa whole-body scanner (Magnex 3T/94 cm bore, GE software rev. ESE_E2M4, GE Healthcare, Milwaukee, WI). To experimentally verify the frequency-dependent inversion characteristics of the BASING pulses, a phantom containing brain metabolites in aqueous solution (Phantom 1) (MRS Head Sphere, GE Healthcare) was scanned in single-voxel mode with the water suppression turned off. The center of the BASING inversion band was stepped over a range of ± 200 Hz relative to water in 10 Hz increments and a proton spectrum was acquired at each step. The spectra were acquired at a TE of 144 ms and a repetition time (TR) of 1.5 s.
For testing the effectiveness of the J-difference lactate editing method, a phantom was constructed which contained lactate and lipid in adjacent compartments (Phantom 2). The phantom consisted of a 60-cc syringe filled with a 10 mM aqueous solution of lactate immersed in a 500-ml glass beaker filled with solidified high-grade animal lard. Phantom 2 was placed in the quadrature head coil, scout images were acquired, and a PRESS voxel was prescribed. The J-difference BASING sequence was applied with TE/TR = 144/1500 ms, respectively, 32 total scans (16 for each cycle, applied alternately), 5 kHz spectral width, 2048 data points, and scan duration =1 min 18 sec. The PRESS volume was 2.3 cm3. Water suppression was achieved through the use of three chemical-shift selective (CHESS) pulses (23).
Chemical Shift Effects and Voxel Overprescription: Phantom Study
To measure the potential loss in lactate signal on our scanner due to chemical shift-induced modulation differences in lactate across the PRESS voxel, Phantom 1 was studied with the J-difference sequence without overprescription of the PRESS volume and with over-prescription by factors of 1.3, 1.5, and 1.7. In the over-prescribed experiments, the desired 2 cm × 2 cm × 2 cm voxel size was increased by 10% in the A/P dimension, which was defined by a 90° slice selection pulse, and by 30%, 50%, or 70% in the R/L and S/I dimensions which were defined by 180° pulses. Overprescription was achieved by decreasing the amplitude of the voxel selection gradients. The bandwidth of the slice-selective 180° pulses was 1385 Hz. Six very selective saturation (VSS) bands (24) of 30 mm thickness were placed at the edges of the initially defined 2 cm × 2 cm × 2 cm voxel to saturate the overprescription regions. The same VSS bands were used with and without voxel overprescription. Scan parameters were: TE/TR = 144/1500 ms, 64 total scans (32 per cycle). PRESS volume = 2.0 cm3, 5 kHz spectral width and 2048 data points. CHESS water suppression was applied.
J-Difference Pulse Sequence Testing: Volunteer Study
A normal volunteer was studied to demonstrate the effectiveness of the J-difference BASING sequence in vivo. The male subject was placed prone on the scanner bed with his calf in the clinical knee coil. A blood pressure cuff was positioned just above the knee, superior to the calf volume of interest. After acquisition of localizer images, a voxel was prescribed which contained predominantly soleus and gastrocnemius lateralis muscle. The blood pressure cuff was inflated and the first J-difference scan immediately commenced. A voxel overprescription factor of 1.7 was used with VSS pulses at each edge of the voxel. Each scan was 1 min 40 s in duration and 11 scans were performed. Scan parameters were: TE/TR = 144/1500 ms, 32 total scans (16 for each cycle, applied alternately), PRESS volume = 8 cm3, 5 kHz spectral width and 2048 data points. CHESS water suppression was applied.
Data Processing
All spectra were processed using SAGE (GE Medical Systems, Milwaukee, WI). For the J-difference edited data, data frames were sorted as either cycle 1 or cycle 2 and all transients for a given cycle were summed. The resulting summed transient for each cycle was then apodized with 3 Hz Lorentzian line broadening and Fourier transformed. Finally, the resulting cycle 1 and cycle 2 spectra were either added, resulting in a spectrum reflecting noncoupled metabolites, or subtracted, resulting in a spectrum reflecting J-coupled metabo-lites. Peak area fitting was performed in the time domain using jMRUI (25). Because lipid was undetectable in the difference spectrum, the absolute lipid suppression factor was calculated as the lipid peak height in cycle (1 + 2) spectrum divided by the noise standard deviation.
RESULTS
The water spectra in Figure 2 demonstrate the experimental profile of the BASING pulse indicating an inversion/suppression bandwidth of approximately 100 Hz (0.78 ppm at 3T). With the BASING pulse offset from water by −50 Hz (cycle 2), the lactate methine peak (−85.8 Hz relative to water) would experience the suppression indicated at −35.8 Hz on this plot. Choline at 3.2 ppm (−198 Hz) relative to water would be located at −148 Hz on the plot which is well in the passband.
Figure 2.
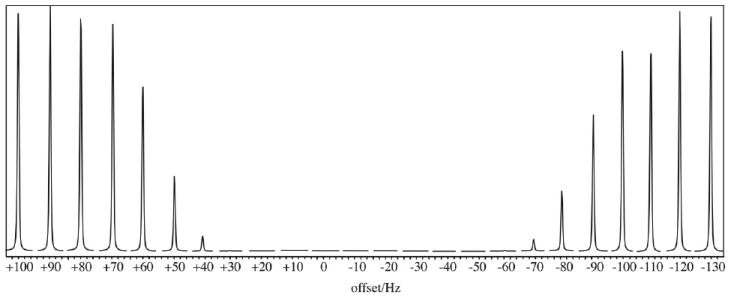
The experimental inversion/suppression profile for the 30-ms BASING waveform presented in Figure 1. The unsuppressed water resonance from a cubic volume in a large spherical phantom is shown as a function of transmitter offset.
Figure 3 demonstrates the effectiveness of the lactate editing within a location in Phantom 2 that contained both lactate and lipid (lard). The first spectrum in the left panel results from the addition of the odd- and even-numbered transients of the acquisition. The second spectrum in the right panel results from the subtraction of the odd from the even-numbered transients. The absolute lipid suppression factor was 4354. The origin of the inverted peaks downfield of the lactate CH3 peak is not known but could be due to coupled triglyc-eride protons in the lard.
Figure 3.
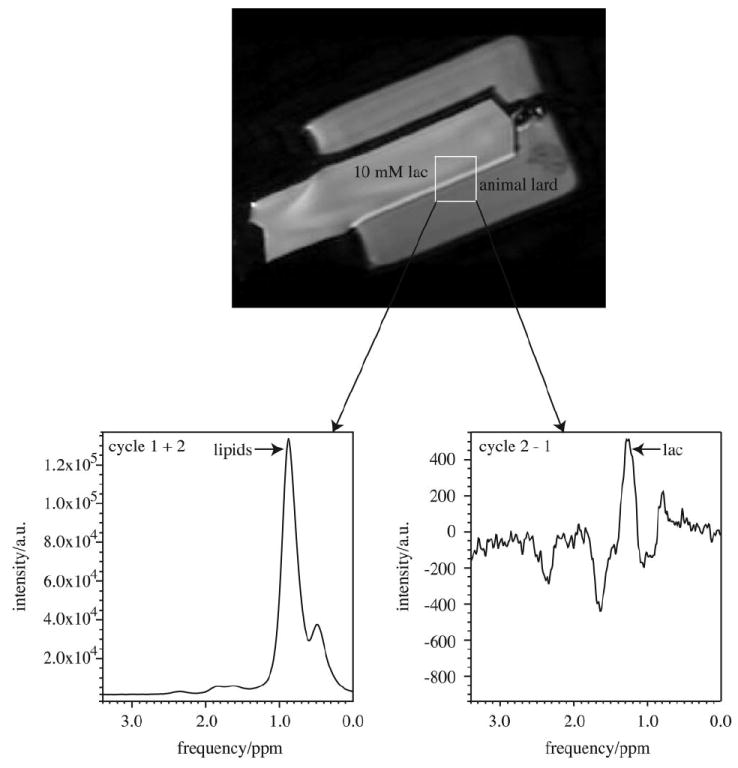
The spectra from a lactate/lipid phantom (Phantom 2) using the BASING-PRESS lactate editing pulse sequence. The 2.3 cm3 voxel encompassed both lactate and lipid-containing regions. The left panel is the spectrum obtained by adding each cycle. The right panel shows the results of subtracting the two cycles. TE/TR = 144/1500 ms, 768 transients (384 per cycle).
The recovery of lactate signal resulting from voxel overprescription using the single-voxel J-difference sequence is demonstrated in Figure 4. Lactate peak areas in normalized units for overprescription factors = 1, 1.3, 1.5, and 1.7 were 3.323 × 104, 5.261 × 104, 5.227 × 104, and 5.795 × 104, respectively. Thus, increases in lactate signal of 58%, 57%, and 74% were measured for overprescription factors of 1.3, 1.5, and 1.7, respectively.
Figure 4.
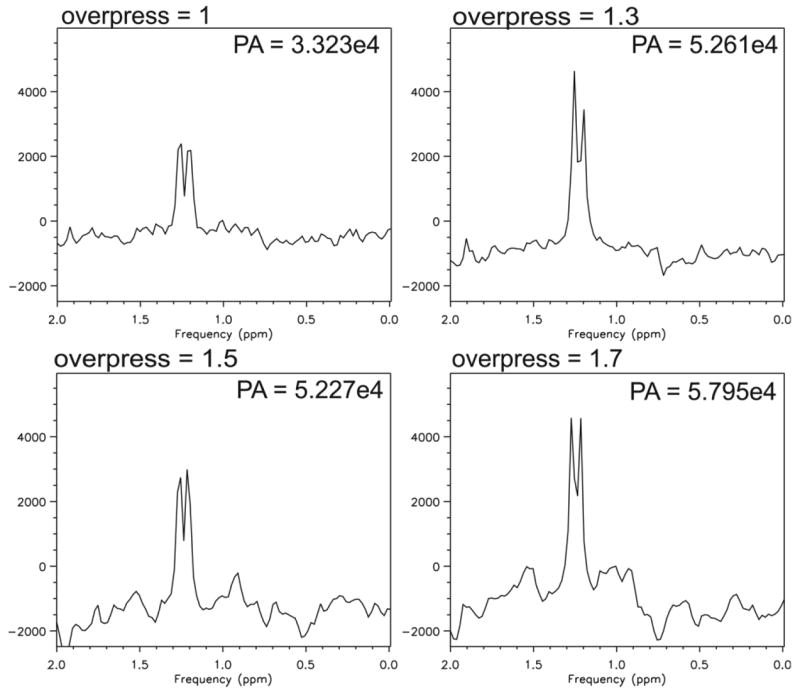
Lactate region of spectra from Phantom 1 containing physiological concentrations of brain metabolites in addition to 5 mM lactate at various voxel overprescrip-tion factors. A: Single voxel subtraction spectrum using the BASING-PRESS editing sequence with voxel overpre-scription factor = 1 (no over-prescription). B–D: Subtraction spectra resulting from over-prescribing the voxel size and adding very-selective saturation bands to the perimeter to define the final 2 cm × 2 cm × 2 cm volume. B: Overprescription factor = 1.3. C: Overprescription factor = 1.5. D: overprescription factor = 1.7. TE/TR = 144/1500 ms, 64 transients (32 per cycle). Data are plotted with identical vertical axis scales. Overpress, voxel overprescription factor; PA, peak area.
The effectiveness of the lactate editing sequence in the presence of lipids in vivo is demonstrated in Figure 5 which contains edited spectra from the time-series acquisition in transiently ischemic muscle. The baseline difference spectrum showed no lactate while spectra at 7 min 30 s and 9 min 10 s showed lactate. The reperfusion spectrum contained a small subtraction artifact probably due to motion when the tourniquet was released. Figure 6 shows the addition and subtraction spectra in muscle after 9 min of ischemia, demonstrating an absolute lipid suppression factor of 251.
Figure 5.
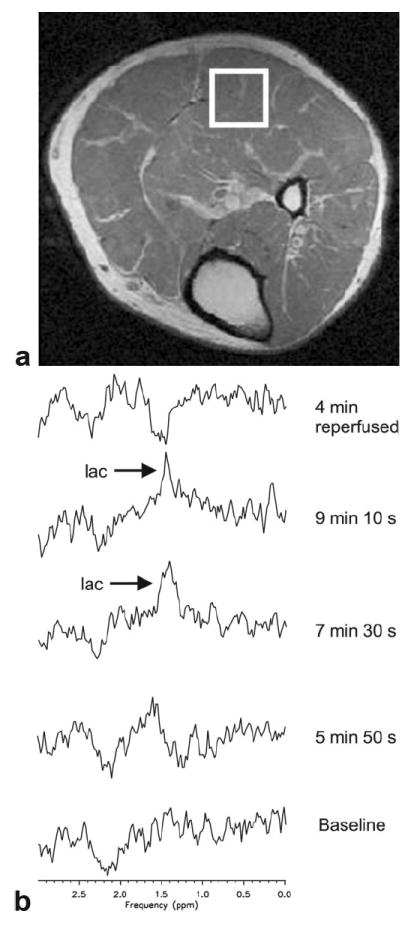
Data from volunteer study of calf muscle under ischemia. The volunteer was positioned prone. A tourniquet (blood pressure cuff) was applied to the leg just above the knee to restrict the subject’s blood flow. A: Prescribed voxel overlaid on corresponding MR image. B: Spectra from serial time series. Each spectrum was acquired in 1 min 40 s. Time points indicate the midpoint of the given acquisition. Spectra were acquired using the J-difference BASING-PRESS sequence with TE/TR = 144/1500 ms, 32 total acquisitions (16 per cycle), voxel overprescription factor = 1.7.
Figure 6.
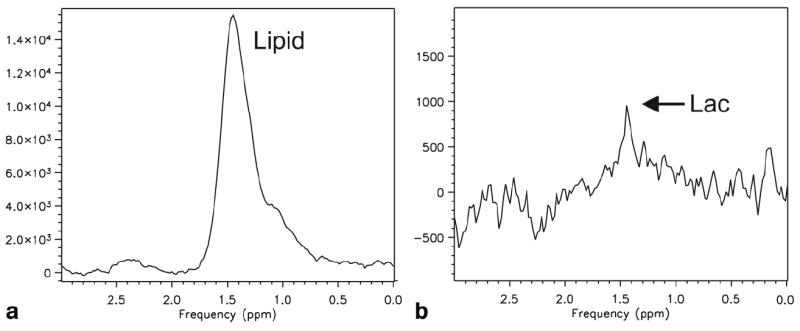
Muscle spectra acquired after 9 min 10 s of ischemia extracted from the time series data set. A: Noncoupled metabolites (cycle 1 + 2). B: Coupled metabolites (cycle 2-1). Acquisition parameters given in Figure 5.
DISCUSSION
The work described here demonstrates the effectiveness of the BASING J-difference editing scheme for detecting lactate in the presence of high lipid concentrations on the 3T MR scanner. Absolute lipid suppression factors of 4354 and 251 were observed in phantom and calf-muscle, respectively. The higher lipid concentration in the phantom gave rise to a higher lipid SNR and thus permitted a more precise measurement of the absolute suppression factor. Thus, this editing scheme can achieve an absolute lipid suppression factor of approximately 4000. Our results also demonstrate the successful incorporation of voxel overprescription and saturation of regions with anomalous coupling in this PRESS-based scheme. Edden et al (12) presented a formula which predicts the expected efficiency for detection of the lactate methyl peak using standard PRESS with no compensation for anomalous coupling. With our acquisition parameters, the predicted standard PRESS efficiency for the lactate methyl peak would be 0.48 at TE = 1/J, and one would expect that implementation of voxel overprescription and saturation would bring the measured efficiency closer to 1. Our implementation of voxel overprescription resulted in 58%, 57% and 74% increases in lactate signal for overprescription factors of 1.3, 1.5, and 1.7, respectively. This improved the lactate detection efficiency to 0.75–0.83, assuming that the standard PRESS efficiency for methyl detection truly was 0.48. It is possible that a larger overprescription factor could compensate for nonideal 180° pulses and further improve the lactate detection efficiency (12).
The pulse sequence generated here is suitable for lactate detection in tumors outside of the brain such as sarcomas and head and neck cancers where hypoxia has been shown to predict outcome. This sequence will be improved with the implementation of phase encoding which will permit the assessment of heterogeneity of tumor lactate.
The current work builds on the BASING J-difference lactate editing work performed at 1.5T (5,15,16) and addresses the complicating factor of coupling anomalies at 3T. BASING has been applied to proton spectroscopy at 3T with the goal of eliminating the anomalous lactate coupling problem in a single-shot scheme not designed to detect lactate in the presence of lipids (11). Voxel overprescription has been demonstrated by others (12) but in a sequence which targets the brain and does not edit lactate from lipid. Two studies have demonstrated lactate editing outside the brain in the presence of lipids at 3T (9,10). Each used a double-quantum editing technique in muscle, and the authors of the latter study noted that their add/subtract scheme addressed the chemical shift effect.
Hypoxia (26 –31) and lactate (3,6,7) have been linked to poor outcome in solid tumors. The technique presented here would permit the investigation of lactate as a noninvasive marker of tumor prognosis. It would also permit comparison of tumor lactate content with other vivo hypoxia markers such as 18F-misonidazole. Finally, because tumor cells in hypoxic environments are known to be resistant to therapy, identification of resistant regions of tumor could be used for planning localized therapies such as brachytherapy, focused ultrasound, and intensity modulated radiation to enhance tumor kill in resistant parts of the tumor.
In conclusion, the results outlined in this communication demonstrate the ability of the BASING J-difference technique to detect lactate in the presence of strong lipid signals outside the brain at 3T. This robust technique should permit noninvasive lactate measurements in human tumors to investigate its potential as a prognostic indicator.
Acknowledgments
Contract grant sponsor: the Byrne Foundation; Contract grant sponsor: National Cancer Institute/National Institutes of Health; Contract grant number: 1 R01 CA115895.
Footnotes
Published online in Wiley InterScience (www.interscience.wiley.com).
References
- 1.Warburg OH. In: The metabolism of tumors; investigations from the Kaiser Wilhelm institute for biology, Berlin-Dahlem. Dickens F, translator; Warburg OH, editor. London: Constable and Company, Ltd; 1930. [Google Scholar]
- 2.Adalsteinsson E, Spielman DM, Pauly JM, et al. Feasibility study of lactate imaging of head and neck tumors. NMR Biomed. 1998;11:360–369. doi: 10.1002/(sici)1099-1492(1998110)11:7<360::aid-nbm518>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 3.Brizel DM, Schroeder T, Scher RL, et al. Elevated tumor lactate concentrations predict for an increased risk of metastases in head-and-neck cancer. Int J Radiat Oncol Biol Phys. 2001;51:349–353. doi: 10.1016/s0360-3016(01)01630-3. [DOI] [PubMed] [Google Scholar]
- 4.Quennet V, Yaromina A, Zips D, et al. Tumor lactate content predicts for response to fractionated irradiation of human squamous cell carcinomas in nude mice. Radiother Oncol. 2006;81:130–135. doi: 10.1016/j.radonc.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 5.Star-Lack J, Spielman D, Adalsteinsson E, et al. In vivo lactate editing with simultaneous detection of choline, creatine, NAA, and lipid singlets at 1.5 T using PRESS excitation with applications to the study of brain and head and neck tumors. J Magn Reson. 1998;133:243–254. doi: 10.1006/jmre.1998.1458. [DOI] [PubMed] [Google Scholar]
- 6.Walenta S, Chau TV, Schroeder T, et al. Metabolic classification of human rectal adenocarcinomas: a novel guideline for clinical oncologists? J Cancer Res Clin Oncol. 2003;129:321–326. doi: 10.1007/s00432-003-0450-x. [DOI] [PubMed] [Google Scholar]
- 7.Walenta S, Wetterling M, Lehrke M, et al. High lactate levels predict likelihood of metastases, tumor recurrence, and restricted patient survival in human cervical cancers. Cancer Res. 2000;60:916–921. [PubMed] [Google Scholar]
- 8.Adalsteinsson E, Spielman DM, Wright GA, et al. Incorporating lactate/lipid discrimination into a spectroscopic imaging sequence. Magn Reson Med. 1993;30:124–130. doi: 10.1002/mrm.1910300119. [DOI] [PubMed] [Google Scholar]
- 9.Jouvensal L, Carlier PG, Bloch G. Practical implementation of single-voxel double-quantum editing on a whole-body NMR spectrometer: localized monitoring of lactate in the human leg during and after exercise. Magn Reson Med. 1996;36:487–490. doi: 10.1002/mrm.1910360325. [DOI] [PubMed] [Google Scholar]
- 10.Meyerspeer M, Kemp GJ, Mlynarik V, et al. Direct noninvasive quantification of lactate and high energy phosphates simultaneously in exercising human skeletal muscle by localized magnetic resonance spectroscopy. Magn Reson Med. 2007;57:654–660. doi: 10.1002/mrm.21188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kelley DA, Wald LL, Star-Lack JM. Lactate detection at 3T: compensating J coupling effects with BASING. J Magn Reson Imaging. 1999;9:732–737. doi: 10.1002/(sici)1522-2586(199905)9:5<732::aid-jmri17>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 12.Edden RA, Schar M, Hillis AE, et al. Optimized detection of lactate at high fields using inner volume saturation. Magn Reson Med. 2006;56:912–917. doi: 10.1002/mrm.21030. [DOI] [PubMed] [Google Scholar]
- 13.Slotboom J, Mehlkopf AF, Bovee WMMJ. The Effects of Frequency-Selective RF Pulses on J-Coupled Spin- Systems. J Magn Reson A. 1994;108:38–50. [Google Scholar]
- 14.Yablonskiy DA, Neil JJ, Raichle ME, et al. Homonuclear J coupling effects in volume localized NMR spectroscopy: pitfalls and solutions. Magn Reson Med. 1998;39:169–178. doi: 10.1002/mrm.1910390202. [DOI] [PubMed] [Google Scholar]
- 15.Star-Lack JM, Adalsteinsson E, Adam MF, et al. In vivo 1H MR spectroscopy of human head and neck lymph node metastasis and comparison with oxygen tension measurements. Am J Neuroradiol. 2000;21:183–193. [PMC free article] [PubMed] [Google Scholar]
- 16.Wang ZJ, Vigneron DB, Miller SP, et al. Brain metabolite levels assessed by lactate-edited MR spectroscopy in premature neonates with and without pentobarbital sedation. AJNR Am J Neuroradiol. 2008;29:798–801. doi: 10.3174/ajnr.A0912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Star-Lack J, Nelson SJ, Kurhanewicz J, et al. Improved water and lipid suppression for 3D PRESS CSI using RF band selective inversion with gradient dephasing (BASING) Magn Reson Med. 1997;38:311–321. doi: 10.1002/mrm.1910380222. [DOI] [PubMed] [Google Scholar]
- 18.Males R, Vigneron D, Star-Lack J, et al. Clinical application of BASING and spectral/spatial water and lipid suppression pulses for prostate cancer staging and localization by in vivo 3D 1H magnetic resonance spectroscopic imaging. Magn Reson Med. 2000;43:17–22. doi: 10.1002/(sici)1522-2594(200001)43:1<17::aid-mrm3>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 19.Bottomley P. Selective volume method for performing localized NMR spectroscopy. 4,480,228. USA patent. 1984
- 20.Lange T, Dydak U, Roberts TP, et al. Pitfalls in lactate measurements at 3T. AJR Am J Neuroradiol. 2006;27:895–901. [PMC free article] [PubMed] [Google Scholar]
- 21.Shinnar M, Bolinger L, Leigh JS. The use of finite impulse response filters in pulse design. Magn Reson Med. 1989;12:81–87. doi: 10.1002/mrm.1910120110. [DOI] [PubMed] [Google Scholar]
- 22.Pauly J, Le Roux P, Nishimura D. Parameter Relations for the Shinnar-Le Roux selective excitation pulse design algorithm. IEEE Trans Med Imaging. 1991;10:53–65. doi: 10.1109/42.75611. [DOI] [PubMed] [Google Scholar]
- 23.Haase A, Frahm J, Hanicke W, et al. 1H NMR Chemical Shift Selective (CHESS) imaging. Phys Med Biol. 1985;30:341–344. doi: 10.1088/0031-9155/30/4/008. [DOI] [PubMed] [Google Scholar]
- 24.Tran T, Vigneron D, Sailasuta N, et al. Very selective suppression pulses for clinical MRSI studies of brain and prostate cancer. Magn Reson Med. 2000;43:23–33. doi: 10.1002/(sici)1522-2594(200001)43:1<23::aid-mrm4>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 25.Naressi A, Couturier C, Castang I, et al. Java-based graphical user interface for MRUI, a software package for quantitation of in vivo/medical magnetic resonance spectroscopy signals. Comput Biol Med. 2001;31:269–286. doi: 10.1016/s0010-4825(01)00006-3. [DOI] [PubMed] [Google Scholar]
- 26.Brizel DM, Sibley GS, Prosnitz LR, et al. Tumor hypoxia adversely affects the prognosis of carcinoma of the head and neck. Int J Radiat Oncol Biol Phys. 1997;38:285–289. doi: 10.1016/s0360-3016(97)00101-6. [DOI] [PubMed] [Google Scholar]
- 27.Ghafar MA, Anastasiadis AG, Chen MW, et al. Acute hypoxia increases the aggressive characteristics and survival properties of prostate cancer cells. Prostate. 2003;54:58–67. doi: 10.1002/pros.10162. [DOI] [PubMed] [Google Scholar]
- 28.Haensgen G, Krause U, Becker A, et al. Tumor hypoxia, p53, and prognosis in cervical cancers. Int J Radiat Oncol Biol Phys. 2001;50:865–872. doi: 10.1016/s0360-3016(01)01523-1. [DOI] [PubMed] [Google Scholar]
- 29.Hockel M, Schlenger K, Aral B, et al. Association between tumor hypoxia and malignant progression in advanced cancer of the uterine cervix. Cancer Res. 1996;56:4509–4515. [PubMed] [Google Scholar]
- 30.Sundfor K, Lyng H, Rofstad EK. Tumour hypoxia and vascular density as predictors of metastasis in squamous cell carcinoma of the uterine cervix. Br J Cancer. 1998;78:822–827. doi: 10.1038/bjc.1998.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nordsmark M, Alsner J, Keller J, et al. Hypoxia in human soft tissue sarcomas: adverse impact on survival and no association with p53 mutations. Br J Cancer. 2001;84:1070–1075. doi: 10.1054/bjoc.2001.1728. [DOI] [PMC free article] [PubMed] [Google Scholar]


