PREFACE
Proteolysis in cellular membranes to liberate effector domains from their transmembrane anchors is a well-studied regulatory mechanism in animal biology and disease. By contrast, the function of intramembrane proteases in unicellular organisms has received little attention. Recent progress has now established that intramembrane proteases execute pivotal roles in a range of pathogens, from regulating Mycobacterium tuberculosis envelope composition, cholera toxin production, bacterial adherence and conjugation, to malaria parasite invasion, fungal virulence, immune evasion by parasitic amoebae and hepatitis C virus assembly. These advances raise the exciting possibility that intramembrane proteases may serve as targets for combating a wide range of infectious diseases. I focus on summarizing the advances, evaluating the limitations and highlighting the promise of this newly emerging field.
Keywords: cell signalling, regulated intramembrane proteolysis, rhomboid, site-2 protease, signal peptide peptidase, presenilin, invasion, malaria, hepatitis, cholera, tuberculosis
INTRODUCTION
Pathogens have evolved mechanisms to adapt quickly to the unreceptive environment of the host and establish a flourishing infection. Many of the events to which pathogens must respond occur at cellular membranes as sites of first contact. Work over the past decade has identified a novel class of membrane proteases that function to liberate effector domains from the membrane by cleaving transmembrane segments1–3. These intramembrane proteases are unusual polytopic enzymes; their active sites are assembled from residues on membrane-spanning segments, which embeds their water-dependent catalytic apparatus beneath the surface of the membrane.
Intramembrane proteases are among the most conserved of all membrane proteins known, with members in all kingdoms of life4–6. Despite being widespread in unicellular organisms, without exception every intramembrane protease family was initially discovered through the study of metazoan biology or disease. Early examples of intramembrane proteolysis in unicellular organisms were relatively rare7–9, but this field has gained momentum, especially in the area of pathogens, and is coalescing into a new and distinct field. It is now established that intramembrane proteolysis is a fundamental biochemical mechanism used by a wide variety of pathogens to coordinate their infective cycles.
In this review, I will focus on the roles that intramembrane proteases have been discovered to play in pathogenic microorganisms, starting with bacterial pathogens, moving to pathogenic eukaryotes including protozoa and fungi, and closing with viruses. In comparing these examples, the challenge is to identify emerging common principles, and to expose promising areas for future investigation.
THE PROTEASE CAST
Molecular dissection of a diverse array of cellular processes over the past ∼12 years has identified dozens of intramembrane proteases, all of which can be grouped into three families based both on sequence similarity as well as shared properties 1–3 (Figure 1).
Figure 1. The three families of intramembrane proteases.
Graphical representation of the general topology of each intramembrane protease (cytoplasm is down). Active site residues are shown in yellow and the zinc ion of S2P in green. Generic substrates are diagramed in black, with a lightning bolt depicting the proteolytic event, arrow for direction of domain release, and activated effector as a star in black and yellow. The core six transmembrane segments of rhomboid are in blue, with an additional transmembrane segment and cytosolic domain that are present in many homologues depicted in red and brown, respectively. The conserved three transmembrane core of S2P is depicted in purple, with variable transmembrane segments and extramembrane domains (frequently a PDZ domain) of some S2P members shown in orange. The domains present in E. coli, human, and Methanocladococcus jannaschii S2Ps are delineated below the topology diagram. SPP is shown in green (note that presenilins also have nine transmembrane segments but the opposite membrane orientation). In the bottom left panel, the crystal structures of membrane core fragments of prokaryotic rhomboid and S2P are shown (colors and orientation correspond to topology diagram). Bottom right illustrates the conservation of each intramembrane protease in different forms of life. Presenilins are conserved only in multicellular organisms (not shown).
The first intramembrane protease to be discovered was site-2 protease (S2P), the enzyme responsible for catalyzing the second cleavage that releases the transmembrane SREBP transcription factor for cholesterol biosynthesis in mammalian cells10. S2P also functions in the unfolded protein response11, which is paralleled by the Escherichia coli RseP/YaeL homologue in the EXTRACYTOPLASMIC FUNCTION12, 13 stress response that also responds to unfolded proteins (see Figure 2). S2P homologues are conserved in all forms of life, from Archaea to humans, although a few organisms lack recognizable members6. S2P enzymes are polytopic membrane proteins consisting of a conserved three transmembrane core with a well-defined HExxH METALLOPROTEASE signature in one transmembrane segment, with the third residue required for zinc coordination (along with the two histidines) supplied by an aspartate on a distal segment6 (Figure 1). Individual S2P members elaborate this core structure through the addition of transmembrane segments preceding and/or following the core, as well as insertion of extra-membranous domains.
Figure 2. S2P circuits and bacterial virulence: variations on a theme.

The E. coli extracytoplasmic function (top left panel) serves as a paradigm for the S2P circuit in bacterial pathogenesis. An input signal (yellow box and arrow), in this case envelope stress, activates the site-1 protease DegS and relief of inhibition by RseB, allowing cleavage of the anti-sigmaE factor RseA at site-1 (top bolt). Intramembrane proteolysis by RseP ensues at site-2 (bottom bolt), resulting in cytoplasmic (bottom) release, degradation of RseA, and liberation of σE to activate the output response, in this case transcription of stress response genes (black arrow). The other S2P circuits involved in regulating virulence of various bacterial pathogens (detailed in the text) are depicted in the same way for the sake of comparison. Components that have been identified are named, while missing components are labeled with a question mark.
Intramembrane ASPARTYL PROTEASES were discovered through the analysis of Alzheimer’s Disease etiology: presenilin protein within the γ-secretase complex catalyzes intramembrane proteolysis to generate the neurotoxic Aβ42 peptide. While γ-secretase is generally absent from unicellular organisms and will not be discussed further, intramembrane aspartyl proteases are widely represented by signal peptide peptidase (SPP), a presenilin-type protein that uses two intramembrane aspartates to catalyze proteolysis14, 15 (Figure 1). Detection of signal peptide cleavage following their removal from proteins by signal peptidase led to the discovery of SPP in human cells. Some of the released fragments act as bioactive peptides with a variety of post-targeting functions16. Unlike γ-secretase, SPPs do not require other protein cofactors for activity17, 18, and are widely conserved as multimember families in multicellular organisms, but they are absent from bacteria14, 19.
Rhomboid proteins are intramembrane SERINE PROTEASES that were identified through the genetic dissection of insect embryogenesis. Drosophila rhomboid initiates cell signalling by cleaving the transmembrane EGF precursor, Spitz, releasing it as an active signal from the membrane20 (also see Figure 3). Rhomboid is conserved in all kingdoms of life, with members sharing a conserved core structure of 6 transmembrane segments that contain the catalytic serine-histidine pair, and a membrane-inserted loop4, 5, 21–24 (Figure 1). Many homologues contain a variable N-terminal domain that protrudes into the cytosol, and/or a seventh transmembrane segment.
Figure 3. Dis-similarity in signalling by rhomboid proteases in Drosophila and a bacterium.

In Drosophila cells (left), the EGF precursor Spitz is transported by Star from the ER to the Golgi apparatus, where intramembrane proteolysis by Rhomboid-1 ensures. The processed Spitz is then secreted from the cell, ready to activate EGF receptor signalling in neighboring cells. In this context, rhomboid directly processes the signalling molecule to activate it from a latent form. Conversely in a bacterium (right), the signal itself is unknown and does not require rhomboid processing. Instead, the rhomboid protease AarA cleaves the TatA component of the twin arginine transporter, presumably to facilitate export of the unknown quorum-sensing signal. Notably, the mechanisms are fundamentally different because AarA does not act on the signal itself. Removal of the N terminal extension of TatA by AarA is shown to be required for TatA oligomerzation to form the export pore (along with other subunits), although this is only one model and other possibilities exist.
In addition to the catalytic mechanism, intramembrane proteases differ with respect to three main properties that allow them to fulfill distinct roles in the cell. First, both S2P and SPP are dependent on a prior, site-1 cleavage that sheds the substrate ectodomain; this cleavage regulates processing of the substrate since intramembrane cleavage follows automatically1, 2. Conversely, rhomboid proteases cleave full-length proteins and in no circumstance require a prior cleavage3, 20. Secondly, the active site of rhomboid enzymes is situated closer to the extracellular face of the membrane, and they generally function to release effector domains to the cell exterior25 (Figure 1). Conversely, both S2P and SPP release proteins into the cytosol, and their active sites are accordingly situated closer to the cytosolic face of the membrane. Lastly, rhomboid and γ-secretase cleave transmembrane segments of type I orientation (N-termini outside the cell), while SPP and S2P cleave transmembrane segments of the opposite, type II orientation (C terminus out).
Recently there have been major advances in understanding the biochemistry of these enzymes, including development of pure enzyme reconstitution assays for all three protease classes17, 18, 26–28, and several crystal structures of both prokaryotic rhomboid21–24 and S2P homologues29 (Figure 1). This has also resulted in defining new steps in the reaction, including insights into how substrates enter the active site via lateral gating mechanisms30–33. No structure has yet been solved for any aspartyl intramembrane protease, although low-resolution cryo-electron microscopy and particle reconstructions of γ-secretase provide an initial glimpse34. These biochemical advances are beyond the scope of the current discussion, but have been the focus of several recent articles33, 35.
ROLES IN PROKARYOTIC PHYSIOLOGY
In addition to central roles in the development and disease of animals, our understanding of intramembrane proteolysis in microbial cell biology is improving (summarized in Table 1). Investigations over the past few years have revealed direct roles for intramembrane proteolysis in bacterial sporulation7, maintenance of cell polarity36, metabolite transport37, 38, stress responses12, 13, 39, 40 and cell division41. It is also noteworthy that all three classes of intramembrane proteases are conserved in Archaea, and some of these homologues have already served as useful models for the study of intramembrane protease biochemistry and structure29. However, while Archaea are widespread ecologically, the biological roles of archaeal intramembrane proteases have not yet been explored in any context, and this topic deserves future investigation. For the sake of clarity, the discussion here will be limited to direct roles of intramembrane proteases in pathogenic events.
TABLE 1.
Currently-defined roles of intramembrane proteases in microorganisms.
| Organism | Protease (Type) | Substrate | Process | Role | Ref |
|---|---|---|---|---|---|
| Bacillus subtilis | SpoIVFB (S2P) | pro-σK | sporulation | cleaves pro-σK to activate gene expression in the mother cell | 7 |
| Bacillus subtilis | YluC/RasP (S2P) | RsiW (anti-sigma) | extracytoplasmic stress response | releases σW to activate stress response | 39 |
| Bacillus subtilis | YluC/RasP (S2P) | FtsL | cell division | negatively regulates cell division by degrading FtsL | 41 |
| Bacillus subtilis | YqgP (Rho) | ??? | cell division? sugar metabolism? | yqgP mutant has filamentous phenotype | 37 |
| Bordetella bronchiseptica | HurP (S2P) | HurR (anti-sigma) | iron uptake | cleaves HurR to activate heme receptor bhuR expression | 38 |
| Caulobacter | MmpA (S2P) | PodJS | cell polarity | preserves cell asymmetry by cleaving PodJS for degradation | 36 |
| Escherichia coli | RseP (S2P) | RseA (anti-sigma) | extracytoplasmic stress response | releases σE to activate stress response genes | 12, 13 |
| Escherichia coli | GlpG (ROM) | ??? | ??? | glpG mutant shows a slight antibiotic resistance | 68 |
| Streptococcus uberis | Eep (S2P) | MtuA | leader peptidase cleavage | compensates for absence of lipoprotein signal peptidase Lsp | 40 |
| Mycobacterium tuberculosis | Rv2869c (S2P) | ??? | virulence | regulates lipid metabolism genes, envelope composition | 46 |
| Pseudomonas aeruginosa | MucP (S2P) | MucA (anti-sigma) | alginate production | regulates alginate biosynthesis genes | 48 |
| Vibrio cholerae | YaeL (S2P) | TcpP | toxin expression | Negatively regulates toxin expression, destabilizes TcpP | 52 |
| Enterococcus faecalis | Eep (S2P) | Cad, CcfA, iAD1, iCF10 | pheromone-induced conjugation, cell aggregation/ virulence | produces multiple octapeptide pheromones and inhibitors by processing latent signal peptides | 9, 57, 58, 62 |
| Providencia stuartii | AarA (ROM) | TatA | quorum sensing | activates TatA, results in density-dependent gene repression and activation | 8, 65, 67 |
| Toxoplasma gondii | TgROM1 | ??? | intracellular growth | knockdown parasites grow less efficiently than wildtype | 72, 88, 89 |
| Toxoplasma gondii | TgROM5 | AMA1 and MIC adhesins | invasion | breaks down moving junction | 72, 74, 75 |
| Plasmodium falciparum or P. berghei | PfROM1/ PbROM1 | AMA1, other adhesins? | invasion, growth, oocyst differentiation | cleaves some adhesins, other substrates unknown | 77, 81, 90 |
| Plasmodium falciparum | PfROM4 | EBLs, RBLs, and TRAP adhesins | invasion | cleaves many families of adhesins known to be essential for invasion | 77, 80 |
| Plasmodium falciparum | PfSPP | ??? | invasion | binds to band 3; protease activity required for invasion and growth of blood stage | 85, 87 |
| Cryptosporidium parvum | CpROM4 | ??? | ??? | protein present in sporozoites | 83 |
| Eimeria tenella | EtROM | ??? | ??? | protein present in sporozoites | 84 |
| Entamoeba histolytica | EhROM1 | Gal/GalNAc lectin heavy subunit | phagocytosis, receptor capping | localizes to phagosomes, and cap neck during immune evasion; exact role(s) in these processes are unknown | 91 |
| Cryptococcus neoformans | Stp1 (S2P) | Sre1 | hypoxia adaptation, lipid metabolism | liberates SREBP homologue Sre1 from the membrane to activate gene expression | 95, 96 |
| Saccharomyces cerevisiae | Pcp1/Rbd1 (ROM) | Mgm1, Ccp1 | mitochondrial fusion, protein import | removes Ccp1 targeting sequence, generates short form of Mgm1 GTPase | 112 |
| Hepatitis C Virus (HCV) | SPP (host) | Core | polyprotein processing | processing facilitates core trafficking to lipid droplets and viral assembly | 99–104 |
| GB Virus B (GBV-B) | SPP (host) | Core | polyprotein processing | processing facilitates core trafficking to lipid droplets and viral assembly | 106 |
| Classical Swine Fever Virus (CSFV) | SPP (host) | Core | polyprotein processing | processing required for viral production | 105 |
| Cytomegalovirus (CMV) | SPP (host) | ??? | immune evasion | MHC class I heavy chain dislocation from ER; unknown if protease activity required | 107 |
NOTES: Only intramembrane proteases that have been studied are listed; most organisms also encode additional intramembrane proteases that have not been analyzed.
Abbreviations: S2P, site-2 protease; ROM, rhomboid; SPP, signal peptide peptidase; AMA1, apical membrane antigen 1; MIC, microneme; EBL, erythrocyte binding-like; RBL, reticulocyte binding-like, TRAP, thrombospondin-related anonymous protein, ER, endoplasmic reticulum Color code: orange, bacterial physiology; blue, bacterial pathogenesis; green, protozoan parasites; red, fungi; yellow, viruses
ROLES IN BACTERIAL PATHOGENESIS
Bacteria have rhomboid proteases that cleave membrane proteins of type I orientation, and S2Ps that cleave type II membrane proteins, but notably lack intramembrane aspartyl proteases such as SPP. Confusingly, there is an activity that is responsible for cleaving bacterial signal peptides, which was historically also named signal peptide peptidase A (also abbreviated SPP)42. However, this bacterial SPP, whose structure has recently been solved, is a serine protease with a single transmembrane segment and an active site that lies in the periplasm43: it bears no sequence or evolutionary similarity to the eukaryotic aspartyl intramembrane SPP.
S2P Circuits and Virulence
The functions of bacterial intramembrane proteases are currently best understood for S2Ps, which, as a rule, regulate membrane-tethered transcription factors: S2Ps cleave transmembrane ANTI-SIGMA FACTORS, or transcription factors that are directly tethered to the membrane. The role of RseP/YaeL in the E. coli extracytoplasmic stress response serves as a well-characterized but non-pathogenic paradigm (Figure 2)12, 13, 44, although the recent implication of this pathway in virulence of some pathogens implies that the pathogenic roles of S2Ps are likely to grow in the near future. Currently S2Ps are known to directly regulate four distinct pathways that contribute to virulence of several deadly bacterial pathogens.
Mycobacterium tuberculosis S2P and lipid metabolism
M. tuberculosis is an obligate human pathogen with complex stages of pathogenicity. While it lacks an outer membrane, virulence is known to depend on the unusual waxy cell wall comprised of a hydrophobic envelope45. Genome sequencing revealed a multitude of lipid biosynthetic genes, but the mechanism by which they are regulated is not well understood. The precedent of S2P regulating membrane composition in animal cells prompted a similar analysis of the S2P homologue in M. tuberculosis. In a striking parallel, the S2P knockout bacteria exhibited altered colony morphology consistent with altered lipid composition of the envelope, a defect that relied on S2P proteolytic activity since a catalytic site mutant failed to rescue the morphology46. Although a biochemical analysis failed to reveal major differences in envelope lipids of the S2P knockout strain, switch of S2P knockout cultures to detergent-free media resulted in altered changes in lipid metabolism, suggesting that it is not lipid synthesis, but responses to changing conditions that are defective in S2P mutants. Microarray analysis revealed complex and multi-faceted changes in lipid biosynthetic gene expression, including those for mycolic acid production, an extractable envelope lipid known to contribute to M. tuberculosis virulence.
Although the exact basis of the dysregulation awaits further investigation, it is important to note that the S2P knockout bacteria exhibited 100-fold lower acute replication, and 10,000-fold lower persistence in an aerosol infection model of mouse tuberculosis. These defects could be rescued by the S2P transgene, but to date the involvement of a site-1 protease or even the identity of the S2P substrate(s) remain unknown (Figure 2). Nevertheless this work underscores the possibility that S2P may be a viable therapeutic target for tuberculosis, by illustrating that removing S2P activity has dramatic and direct, although not well understood, effects on several phases of the disease.
Pseudomonas aeruginosa S2P and exopolysaccharides
The S2P circuit has been characterized in greater detail in Pseudomonas aeruginosa, a Gram negative opportunistic pathogen that colonizes the lungs of cystic fibrosis (CF) patients. Alginate is an EXOPOLYSACCHARIDE that is produced by P. aeruginosa and confers a mucoid morphology on bacterial colonies grown in culture, and in the lungs of CF patients renders the bacteria more adherent, less sensitive to antibiotics and host defenses, and contributes to biofilm formation47. During colonization, conversion of P. aeruginosa strains from non-mucoid to a ‘slimy’ mucoid appearance correlates with a poor prognosis in CF patients.
The precise molecular pathway regulating alginate biosynthesis was recently revealed to be regulated by an intramembrane proteolysis circuit similar to the E. coli extracytoplasmic stress response48 (Figure 2). Pathway activation is subject to multiple environmental inputs and intersects with other pathways, but the core of the circuit is sequential site-1 and site-2 protease mediated cleavage of the anti-sigma factor MucA (RseA homologue)48. The S1P is AlgW and the S2P is MucP, which is ultimately responsible for cleaving MucA, thereby releasing the σE homologue AlgU from the inner membrane to activate the alginate biosynthesis operon. However, unlike RseP/YaeL in E. coli, MucP is not an essential gene.
Clinically, MucA is a hot spot for mutations that confer a stable mucoid phenotype to CF patient isolates; as high as 90% of mucoid isolates can carry MucA mutations, the majority of which are nonsense or frameshift mutations that render MucA unable to sequester AlgU49. Cell wall-targeting antibiotics also activate the pathway, resulting in alginate production and expression of other factors including cell wall repair enzymes50. Although this may be a natural defense mechanism against antibiotic-secreting microbes P. aeruginosa encounters in its native soil environment, this response also has implications for treating P. aeruginosa infections. It is presently unclear if biosynthesis of exopolysaccharides is generally regulated by intramembrane proteolysis, as alginates themselves are known to be synthesized by only two bacterial genera, Pseudomonas and Azotobacter.
Vibrio cholerae S2P and toxin production
It is noteworthy that the opposite situation, although less common, also exists: some transmembrane transcription factors are competent for transcriptional activation while integral to the membrane, but proteolytic release renders them inactive. V. cholerae is a devastating diarrhoeal pathogen that has killed millions of people in eight documented pandemics throughout history. Virulence gene expression must be carefully controlled by such a pathogen that also exhibits an aquatic existence outside the human host51. Intramembrane proteolysis has been implicated in limiting virulence gene expression52. The master virulence gene activator ToxT directly activates transcription of the canonical virulence factors cholera toxin and the toxin co-regulated pilus51. A myriad of inputs including environmental and quorum-sensing signals converge on toxT transcription ultimately through two transcription factors, ToxR and TcpP. TcpP appears to be the dominant factor, and is rendered unstable under non-virulence or unfavorable conditions.
A genetic screen for the protease responsible for TcpP instability implicated the S2P protein YaeL52 (Figure 2). The stabilized TcpP in a YaeL mutant was smaller, consistent with site-1 cleavage, but despite testing four candidates the identity of the site-1 protease remains elusive. Moreover, the signals that induce degradation of TcpP by activating site-1 cleavage also remain unclear. Like with M. tuberculosis, the involvement of an S2P in virulence is recent and the precise molecular pathway has yet to be elucidated, although in this case blocking S2P would be expected to induce virulence genes.
Enterococcus faecalis S2P and pheromones
A rare exception to S2P regulating transcription factor activity is exemplified in mating pheromone production by E. faecalis. This Gram positive bacterium is usually a commensal of human intestinal flora, but has also emerged as an important source of NOSOCOMIAL infections. Moreover, E. faecalis poses a serious threat beyond its own infective capabilities, because antibiotic resistance is endemic to E. faecalis, and vancomycin resistance has now become common in Enterococci through horizontal gene transfer53. This has made understanding Enterococcal plasmid transfer an important area of research54, 55.
E. faecalis plasmid transfer is regulated by intricate cell-to-cell signalling via pheromones that are produced by plasmid-free, recipient cells54, 55. The pheromones are hepta or octapeptides that are produced by intramembrane proteolysis of latent signal peptides from chromosomally encoded lipoproteins (which are themselves not involved in the process)56–58(Figure 2). These pheromones enter the plasmid-bearing, donor cells and activate transcription of CONJUGATION genes from the plasmid, including an aggregation substance that aids cell clumping, to promote plasmid transfer. Intriguingly, once transferred, the plasmid encodes an inhibiting pheromone precursor of 21 to 23 residues that still relies on intramembrane processing to the active hepta or octapeptide inhibitor to prevent self-induction. A search for the processing enzyme isolated a site-2 protease termed Eep (enhanced expression of pheromone)9. Strikingly, eep null cells were found to reduce transfer of multiple (but not all) plasmids that use different pheromone systems, indicating that the processing enzyme itself is a common link between multiple pheromone systems.
But recently it has become clear that this form of intramembrane proteolysis also plays a direct role in virulence of this nocosomial pathogen59–61. Induction of conjugal transfer genes was discovered to be triggered upon exposure to human plasma, and the resulting production of the Asc10 aggregation substance increased resistance to phagocyte killing, as well as bacterial adherence and invasion of epithelial cells59–61. Importantly, Asc10 biosynthesis required pheromone production by the bacterial cells, while serum albumin was found to bind the inhibiting pheromone62. Thus, differential sequestration of the inhibitor by albumin alters the pheromone:inhibitor balance, resulting in self-induction and production of the Asc10 aggregation substance. An important implication of these observations is that the autocrine signalling loop that triggers virulence factor production in plasma requires production of the pheromone by the intramembrane protease Eep (Figure 2).
Rhomboid and quorum-sensing
The role of Eep in cell-to-cell communication is exceptional, being the only described case in any organism of an S2P that cleaves proteins to release polypeptides to the cell exterior. This function is normally ascribed to rhomboid proteases, whose active site lies close to the external membrane face. Rhomboid was implicated in cleaving growth factor signals in Drosophila development3, and subsequently in QUORUM SENSING by Providencia stuartii8, a Gram negative pathogen that causes urinary tract infections. Two separate genetic screens isolated the rhomboid homologue AarA (aminoglycoside acetyltransferase regulator A) of P. stuartii as a factor involved in density-dependent repression of an aminoglycoside acetyltransferase, and activation of other, uncharacterized genes8, 63. Subsequent study established that proteolytic mechanism and specificity were conserved between Drosophila rhomboid and AarA64, and remarkably, interchanging their genes could rescue fly and bacterial signalling65. Moreover, although its identity remained unknown, the bacterial signal had biochemical properties consistent with being a small peptide66. Taken together, these observations dramatically suggested that activation of cell-to-cell signalling through rhomboid-mediated processing of precursor proteins might be the first signalling circuit known to be conserved between animals and bacteria65.
Contrary to expectation, this turned out to be both a rare and superficial similarity (Figure 3). In an attempt to clone additional rhomboid homologues, the aarA mutant of P. stuartii was found to be rescued by the TatA subunit of the twin arginine translocase from several other bacteria, but mysteriously not from P. stuartii itself67. A sequence comparison revealed that the Providencia TatA contains a 7 residue extension on its N terminus. This observation immediately suggested that this extension is not the signal, as occurs in lipoprotein processing by Eep, but rather that this extension acted negatively to prevent TatA activity prior to its proteolytic removal. The role of AarA was clarified in an elegant genetic experiment: production of the signal for quorum-sensing, as well as all known PLEIOTROPHIC phenotypes of aarA, were restored in an aarA mutant expressing an experimentally truncated TatA lacking the 7 residue extension67. Moreover AarA was shown to cleave TatA. Understanding this unusual quorum-sensing system now awaits the identification of the signaling molecule, but it is clear that the only apparent role of AarA in P. stuartii is in activating TatA, presumably for signal export, and has no further role in production of the signal itself.
Apart from P. stuartii, our knowledge of rhomboid function in bacterial physiology or pathogenesis remains absent. The only known bacterial substrate for rhomboid is TatA, yet only a few bacteria, perhaps less than 1%, encode the rhomboid-dependent form of TatA. This implies that the role of AarA is a rare adaptation rather than a generally applicable paradigm. To date, the genetic deletion of the E. coli rhomboid GlpG has no discernible phenotype even when interrogated using a BIOLOG approach68, while a filamentous phenotype has been reported in a B. subtilis rhomboid knockout, although its molecular basis is unclear37.
It is also at present unclear whether bacterial rhomboid enzymes might constitute potential therapeutic targets. While the Tat system itself has documented roles in pathogenesis in some bacteria and it is possible that AarA could thus turn out to be a target in P. stuartii, the rarity of extended TatA forms requiring rhomboid processing in other pathogens precludes widespread utility69. Moreover, the few available examples of rhomboid-deficient bacteria display a modest increase in antibiotic resistance63, 68, suggesting that inhibiting rhomboid might not be a good therapeutic strategy. Conversely, recent identification of rhomboid protease mutants with increased activity might suggest that if this increase can be mimicked pharmacologically, it could lead to a new strategy for lowering antibiotic resistance by stimulating rhomboid activity30, 32.
ROLES IN PROTOZOAN PARASITES
Eukaryotic protozoan parasites are among the most successful and devastating pathogens today. Important non-signalling roles for intramembrane proteases in these pathogens have recently been identified that offer promising targets for therapeutic intervention.
Apicomplexan Parasites
Protozoan parasites of the phylum Apicomplexa include Plasmodium, which causes malaria, afflicting 10–40% of the world’s population and claiming 2 million lives annually (mainly children). Toxoplasma gondii and Cryptosporidium parvum are major pathogens of immuno-compromised individuals including AIDS patients. The problems that apicomplexans cause are not limited to human health: Eimeria tenella infects poultry and causes annual losses in revenue totaling nearly a billion dollars.
Without exception apicomplexans are obligate intracellular pathogens, and this dependence on living inside host cells has focused attention on understanding their invasion mechanism (Box 1). Molecular dissection of Toxoplasma invasion revealed that proteolytic shedding of adhesins from the parasite surface is required for invasion70, 71 (Figure 4). Although the protease initially eluded identification, many lines of evidence now suggest that a parasite-encoded rhomboid enzyme is responsible. Toxoplasma encodes six rhomboid enzymes, four of which are proteolytically active72, 73, but only one, TgROM5, has the capacity to cleave parasite adhesins directly in a defined heterologous assay72. Importantly, the cleavage site of numerous adhesins during invasion have been mapped to their transmembrane segments74–76. Lastly, TgROM5 resides on the parasite surface, enriched towards the posterior end, the expected site of adhesin cleavage72. Although these observations provide strong, correlative evidence that TgROM5 is the protease that dissolves the moving junction at the end of the invasion program, they should not be viewed as causative. It is important that genetic tests be applied to validate the importance of TgROM5 in invasion, but due to the difficulty of the system this has not yet been possible.
BOX 1.
Unlike most invasive pathogens, apicomplexa do not enter by triggering endocytic uptake by the host, but rather employ an intricate form of parasite motility to force their way into host cells108, 109 (Figure 4). Key molecules in the invasion process are parasite-encoded adhesins that form the interface between the parasite and host. These transmembrane proteins act as bridging molecules that form strong and specific interactions with a variety of receptors on the host surface, and link to the actomyosin contractile system inside the parasite108, 109. Parasites encode a diverse arsenal of adhesins, some of which are specific for invasion of different cell types, others define different entry routes into the same cell, while others may act in concert to facilitate invasion82, 110. The result is formation of a MOVING JUNCTION between the parasite and host surface that sweeps across the parasite from apical to posterior end, propelling the parasite into the host and causing invagination of the host membrane to form the nascent parasitophorous vacuole111 (Figure 4). While the moving junction forms the essential tight association that drives invasion, it must be dismantled at the end of the invasion program for parasite internalization and sealing of the host membrane70, 71.
Figure 4. Function of rhomboid proteases in two different protozoan parasites.

An example of parasitism of a human red blood cell by two different protozoan pathogens is shown. In the top panel, a Plasmodium merozoite invading the red blood cell using rhomboid enzymes PfROM4 (green) and PfROM1 (red, initially in mononeme) to breakdown the moving junction (formed between parasite adhesins and host receptors) at the end of invasion. The steps and roles of rhomboid enzymes are detailed in the text, but lead to malaria in the host. The invasion mechanism and role of rhomboid is similar in Toxoplasma. Bottom, an Entamoeba histolytica trophozoite is in the process of internalizing a red blood cell via phagocytosis. The rhomboid EhROM1 (green) relocalizes from the secretory pathway to the phagosome, colocalizing with its putative substrate the Gal/GalNAc lectin (blue). On the right, the amoeba translocates antibody-bound surface proteins to the cap, a membraneous vesicle-like structure that it will jettison to evade immune system attack. EhROM1 colocalizes to the neck of the cap during the capping process. Penetration of the intestine by E. histolytica and evasion of the immune system can lead to life-threatening abcesses.
Extension of these observations to Plasmodium falciparum, which causes the lethal form of malaria, met unexpected complexity. Although P. falciparum encodes up to eight rhomboid proteins, it does not have a direct orthologue of TgROM577, 78. Moreover, most Plasmodium adhesins contain aromatic residues within their transmembrane segments and as a result are refractory to cleavage by typical rhomboid enzymes77. While it is clear that other proteases are also involved in invasion, including the membrane-tethered subtilisin PfSUB279, a direct enzymatic analysis of Plasmodium rhomboid proteases revealed that one rhomboid, PfROM4, displays an atypical mode of specificity specialized for cleaving Plasmodium transmembrane adhesins that cannot be processed by PfSUB2 or canonical rhomboid enzymes77, 80. Indeed, PfROM4 is unable to cleave typical rhomboid substrates, but a second enzyme, PfROM1, provides this activity77. Intriguingly, TgROM5 cleaves both standard rhomboid substrates, as well as adhesins with aromatic residues within their transmembrane segments, suggesting that it functions as a ‘dual specificity’ rhomboid77. As such, it is tempting to speculate that when P. falciparum lost a TgROM5 homologue, it co-opted two rhomboid proteases with different substrate specificities that together could cover the repertoire of substrates that were normally cleaved by TgROM5 alone.
Further analysis of over a dozen P. falciparum adhesins known to be involved in invasion revealed that they all could be processed by PfROM4, or less commonly by PfROM1, or at times both77. These observations potentially broaden the role of rhomboid enzymes to all invasive stages of the parasite life cycle, both in the human host and in the mosquito vector. Analysis of PfROM4 and PfROM1 directly in MEROZOITES, the parasite form that causes the disease, further revealed that they are at the right place at the right time to execute their hypothesized roles during invasion (Figure 4). PfROM4 was found to be on the surface of the merozoite80, while PfROM1 was localized internally and defined the existence of a new organelle termed the mononeme81. However, consistent with a role in adhesin processing, PfROM1 was found to be released onto the merozoite surface prior to or during invasion81.
Importantly, different adhesins are known to define alternative invasion routes into a host cell, and parasites are able to switch pathways when one becomes blocked82. An implication of these early observations is that rhomboid cleavage of most transmembrane adhesins provides a convergence point that might be a weakness of the parasite invasion strategy. In both P. falciparum and T. gondii, difficulty of working with haploid organisms has thus far impeded genetic tests of this model. Perhaps the most compelling observation currently is the discovery that at least one essential adhesin is cleaved in its transmembrane segment during merozoite invasion80. Moreover, a further encouraging observation is failure to generate transgenic parasites harboring a rhomboid-uncleavable adhesin80, but this provides only indirect evidence that rhomboid proteolysis is essential. Other apicomplexan parasites also encode rhomboid proteases, including C. parvum and E. tenella, and although their functions are not yet assigned, they have been identified in several antigen expression cloning screens from specific stages of the parasite lifecycle83, 84. Analogous roles during parasite invasion are anticipated but untested.
In addition to rhomboid proteases, an SPP homologue has recently been implicated in invasion of erythrocytes by P. falciparum85. Unusual for an intramembrane protease, PfSPP was identified through a yeast two-hybrid screen for parasite proteins that interact with BAND 3 of human erythrocytes. Contrary to metazoan SPPs, PfSPP was not localized in the ER, but internally in MICRONEMES and on the apical surface of the parasite, and an antibody to the surface-exposed region of PfSPP blocked parasite invasion of erythrocytes. Moreover, intramembrane protease activity of PfSPP was shown to be similar to its human counterpart as assayed in a cell-based assay86, and two general SPP inhibitors were recently found to block merozoite invasion of erythrocytes, and subsequent parasite multiplication87. These observations suggest that PfSPP may have multiple essential proteolytic functions in the erythrocytic stage. PfSPP is also expressed in other phases of the parasite lifecycle, and thus could provide proteolytic activity for other cellular processes. Identifying PfSPP substrates, which based on its topology would be expected to be type II membrane proteins, should help clarify its function(s).
Rhomboid functions beyond invasion
Many parasites including T. gondii and Plasmodium spp. encode multiple rhomboid enzymes not thought to be involved in adhesin cleavage. Functions beyond invasion are also suggested by the finding that several TgROMs are localized to different organelles88, 89. What are the roles of these rhomboid enzymes?
Recent analyses have started to focus on this question: TgROM1 knockdown revealed no defects in T. gondii invasion, but rather impaired multiplication of parasites within host cells89. Similarly, ROM1 knockout in the rodent malaria P. berghei yielded parasites that were able to complete the entire lifecycle, both in the mosquito and mouse hosts, although several stages were delayed, or resulted in production of fewer parasites90. Removal of PbROM1 activity may thus lead to a general ‘unfitness’ of parasites, consistent with analysis in T. gondii. However, it should be noted that while TgROM1 knockdown parasites displayed mild phenotypes, repeated attempts to generate TgROM1 knockout T. gondii failed89. This observation raises the possibility that mechanisms partly compensating for ROM1 loss could have emerged in the knockdown parasites. It remains to be seen whether P. berghei could also have adapted to PbROM1 loss during the knockout procedure, as has been observed for some genes82. Therefore, genetic approaches may not yet have revealed the full spectrum of ROM1 function, and equally the substrate(s) that ROM1 processes in these events also remain unknown.
Despite these limitations, infection of mice with PbROM1 knockout parasites resulted in clearance by the immune system, and protective immunity against subsequent wildtype parasite infection90. Collectively, these experiments provide direct evidence that, while some rhomboid enzymes could have broad but not essential roles in the lifecycle of apicomplexan parasites, blocking their activity may still prove to be a successful strategy for anti-parasite therapy.
Role of rhomboid in parasitic amoeba
Intramembrane proteases are also widely conserved in protozoa that do not invade cells. Thus, a direct role in invasion probably reflects a specialization rather than revealing a common function. What is the role of intramembrane proteases in non-invasive pathogens? Entamoeba histolytica is an enteric pathogen that causes amoebic dysentery, but can also burrow through the intestine and spread to other organs, causing life-threatening abcesses. Unlike most eukaryotic cells, the amoeba genome encodes only one active rhomboid protease, termed EhROM1, making analyzing non-invasive functions of rhomboid enzymes attractive in this exclusively extracellular pathogen91.
Biochemical analysis of EhROM1 revealed it to possess atypical substrate specificity analogous to PfROM491. But the recent excitement over EhROM1 stems primarily from the protein that it cuts: a substrate search identified a heavy subunit of a family of surface Gal/GalNAc LECTINS known to play decisive roles in parasite virulence92. These lectins sit at the crossroads between different pathogenic modes, from establishing a benign, commensal infection, to initiating host attack via cell death and lysis, or differentiation into a dormant cyst. In fact, the lectins themselves are thought to determine the route taken, but how these decisions are made is not clear. At the amoeba surface, lectins contact host cells directly, which is essential both for host cell killing and phagocytosis of erythrocytes92 (Figure 4). EhROM1 is also localized to the surface and in undefined internal structures, but relocalizes to phagosomes along with lectins during phagocytosis91. During some of these events lectins are cleaved, but the role of the proteolysis and the identity of the responsible protease had remained unclear. It is tempting to speculate that lectin cleavage by EhROM1 may play an instructive role in some of these important events. Interestingly, the function of the amebic rhomboid enzyme again is to cleave adhesins like in apicomplexans, although this time the outcome isn’t host-cell invasion, but rather phagocytosis and virulence.
Analysis of EhROM1 function also revealed a new role for rhomboid enzymes. E. histolytica is an extracellular pathogen, and with important proteins such as its lectins prominently displayed on its surface, it must evade the host immune system. One amoebic mechanism for immune evasion is receptor capping; surface proteins that are attacked by the host immune system are translocated to the posterior of the parasite, and jettisoned on a membraneous ball93 (Figure 4). Intriguingly, inducing receptor capping experimentally caused EhROM1 to redistribute to the neck of the nascent cap, implying a role in cap release91 (Figure 4). However, while these intriguing correlations are striking, blocking EhROM1 activity, either via genetic ablation or pharmacologic interference, is required to test the role EhROM1 might play in these important events. These lines of investigation are challenging, since genetic techniques are rudimentary in amoeba, and no rhomboid inhibitors are yet available.
ROLES IN PATHOGENIC FUNGI
The functions of intramembrane proteolysis in pathogenic fungi have only recently been explored. The only work in this area stems from the study of adaptation to hypoxia by Cryptococcus neoformans, an inhaled human pathogen that disseminates to the brain where it causes the most common form of fungal meningitis (Figure 5). C. neoformans is an obligate aerobe that is exquisitely sensitive to oxygen levels for growth, and must therefore adapt to very low oxygen during pathogenesis in body tissues. Recent work discovered that the classical SREBP pathway is responsive to oxygen levels, and SREBP itself signals hypoxic adaptation in Schizosaccharomyces pombe94.
Figure 5. S2P mediates adaptation to hypoxia during fungal dissemination.

Outside the body, Cryptococcus (drawn in blue throughout) experiences a highly aerobic environment, but encounters hypoxia upon inhalation and dissemination into host tissues. Hypoxia is thought to lower sterol production, since biosynthesis requires oxygen. As in human cells, lower sterol levels are thought to result in release of the Scp1:Sre1 complex (SCAP:SREBP in humans) from the ER to the Golgi apparatus (depicted on the right), followed by sequential cleavage by an unidentified Cryptococcal site-1 protease (S1P in humans) and Stp1 (S2P in humans). The site-2 cleavage releases the Sre1 transcription factor domain from the membrane, allowing it to enter the nucleus and activate biosynthetic and hypoxia response genes. In humans, SREBP does not participate in a hypoxic response, but if Stp1 activity is blocked by mutation, Cryptococcus is unable to adapt to hypoxic conditions and cannot survive in host tissues.
Screens for mutants defective in hypoxic growth95, and extension of the SREBP paradigm to C. neoformans identified cleavage of its SREBP96, Sre1, as a response to low oxygen levels (Figure 5). Microarray analyses identified 50–100 genes regulated by Sre1 in response to oxygen. During infection of a mouse model, fungi harboring deletions of Sre1, or the S2P homologue Stp1 yielded greatly reduced growth in animal tissues, resulting in not only improvement of gross pathology, but a striking increase in survival of infected animals. Thus, S2P–mediated proteolysis plays a key role in virulence of C. neoformans by facilitating aclimatization of the fungi to the hypoxic nature of host tissues. Another particularly encouraging observation is that C. neoformans mutant for Stp1 is 100-fold more sensitive to common anti-fungal AZOLE DRUGS, suggesting that targeting S2P may at least provide synergistic effects in combination therapy with current drug regimens, if not being an improved regimen on its own. This awaits future investigation, as does identification of other components of this pathway, including a possible site-1 protease.
It should be noted that recognizable S2P homologues appear to be absent in many fungi, some of which may compensate by releasing SREBP using alternative pathways such as RUP97. As such, S2Ps may not prove to be broadly applicable as anti-fungal targets. However, most fungi encode other intramembrane proteases, with rhomboid proteases being particularly common, that have not been explored in the context of either biological regulation or fungal pathogenesis. This remains a fertile area of research for the future.
ROLES IN VIRAL MATURATION
Although there are currently no known viral intramembrane protease, several Flaviviridae have been found to rely on host-encoded SPP to provide essential functions for viral propagation. Most research has centered on assembly of hepatitis C virus (HCV), a positive strand RNA virus that infects over 150 million people globally and constitutes a major cause of cirrhosis and hepatocellular carcinoma.
The HCV genome consists of a single ORF that codes for one polyprotein of ∼3000 residues from which all individual viral factors must be excised98. As such, the virus relies heavily on proteases, both host and viral, to process the polyprotein into functional viral components. The N terminal one third contains the structural capsid core protein followed by two envelope proteins. The first signal peptide between core and E1 directs insertion into the ER membrane, and cellular signal peptidase releases the signal peptide from E1. But as a consequence of polyprotein continuity, the released signal peptide of E1 becomes a de facto C terminal transmembrane segment of core protein (Figure 6). This signal peptide is processed further by SPP activity, which was recognized well before the gene for SPP was identified99. The SPP-processed core remains membrane associated, but now is able to traffic to LIPID DROPLETS for virus assembly100. Although the existence of the C terminal, core-processing event was widely documented, whether it was essential for virus production remained unclear until earlier last year.
Figure 6. SPP processes core protein for hepatitis C virus assembly.
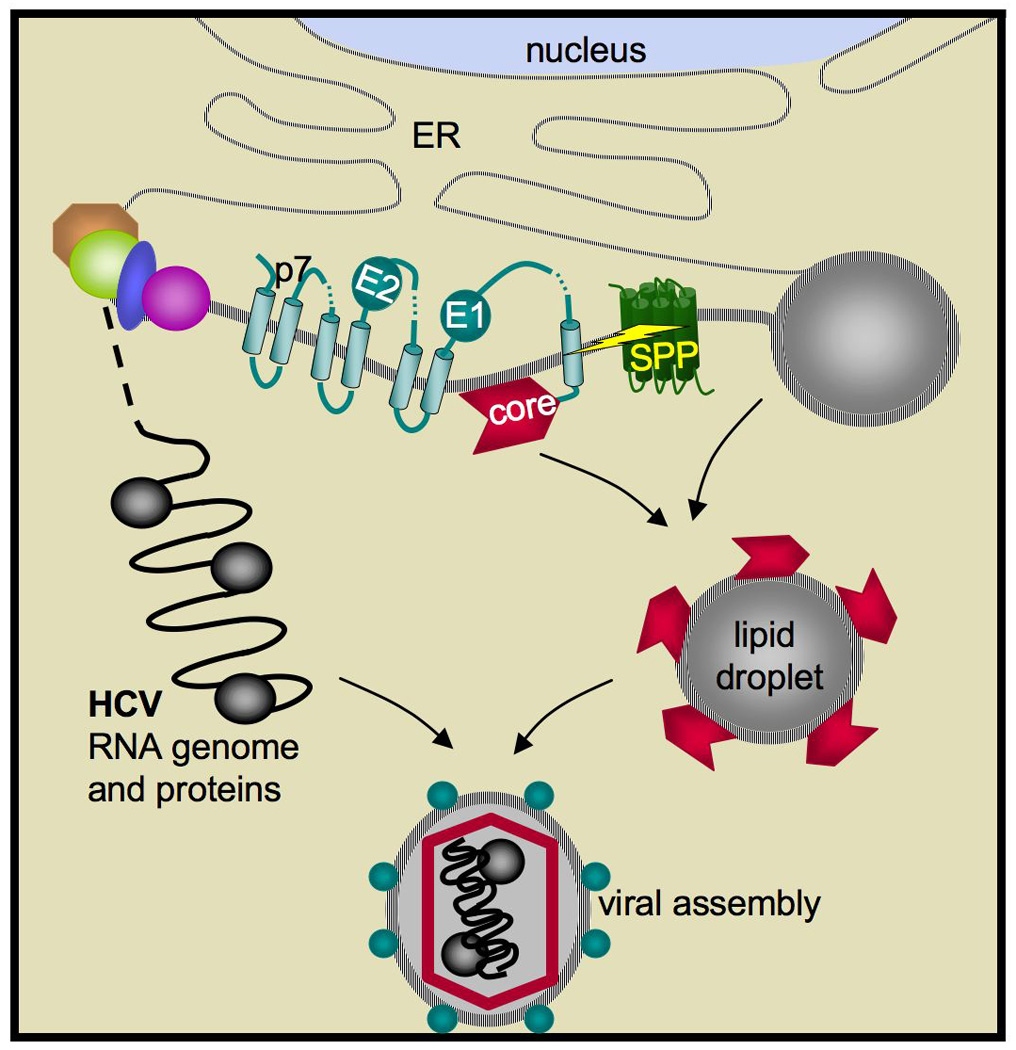
The structural HCV components E1 and E2 envelope proteins, core, and p7 are shown inserted into the ER membrane and already processed by signal peptidase. SPP is subsequently responsible for cleaving the first signal peptide, which following signal peptidase cleavage is attached to the C terminus of core protein. SPP cleavage (depicted by the yellow bolt) is required for core trafficking to lipid droplets (right) and recruitment of viral genomic RNA and factors (left) for capsid assembly. Note the SPP is not encoded by the virus, but rather is an ER-resident protein of the host cell.
Initial attempts to address this issue by making core mutants that cannot be processed by SPP yielded conflicting results, which have now been tracked to core polymorphisms among different HCV serotypes. A second hurdle was developing methodologies to assess impact directly on virus production and infectivity. With these advances in hand, several lines of evidence, including siRNA knockdown of SPP101, use of core mutants102, and SPP inhibitors and dominant negative mutants103, all agree that reducing core processing lowers released virus titre, and hinders HCV propagation and infectivity up to 10-fold. Mechanistically, unprocessed core was delayed in trafficking to lipid droplets and subsequent recruitment of replicated viral RNA and non-structural factors to virions102 (Figure 6). Moreover, capsids self-assembled from unprocessed core expressed in a heterologous system yielded unstable virus-like particles104.
These studies provide proof-of-principle that inhibiting SPP hinders HCV virus propagation, but it is not clear if the observed reduction is enough to limit or cure a chronic infection, for which a widely effective therapy is eagerly sought. Indeed, early observations suggest that inhibiting SPP results not in a block, but rather a delay, in virus assembly102. Animal models, which add pharmacokinetic and immune dimensions to the studies, are required to evaluate this possibility further. Recent advances should help to motivate this challenging approach.
Two other Flaviviridae of limited medical importance have also been shown to require core processing by SPP105, 106 (Table 1), and SPP also plays a role in the infective cycle of an unrelated virus, the herpes virus cytomegalovirus107. In this context, SPP is required for the dislocation of MHC proteins from the ER as part of a larger viral immune evasion strategy. However, whether SPP requires proteolytic activity for this function, or the impact of blocking SPP on virus infection, remain unexplored.
Notably, in these contexts it is the host SPP enzyme, rather than a viral enzyme, that would serve as a target for antiviral therapy. This has two main implications: first, it could confound therapy with side-effects due to the other roles SPP has in cells, especially in the immune system. Secondly, and on a positive note, targeting host SPP could limit the scope for emergence of drug resistance (since it would be limited to the host cell rather than the virus).
EMERGING PRINCIPLES AND FUTURE DIRECTIONS
Intramembrane proteolysis as a field has its origins in animal biology and disease, but intramembrane proteases are widely conserved in unicellular organisms. The past few years have witnessed major advances in our understanding of the roles these enzymes play in pathogenic organisms of all types. While this can now be viewed as a firmly-established and distinct new field, key challenges remain both in our understanding of the roles these enzymes play, and in evaluating their potential as therapeutic targets.
An important goal is to gain a representative picture of the roles intramembrane proteolysis plays in pathogenic organisms across evolution. Currently the majority of known roles for intramembrane proteases generally fall within pathogen types; S2P figures prominently in bacterial pathogens, rhomboid proteases play central roles in protozoan parasites, and SPP is the only intramembrane protease implicated in viral assembly. Certainly the groupings are not exclusive and at least partly due to the consequence of limited study, as illustrated by well-defined exceptional cases. But it will be important to see to what degree this general trend holds as more examples become characterized. Another major gap is the lack of any information on intramembrane proteolysis in pathogens that infect plants, probably because these pathogens do not receive comparable levels of effort. Collectively, this information is key to understanding the specialization and evolutionary origins of intramembrane proteolysis in pathogenic microbes.
Secondly, despite the demonstrated importance of intramembrane proteolysis in multiple pathogens, not a single intramembrane protease has been targeted with inhibitors in the context of disease models. Currently this approach is hampered by the difficulty in developing specific inhibitors for both rhomboid and S2P families, but strategies to overcome hurdles should emerge as our biochemical and structural understanding of these enzymes increases. Another potential challenge is the conservation of intramembrane proteases in animal hosts, which could cross-react with inhibitors. While this is a major issue, it is possible that a window of opportunity exists because often these are distantly-related homologues, and because infections tend to require acute rather than chronic treatment. These lines of investigation present tremendous challenges and opportunities for the future, and given the attractive nature of proteases and membrane proteins as drug targets, could even culminate in new strategies for therapeutic intervention into a variety of infectious disease.
ACKNOWLEDGEMENTS
I sincerely apologize to those scientists whose work could not be discussed or cited as a result of space limitations. A special thanks to Rosanna Baker for expert help with rendering the illustrations. Work in the Urban lab is supported by NIH grant R01AI066025, a career award from the Burroughs-Wellcome Fund, and a Packard Foundation Fellowship for Science and Engineering.
GLOSSARY
- SREBP
sterol regulatory element binding protein, a transcription factor that actives expression of genes required for cholesterol and lipid biosynthesis in animals
- Extracytoplasmic function
a variety of conditions that are commonly detected by unfolding of outer membrane proteins, which stimulates proteases that liberate σE from the membrane to activate response genes expression
- Metalloprotease
an enzyme that cleaves peptide bonds using bound zinc ion to facilitate hydrolysis. The zinc is held in place usually by two conserved histidines and one acidic residue
- Aspartyl protease
a hydrolytic enzyme that uses two aspartate residues to activate water for cleaving peptide bonds
- Serine protease
an enzyme that uses a serine as a nucleophile for cleavage of peptide bonds. The serine is usually activated by a basic residue, and forms a covalent intermediate with the substrate that is released through attack by water.
- Anti-sigma factor
a protein that binds and hinders the transcription-activating function of a bacterial sigma factor
- Exopolysaccharide
a linear polymer of modified sugars that is not generally attached to a bacterium; act as an extracellular matrix
- Nosocomial
an infection acquired in a hospital
- Conjugation
a physical joining of two bacterial cells for the purpose of transferring genetic material
- Quorum sensing
a cell-to-cell signalling mechanism by which bacteria monitor their population size and react to it accordingly
- Pleiotrophic
having multiple, and seemingly unrelated, phenotypes
- Biolog
a commercial, phenotypic testing method conducted using a large number of standardized conditions
- Moving junction
a specialized adhesive point of contact between the parasite and host that appears as an electron-dense structure in EM analysis and traverses the surface of the parasite as a tight ring during invasion of the host
- Merozoite
the parasite form specialized for invasion of and replication within erythrocytes
- Band 3
an integral membrane protein responsible for chloride/bicarbonate exchange on erythrocytes
- Micronemes
specialized, apical, tiny, vesicle-like organelles of apicomplexan parasites that house adhesins and other proteins for secretion during invasion
- Gal/GalNAc lectin
a protein specialized for binding sugars, in this case containing galactose and N-acetylgalactosamine
- Azole drugs
a class of anti-fugnal drugs that inhibit the enzyme lanosterol-14a–deme-thylase required for ergoesterol biosynthesis
- RUP
‘regulated ubiquitin/proteasome-dependent processing’, a form of proteasome degradation that releases intact domains from the membrane into the cytosol
- Lipid droplets
lipid storage organelles of cells, in which triacylglycerides in the form of a drop are surrounded by a single leaflet of membrane phospholipid
REFERENCES
- 1.Brown MS, Ye J, Rawson RB, Goldstein JL. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell. 2000;100:391–398. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- 2.Wolfe MS, Kopan R. Intramembrane proteolysis: theme and variations. Science. 2004;305:1119–1123. doi: 10.1126/science.1096187. [DOI] [PubMed] [Google Scholar]
- 3.Urban S. Rhomboid proteins: conserved membrane proteases with divergent biological functions. Genes Dev. 2006;20:3054–3068. doi: 10.1101/gad.1488606. [DOI] [PubMed] [Google Scholar]
- 4.Koonin EV, et al. The rhomboids: a nearly ubiquitous family of intramembrane serine proteases that probably evolved by multiple ancient horizontal gene transfers. Genome Biol. 2003;4:R19. doi: 10.1186/gb-2003-4-3-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lemberg MK, Freeman M. Functional and evolutionary implications of enhanced genomic analysis of rhomboid intramembrane proteases. Genome Res. 2007;17:1634–1646. doi: 10.1101/gr.6425307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kinch LN, Ginalski K, Grishin NV. Site-2 protease regulated intramembrane proteolysis: sequence homologs suggest an ancient signaling cascade. Protein Sci. 2006;15:84–93. doi: 10.1110/ps.051766506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rudner DZ, Fawcett P, Losick R. A family of membrane-embedded metalloproteases involved in regulated proteolysis of membrane-associated transcription factors. Proc Natl Acad Sci U S A. 1999;96:14765–14770. doi: 10.1073/pnas.96.26.14765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rather PN, Ding X, Baca-DeLancey RR, Siddiqui S. Providencia stuartii genes activated by cell-to-cell signaling and identification of a gene required for production or activity of an extracellular factor. J Bacteriol. 1999;181:7185–7191. doi: 10.1128/jb.181.23.7185-7191.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.An FY, Sulavik MC, Clewell DB. Identification and characterization of a determinant (eep) on the Enterococcus faecalis chromosome that is involved in production of the peptide sex pheromone cAD1. J Bacteriol. 1999;181:5915–5921. doi: 10.1128/jb.181.19.5915-5921.1999.A library over-expression screen identified an S2P homologue as the protease required for producing several different pheromones in Enterococcus conjugation.
- 10.Rawson RB, et al. Complementation cloning of S2P, a gene encoding a putative metalloprotease required for intramembrane cleavage of SREBPs. Mol Cell. 1997;1:47–57. doi: 10.1016/s1097-2765(00)80006-4. [DOI] [PubMed] [Google Scholar]
- 11.Ye J, et al. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell. 2000;6:1355–1364. doi: 10.1016/s1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- 12.Kanehara K, Ito K, Akiyama Y. YaeL (EcfE) activates the sigma(E) pathway of stress response through a site-2 cleavage of anti-sigma(E), RseA. Genes Dev. 2002;16:2147–2155. doi: 10.1101/gad.1002302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alba BM, Leeds JA, Onufryk C, Lu CZ, Gross CA. DegS and YaeL participate sequentially in the cleavage of RseA to activate the sigma(E)-dependent extracytoplasmic stress response. Genes Dev. 2002;16:2156–2168. doi: 10.1101/gad.1008902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weihofen A, Binns K, Lemberg MK, Ashman K, Martoglio B. Identification of signal peptide paptidase, a presenilin-type aspartic protease. Science. 2002;296:2215–2218. doi: 10.1126/science.1070925. [DOI] [PubMed] [Google Scholar]
- 15.Fluhrer R, Haass C. Signal peptide peptidases and gamma-secretase: cousins of the same protease family? Neurodegener Dis. 2007;4:112–116. doi: 10.1159/000101835. [DOI] [PubMed] [Google Scholar]
- 16.Weihofen A, Martoglio B. Intramembrane-cleaving proteases: controlled liberation of proteins and bioactive peptides. Trends Cell Biol. 2003;13:71–78. doi: 10.1016/s0962-8924(02)00041-7. [DOI] [PubMed] [Google Scholar]
- 17.Sato T, et al. Signal peptide peptidase: biochemical properties and modulation by nonsteroidal antiinflammatory drugs. Biochemistry. 2006;45:8649–8656. doi: 10.1021/bi060597g. [DOI] [PubMed] [Google Scholar]
- 18.Narayanan S, Sato T, Wolfe MS. A C-terminal region of signal peptide peptidase defines a functional domain for intramembrane aspartic protease catalysis. J Biol Chem. 2007;282:20172–20179. doi: 10.1074/jbc.M701536200. [DOI] [PubMed] [Google Scholar]
- 19.Ponting CP, et al. Identification of a novel family of presenilin homologues. Hum Mol Genet. 2002;11:1037–1044. doi: 10.1093/hmg/11.9.1037. [DOI] [PubMed] [Google Scholar]
- 20.Urban S, Lee JR, Freeman M. Drosophila rhomboid-1 defines a family of putative intramembrane serine proteases. Cell. 2001;107:173–182. doi: 10.1016/s0092-8674(01)00525-6. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y, Zhang Y, Ha Y. Crystal structure of a rhomboid family intramembrane protease. Nature. 2006;444:179–180. doi: 10.1038/nature05255. [DOI] [PubMed] [Google Scholar]
- 22.Wu Z, et al. Structural analysis of a rhomboid family intramembrane protease reveals a gating mechanism for substrate entry. Nat Struct Mol Biol. 2006;13:1084–1091. doi: 10.1038/nsmb1179. [DOI] [PubMed] [Google Scholar]
- 23.Ben-Shem A, Fass D, Bibi E. Structural basis for intramembrane proteolysis by rhomboid serine proteases. Proc Natl Acad Sci U S A. 2007;104:462–466. doi: 10.1073/pnas.0609773104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemieux MJ, Fischer SJ, Cherney MM, Bateman KS, James MN. The crystal structure of the rhomboid peptidase from Haemophilus influenzae provides insight into intramembrane proteolysis. Proc Natl Acad Sci U S A. 2007;104:750–754. doi: 10.1073/pnas.0609981104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Urban S, Freeman M. Intramembrane proteolysis controls diverse signalling pathways throughout evolution. Curr Opin Genet Dev. 2002;12:512–518. doi: 10.1016/s0959-437x(02)00334-9. [DOI] [PubMed] [Google Scholar]
- 26.Akiyama Y, Kanehara K, Ito K. RseP (YaeL), an Escherichia coli RIP protease, cleaves transmembrane sequences. Embo J. 2004;23:4434–4442. doi: 10.1038/sj.emboj.7600449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Urban S, Wolfe MS. Reconstitution of intramembrane proteolysis in vitro reveals that pure rhomboid is sufficient for catalysis and specificity. Proc Natl Acad Sci U S A. 2005;102:1883–1888. doi: 10.1073/pnas.0408306102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maegawa S, Ito K, Akiyama Y. Proteolytic action of GlpG, a rhomboid protease in the Escherichia coli cytoplasmic membrane. Biochemistry. 2005;44:13543–13552. doi: 10.1021/bi051363k. [DOI] [PubMed] [Google Scholar]
- 29.Feng L, et al. Structure of a site-2 protease family intramembrane metalloprotease. Science. 2007;318:1608–1612. doi: 10.1126/science.1150755. [DOI] [PubMed] [Google Scholar]
- 30.Baker RP, Young K, Feng L, Shi Y, Urban S. Enzymatic analysis of a rhomboid intramembrane protease implicates transmembrane helix 5 as the lateral substrate gate. Proc Natl Acad Sci U S A. 2007;104:8257–8262. doi: 10.1073/pnas.0700814104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y, Ha Y. Open-cap conformation of intramembrane protease GlpG. Proc Natl Acad Sci U S A. 2007;104:2098–2102. doi: 10.1073/pnas.0611080104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Urban S, Baker RP. In vivo analysis reveals substrate-gating mutants of a rhomboid intramembrane protease display increased activity in living cells. Biol Chem. 2008;389:1107–1115. doi: 10.1515/BC.2008.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Urban S, Shi Y. Core principles of intramembrane proteolysis: comparison of rhomboid and site-2 family proteases. Curr Opin Struct Biol. 2008;18:432–441. doi: 10.1016/j.sbi.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Osenkowski P, et al. Cryoelectron Microscopy Structure of Purified gamma-Secretase at 12 A Resolution. J Mol Biol. 2008 doi: 10.1016/j.jmb.2008.10.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lemberg MK, Freeman M. Cutting proteins within lipid bilayers: rhomboid structure and mechanism. Mol Cell. 2007;28:930–940. doi: 10.1016/j.molcel.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 36.Chen JC, Viollier PH, Shapiro L. A membrane metalloprotease participates in the sequential degradation of a Caulobacter polarity determinant. Mol Microbiol. 2005;55:1085–1103. doi: 10.1111/j.1365-2958.2004.04443.x. [DOI] [PubMed] [Google Scholar]
- 37.Mesak LR, Mesak FM, Dahl MK. Expression of a novel gene, gluP, is essential for normal Bacillus subtilis cell division and contributes to glucose export. BMC Microbiol. 2004;4:13. doi: 10.1186/1471-2180-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.King-Lyons ND, Smith KF, Connell TD. Expression of hurP, a gene encoding a prospective site 2 protease, is essential for heme-dependent induction of bhuR in Bordetella bronchiseptica. J Bacteriol. 2007;189:6266–6275. doi: 10.1128/JB.00629-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schobel S, Zellmeier S, Schumann W, Wiegert T. The Bacillus subtilis sigmaW anti-sigma factor RsiW is degraded by intramembrane proteolysis through YluC. Mol Microbiol. 2004;52:1091–1105. doi: 10.1111/j.1365-2958.2004.04031.x. [DOI] [PubMed] [Google Scholar]
- 40.Denham EL, Ward PN, Leigh JA. Lipoprotein signal peptides are processed by Lsp and Eep of Streptococcus uberis. J Bacteriol. 2008;190:4641–4647. doi: 10.1128/JB.00287-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bramkamp M, Weston L, Daniel RA, Errington J. Regulated intramembrane proteolysis of FtsL protein and the control of cell division in Bacillus subtilis. Mol Microbiol. 2006;62:580–591. doi: 10.1111/j.1365-2958.2006.05402.x. [DOI] [PubMed] [Google Scholar]
- 42.Hussain M, Ichihara S, Mizushima S. Mechanism of signal peptide cleavage in the biosynthesis of the major lipoprotein of the Escherichia coli outer membrane. J Biol Chem. 1982;257:5177–5182. [PubMed] [Google Scholar]
- 43.Kim AC, Oliver DC, Paetzel M. Crystal structure of a bacterial signal Peptide peptidase. J Mol Biol. 2008;376:352–366. doi: 10.1016/j.jmb.2007.11.080. [DOI] [PubMed] [Google Scholar]
- 44.Grigorova IL, et al. Fine-tuning of the Escherichia coli sigmaE envelope stress response relies on multiple mechanisms to inhibit signal-independent proteolysis of the transmembrane anti-sigma factor, RseA. Genes Dev. 2004;18:2686–2697. doi: 10.1101/gad.1238604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Glickman MS, Jacobs WR., Jr Microbial pathogenesis of Mycobacterium tuberculosis: dawn of a discipline. Cell. 2001;104:477–485. doi: 10.1016/s0092-8674(01)00236-7. [DOI] [PubMed] [Google Scholar]
- 46.Makinoshima H, Glickman MS. Regulation of Mycobacterium tuberculosis cell envelope composition and virulence by intramembrane proteolysis. Nature. 2005;436:406–409. doi: 10.1038/nature03713.Targeted deletion of an S2P and assessment in an animal model of tuberculosis revealed dysregulation of envelope composition and a dramatic decrease in replication and persistence
- 47.Ramsey DM, Wozniak DJ. Understanding the control of Pseudomonas aeruginosa alginate synthesis and the prospects for management of chronic infections in cystic fibrosis. Mol Microbiol. 2005;56:309–322. doi: 10.1111/j.1365-2958.2005.04552.x. [DOI] [PubMed] [Google Scholar]
- 48.Qiu D, Eisinger VM, Rowen DW, Yu HD. Regulated proteolysis controls mucoid conversion in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2007;104:8107–8112. doi: 10.1073/pnas.0702660104.A screen and subsequent genetic analyses revealed a multi-component S2P pathway underlying regulation of alginate biosynthesis
- 49.Bragonzi A, et al. Sequence diversity of the mucABD locus in Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Microbiology. 2006;152:3261–3269. doi: 10.1099/mic.0.29175-0. [DOI] [PubMed] [Google Scholar]
- 50.Wood LF, Ohman DE. Use of cell wall stress to characterize sigma(22) (AlgT/U) activation by regulated proteolysis and its regulon in Pseudomonas aeruginosa. Mol Microbiol. 2009 doi: 10.1111/j.1365-2958.2009.06635.x. [DOI] [PubMed] [Google Scholar]
- 51.Matson JS, Withey JH, DiRita VJ. Regulatory networks controlling Vibrio cholerae virulence gene expression. Infect Immun. 2007;75:5542–5549. doi: 10.1128/IAI.01094-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matson JS, DiRita VJ. Degradation of the membrane-localized virulence activator TcpP by the YaeL protease in Vibrio cholerae. Proc Natl Acad Sci U S A. 2005;102:16403–16408. doi: 10.1073/pnas.0505818102.A genetic screen identified an S2P in the degradation of the transcription factor TcpP as a mechanism for down-regulating cholera toxin production
- 53.Weigel LM, et al. Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science. 2003;302:1569–1571. doi: 10.1126/science.1090956. [DOI] [PubMed] [Google Scholar]
- 54.Clewell DB, Francia MV, Flannagan SE, An FY. Enterococcal plasmid transfer: sex pheromones, transfer origins, relaxases, and the Staphylococcus aureus issue. Plasmid. 2002;48:193–201. doi: 10.1016/s0147-619x(02)00113-0. [DOI] [PubMed] [Google Scholar]
- 55.Chandler JR, Dunny GM. Enterococcal peptide sex pheromones: synthesis and control of biological activity. Peptides. 2004;25:1377–1388. doi: 10.1016/j.peptides.2003.10.020. [DOI] [PubMed] [Google Scholar]
- 56.Flannagan SE, Clewell DB. Identification and characterization of genes encoding sex pheromone cAM373 activity in Enterococcus faecalis and Staphylococcus aureus. Mol Microbiol. 2002;44:803–817. doi: 10.1046/j.1365-2958.2002.02922.x. [DOI] [PubMed] [Google Scholar]
- 57.An FY, Clewell DB. Identification of the cAD1 sex pheromone precursor in Enterococcus faecalis. J Bacteriol. 2002;184:1880–1887. doi: 10.1128/JB.184.7.1880-1887.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Antiporta MH, Dunny GM. ccfA, the genetic determinant for the cCF10 peptide pheromone in Enterococcus faecalis OG1RF. J Bacteriol. 2002;184:1155–1162. doi: 10.1128/jb.184.4.1155-1162.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Waters CM, Wells CL, Dunny GM. The aggregation domain of aggregation substance, not the RGD motifs, is critical for efficient internalization by HT-29 enterocytes. Infect Immun. 2003;71:5682–5689. doi: 10.1128/IAI.71.10.5682-5689.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hirt H, Schlievert PM, Dunny GM. In vivo induction of virulence and antibiotic resistance transfer in Enterococcus faecalis mediated by the sex pheromone-sensing system of pCF10. Infect Immun. 2002;70:716–723. doi: 10.1128/iai.70.2.716-723.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schlievert PM, et al. Aggregation and binding substances enhance pathogenicity in rabbit models of Enterococcus faecalis endocarditis. Infect Immun. 1998;66:218–223. doi: 10.1128/iai.66.1.218-223.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chandler JR, Hirt H, Dunny GM. A paracrine peptide sex pheromone also acts as an autocrine signal to induce plasmid transfer and virulence factor expression in vivo. Proc Natl Acad Sci U S A. 2005;102:15617–15622. doi: 10.1073/pnas.0505545102.Genetic ablation of a pheromone signal was used to clarify how dysregulation of the pheromone pathway leading to virulence occurs upon exposure to human serum
- 63.Rather PN, Orosz E. Characterization of aarA, a pleiotrophic negative regulator of the 2'-N-acetyltransferase in Providencia stuartii. J Bacteriol. 1994;176:5140–5144. doi: 10.1128/jb.176.16.5140-5144.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Urban S, Schlieper D, Freeman M. Conservation of intramembrane proteolytic activity and substrate specificity in eukaryotic and prokaryotic Rhomboids. Current Biology. 2002;12:1507–1512. doi: 10.1016/s0960-9822(02)01092-8. [DOI] [PubMed] [Google Scholar]
- 65.Gallio M, Sturgill G, Rather P, Kylsten P. A conserved mechanism for extracellular signaling in eukaryotes and prokaryotes. Proc Natl Acad Sci U S A. 2002;99:12208–12213. doi: 10.1073/pnas.192138799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rather PN, Parojcic MM, Paradise MR. An extracellular factor regulating expression of the chromosomal aminoglycoside 2'-N-acetyltransferase of Providencia stuartii. Antimicrob Agents Chemother. 1997;41:1749–1754. doi: 10.1128/aac.41.8.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stevenson LG, et al. Rhomboid protease AarA mediates quorum-sensing in Providencia stuartii by activating TatA of the twin-arginine translocase. Proc Natl Acad Sci U S A. 2007;104:1003–1008. doi: 10.1073/pnas.0608140104.Genetic and biochemical identification of TatA as a substrate, and subsequent epistasis analysis, revealed the role of rhomboid in Providencia quorum-sensing to be in activating TatA
- 68.Clemmer KM, Sturgill GM, Veenstra A, Rather PN. Functional characterization of Escherichia coli GlpG and additional rhomboid proteins using an aarA mutant of Providencia stuartii. J Bacteriol. 2006;188:3415–3419. doi: 10.1128/JB.188.9.3415-3419.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Berks BC, Palmer T, Sargent F. The Tat protein translocation pathway and its role in microbial physiology. Adv Microb Physiol. 2003;47:187–254. doi: 10.1016/s0065-2911(03)47004-5. [DOI] [PubMed] [Google Scholar]
- 70.Brossier F, Jewett TJ, Lovett JL, Sibley LD. C-terminal processing of the toxoplasma protein MIC2 is essential for invasion into host cells. J Biol Chem. 2003;278:6229–6234. doi: 10.1074/jbc.M209837200. [DOI] [PubMed] [Google Scholar]
- 71.Carruthers VB, Sherman GD, Sibley LD. The Toxoplasma adhesive protein MIC2 is proteolytically processed at multiple sites by two parasite-derived proteases. J Biol Chem. 2000;275:14346–14353. doi: 10.1074/jbc.275.19.14346. [DOI] [PubMed] [Google Scholar]
- 72.Brossier F, Jewett TJ, Sibley LD, Urban S. A spatially localized rhomboid protease cleaves cell surface adhesins essential for invasion by Toxoplasma. Proc Natl Acad Sci U S A. 2005;102:4146–4151. doi: 10.1073/pnas.0407918102.Biochemical, expression and subcellular localization analysis revealed a role for one Toxoplasma rhomboid protease in cleaving adhesins during host-cell invasion
- 73.Dowse TJ, Pascall JC, Brown KD, Soldati D. Apicomplexan rhomboids have a potential role in microneme protein cleavage during host cell invasion. Int J Parasitol. 2005;35:747–756. doi: 10.1016/j.ijpara.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 74.Opitz C, et al. Intramembrane cleavage of microneme proteins at the surface of the apicomplexan parasite Toxoplasma gondii. Embo J. 2002;21:1577–1585. doi: 10.1093/emboj/21.7.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Howell SA, et al. Distinct mechanisms govern proteolytic shedding of a key invasion protein in apicomplexan pathogens. Mol Microbiol. 2005;57:1342–1356. doi: 10.1111/j.1365-2958.2005.04772.x. [DOI] [PubMed] [Google Scholar]
- 76.Zhou XW, Blackman MJ, Howell SA, Carruthers VB. Proteomic analysis of cleavage events reveals a dynamic two-step mechanism for proteolysis of a key parasite adhesive complex. Mol Cell Proteomics. 2004;3:565–576. doi: 10.1074/mcp.M300123-MCP200. [DOI] [PubMed] [Google Scholar]
- 77.Baker RP, Wijetilaka R, Urban S. Two Plasmodium Rhomboid Proteases Preferentially Cleave Different Adhesins Implicated in All Invasive Stages of Malaria. PLoS Pathog. 2006;2:922–932. doi: 10.1371/journal.ppat.0020113.Analysis of rhomboid protease activity revealed two Plasmodium rhomboid proteases cleave adhesins implicated in all stages of parasite invasion
- 78.Dowse TJ, Soldati D. Rhomboid-like proteins in Apicomplexa: phylogeny and nomenclature. Trends Parasitol. 2005;21:254–258. doi: 10.1016/j.pt.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 79.Harris PK, et al. Molecular identification of a malaria merozoite surface sheddase. PLoS Pathog. 2005;1:241–251. doi: 10.1371/journal.ppat.0010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.O'Donnell RA, et al. Intramembrane proteolysis mediates shedding of a key adhesin during erythrocyte invasion by the malaria parasite. J Cell Biol. 2006;174:1023–1033. doi: 10.1083/jcb.200604136.PfROM4 subcellular localization, cleavage site mapping, and transgenic expression of adhesin mutants together implicate PfROM4 in merozoite invasion
- 81.Singh S, Plassmeyer M, Gaur D, Miller LH. Mononeme: a new secretory organelle in Plasmodium falciparum merozoites identified by localization of rhomboid-1 protease. Proc Natl Acad Sci U S A. 2007;104 doi: 10.1073/pnas.0709999104. 20043-20008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stubbs J, et al. Molecular mechanism for switching of P. falciparum invasion pathways into human erythrocytes. Science. 2005;309:1384–1387. doi: 10.1126/science.1115257. [DOI] [PubMed] [Google Scholar]
- 83.Trasarti E, Pizzi E, Pozio E, Tosini F. The immunological selection of recombinant peptides from Cryptosporidium parvum reveals 14 proteins expressed at the sporozoite stage, 7 of which are conserved in other apicomplexa. Mol Biochem Parasitol. 2007;152:159–169. doi: 10.1016/j.molbiopara.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 84.Li J, Zhang X, Liu Q, Yin J, Yang J. Eimeria tenella: cloning of a novel Eimeria tenella cDNA encoding a protein related to rhomboid family from F2 hybrid strain. Exp Parasitol. 2006;113:215–220. doi: 10.1016/j.exppara.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 85.Li X, Chen H, Oh SS, Chishti AH. A Presenilin-like protease associated with Plasmodium falciparum micronemes is involved in erythrocyte invasion. Mol Biochem Parasitol. 2008;158:22–31. doi: 10.1016/j.molbiopara.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nyborg AC, Ladd TB, Jansen K, Kukar T, Golde TE. Intramembrane proteolytic cleavage by human signal peptide peptidase like 3 and malaria signal peptide peptidase. Faseb J. 2006;20:1671–1679. doi: 10.1096/fj.06-5762com. [DOI] [PubMed] [Google Scholar]
- 87.Li X, et al. Plasmodium falciparum signal peptide peptidase is a promising drug target against blood stage malaria. Biochem Biophys Res Commun. 2009 doi: 10.1016/j.bbrc.2009.01.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sheiner L, Dowse TJ, Soldati-Favre D. Identification of trafficking determinants for polytopic rhomboid proteases in Toxoplasma gondii. Traffic. 2008;9:665–677. doi: 10.1111/j.1600-0854.2008.00736.x. [DOI] [PubMed] [Google Scholar]
- 89.Brossier F, Starnes GL, Beatty WL, Sibley LD. Microneme rhomboid protease TgROM1 is required for efficient intracellular growth of Toxoplasma gondii. Eukaryot Cell. 2008;7:664–674. doi: 10.1128/EC.00331-07.The first genetic knockdown of a rhomboid in any eukaryotic pathogen demonstrates a function for a non-adhesin-processing rhomboid family member in intracellular growth
- 90.Srinivasan P, Coppens I, Jacobs-Lorena M. Distinct roles of Plasmodium rhomboid 1 in parasite development and malaria pathogenesis. PLoS Pathog. 2009;5:e1000262. doi: 10.1371/journal.ppat.1000262.Targeted deletion of PbROM1 revealed parasites complete the entire lifecycle, but with decreased efficiency at multiple steps, were less virulent in rodents, and provided protective immunity against subsequent infection
- 91.Baxt LA, Baker RP, Singh U, Urban S. An Entamoeba histolytica rhomboid protease with atypical specificity cleaves a surface lectin involved in phagocytosis and immune evasion. Genes Dev. 2008;22:1636–1646. doi: 10.1101/gad.1667708.Biochemical and subcellular localization suggest that the sole ameba rhomboid protease functions in lectin shedding, phagocytosis and immune evasion
- 92.Petri WA, Jr, Haque R, Mann BJ. The bittersweet interface of parasite and host: lectin-carbohydrate interactions during human invasion by the parasite Entamoeba histolytica. Annu Rev Microbiol. 2002;56:39–64. doi: 10.1146/annurev.micro.56.012302.160959. [DOI] [PubMed] [Google Scholar]
- 93.Espinosa-Cantellano M, Martinez-Palomo A. Entamoeba histolytica: mechanism of surface receptor capping. Exp Parasitol. 1994;79:424–435. doi: 10.1006/expr.1994.1104. [DOI] [PubMed] [Google Scholar]
- 94.Hughes AL, Todd BL, Espenshade PJ. SREBP pathway responds to sterols and functions as an oxygen sensor in fission yeast. Cell. 2005;120:831–842. doi: 10.1016/j.cell.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 95.Chun CD, Liu OW, Madhani HD. A link between virulence and homeostatic responses to hypoxia during infection by the human fungal pathogen Cryptococcus neoformans. PLoS Pathog. 2007;3:e22. doi: 10.1371/journal.ppat.0030022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chang YC, Bien CM, Lee H, Espenshade PJ, Kwon-Chung KJ. Sre1p, a regulator of oxygen sensing and sterol homeostasis, is required for virulence in Cryptococcus neoformans. Mol Microbiol. 2007;64:614–629. doi: 10.1111/j.1365-2958.2007.05676.x.Above two studies demonstrate a role for the SREBP pathway including the S2P homolog in Cryptococcus hypoxia response and virulence in animal models
- 97.Hoppe T, Rape M, Jentsch S. Membrane-bound transcription factors: regulated release by RIP or RUP. Curr Opin Cell Biol. 2001;13:344–348. doi: 10.1016/s0955-0674(00)00218-0. [DOI] [PubMed] [Google Scholar]
- 98.Moradpour D, Penin F, Rice CM. Replication of hepatitis C virus. Nat Rev Microbiol. 2007;5:453–463. doi: 10.1038/nrmicro1645. [DOI] [PubMed] [Google Scholar]
- 99.Hussy P, Langen H, Mous J, Jacobsen H. Hepatitis C virus core protein: carboxy-terminal boundaries of two processed species suggest cleavage by a signal peptide peptidase. Virology. 1996;224:93–104. doi: 10.1006/viro.1996.0510. [DOI] [PubMed] [Google Scholar]
- 100.McLauchlan J, Lemberg MK, Hope G, Martoglio B. Intramembrane proteolysis promotes trafficking of hepatitis C virus core protein to lipid droplets. Embo J. 2002;21:3980–3988. doi: 10.1093/emboj/cdf414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Randall G, et al. Cellular cofactors affecting hepatitis C virus infection and replication. Proc Natl Acad Sci U S A. 2007;104:12884–12889. doi: 10.1073/pnas.0704894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Targett-Adams P, Hope G, Boulant S, McLauchlan J. Maturation of hepatitis C virus core protein by signal peptide peptidase is required for virus production. J Biol Chem. 2008;283:16850–16859. doi: 10.1074/jbc.M802273200. [DOI] [PubMed] [Google Scholar]
- 103.Okamoto K, et al. Intramembrane processing by signal peptide peptidase regulates the membrane localization of hepatitis C virus core protein and viral propagation. J Virol. 2008;82:8349–8361. doi: 10.1128/JVI.00306-08.These two complementary studies used a series of different approaches and overcame prior strain inconsistencies to demonstrate a direct requirement for core processing by SPP in HCV virus assembly and propagation
- 104.Vauloup-Fellous C, et al. Signal peptide peptidase-catalyzed cleavage of hepatitis C virus core protein is dispensable for virus budding but destabilizes the viral capsid. J Biol Chem. 2006;281:27679–27692. doi: 10.1074/jbc.M602587200. [DOI] [PubMed] [Google Scholar]
- 105.Heimann M, Roman-Sosa G, Martoglio B, Thiel HJ, Rumenapf T. Core protein of pestiviruses is processed at the C terminus by signal peptide peptidase. J Virol. 2006;80:1915–1921. doi: 10.1128/JVI.80.4.1915-1921.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Targett-Adams P, et al. Signal peptide peptidase cleavage of GB virus B core protein is required for productive infection in vivo. J Biol Chem. 2006;281:29221–29227. doi: 10.1074/jbc.M605373200. [DOI] [PubMed] [Google Scholar]
- 107.Loureiro J, et al. Signal peptide peptidase is required for dislocation from the endoplasmic reticulum. Nature. 2006;441:894–897. doi: 10.1038/nature04830. [DOI] [PubMed] [Google Scholar]
- 108.Sibley LD. Intracellular parasite invasion strategies. Science. 2004;304:248–253. doi: 10.1126/science.1094717. [DOI] [PubMed] [Google Scholar]
- 109.Opitz C, Soldati D. "The glideosome": a dynamic complex powering gliding motion and host cell invasion by Toxoplasma gondii. Mol Microbiol. 2002;45:597–604. doi: 10.1046/j.1365-2958.2002.03056.x. [DOI] [PubMed] [Google Scholar]
- 110.Carruthers V, Boothroyd JC. Pulling together: an integrated model of Toxoplasma cell invasion. Curr Opin Microbiol. 2006 doi: 10.1016/j.mib.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 111.Alexander DL, Arastu-Kapur S, Dubremetz JF, Boothroyd JC. Plasmodium falciparum AMA1 Binds a Rhoptry Neck Protein Homologous to TgRON4, a Component of the Moving Junction in Toxoplasma gondii. Eukaryot Cell. 2006;5:1169–1173. doi: 10.1128/EC.00040-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.McQuibban GA, Saurya S, Freeman M. Mitochondrial membrane remodelling regulated by a conserved rhomboid protease. Nature. 2003;423:537–541. doi: 10.1038/nature01633. [DOI] [PubMed] [Google Scholar]



