Abstract
Voxelation creates expression atlases by high-throughput analysis of spatially registered cubes or voxels harvested from the brain. The modality independence of voxelation allows a variety of bioanalytical techniques to be used to map abundance. Protein expression patterns in the brain can be obtained using liquid chromatography (LC) combined with mass spectrometry (MS). Here we describe the methodology of voxelation as it pertains particularly to LC-MS proteomic analysis: sample preparation, instrumental set up and analysis, peptide identification and protein relative abundance quantitation. We also briefly describe some of the advantages, limitations and insights into the brain that can be obtained using combined proteomic and transcriptomic maps.
Keywords: Brain atlas, Brain mapping, Mass spectrometry, Proteomics, Transcriptomics, Voxelation
1. Introduction
Global 3D mapping of gene expression patterns is a useful starting point to understand the molecular basis of the mouse brain. A number of fruitful approaches have been developed to achieve this goal. The most complete datasets have used in situ hybridization (ISH), radioactive and non-radioactive. The Allen Brain Atlas employed non-radioactive ISH to analyze the adult brain and covers nearly all known genes [1]. In contrast, the St Jude Brain Gene Expression Map (BGEM) used radioactive ISH to analyze both the adult and embryonic brain [2]. The BGEM database currently has about 3,300 genes. There are a number of other useful brain expression atlases available, all of which use ISH (for a recent review, see [3].)
A unique approach to mapping 3D expression patterns employs large insert mouse transgenes tagged with enhanced green fluorescent protein (EGFP). This effort, the Gene Expression Nervous System Atlas or GENSAT, has resulted in high-resolution expression images for >400 genes [4].
While ISH provides single cell resolution, the expression patterns are acquired serially. Creating these databases is therefore extremely labor intensive. In contrast, microarrays allow multiplex acquisition of gene expression levels, greatly increasing speed and decreasing expense, but at the cost of decreased spatial resolution. This technology has been used in a large number of studies to examine expression changes in various brain regions as a result of differing environments and disease models [3].
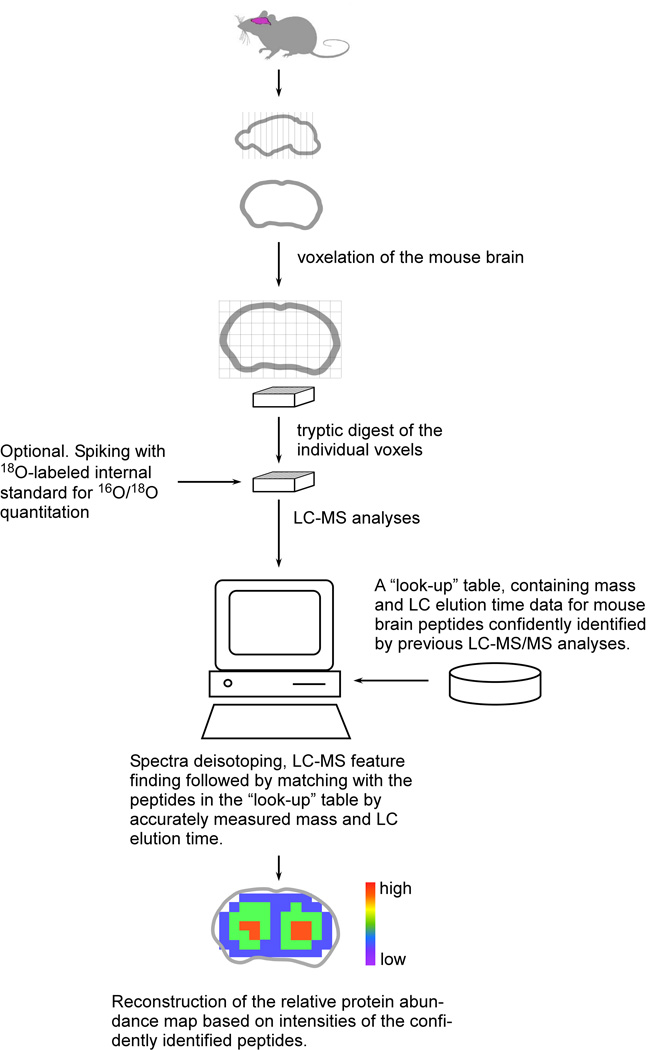
In terms of speed and resolution, voxelation can provide a compromise between high-resolution serial ISH and regional highly multiplexed microarray studies [5,6]. In the voxelation strategy, inspired by biomedical imaging technologies, spatially registered cubes or voxels are harvested from the brain. These voxels can then be analyzed using high-throughput methods and the data used in parallel to reconstruct 3D images.
One advantage of voxelation is that it is modality independent, since the voxels can be interrogated using multiple high-throughput techniques, including liquid chromatography (LC)-mass spectrometry (MS) (Fig. 1). Recently, high-throughput LC-MS combined with voxelation at a resolution of 1 mm3 has been used to obtain protein expression patterns for a coronal section of the mouse brain at the level of the striatum which had been previously studied using voxelation and microarrays [7]. Abundance maps were obtained for over a 1,000 proteins, a daunting task for immunohistochemistry. The comparability of these protein expression patterns with RNA expression patterns from the Allen Brain Atlas and GENSAT was high, ranging from 50–100% for expression patterns enriched in various brain regions. Overall, with the exception of histone proteins, the level of agreement was surprisingly robust, considering previous studies had reported much poorer levels of consistency between RNA and protein levels. The voxelation LC-MS approach hence promises opportunities to better understand the finer roles of translational control and post-translational modifications in the brain.
Fig. 1.
An experimental strategy for spatial mapping of protein abundance in the mouse brain.
Mass spectrometry is the technology of choice for parallel protein abundance measurements in complex samples due to its sensitivity, dynamic range and resolution on advanced instrumentation. The technique has been used in brain imaging studies for a number of years and can be divided into three categories: secondary ion (SIMS) based ionization coupled with a position sensitive detector or the so-called molecular microscope [8], matrix assisted laser desorption ionization (MALDI) imaging MS [9]; and the voxelation-based proteomic approach using high-throughput LC-MS.
SIMS provides very high speed data acquisition and very high spatial resolution (1 nm or so); however SIMS ionization is not very suitable for imaging of proteins and most peptides, other than light peptides, although it works well for low molecular weight compounds e.g. neurotransmitters. The spatial resolution of MADLI-MS is about 10–100 µm and is capable of detecting a few hundred chemical substances, presumably low molecular proteins and peptides.
The voxelation based proteomic approach we describe here offers relatively low spatial resolution (1 mm3); however, it has the advantages of providing extensive proteome coverage and abundance quantification by applying high-throughput LC-MS. Currently, we have mapped over 1,000 of the most abundant proteins. Given the continued advances of LC-MS for proteome profiling, the expected achievable coverage will be significantly expanded in the future.
A previous review described in depth the methodology behind voxelation mapping of transcripts in the mouse brain [5]. A general review of proteomics [10] and high mass measurement accuracy in mass spectrometry [11] can also be found elsewhere. However, there is a wide variety of proteomic applications, ranging from top-down analysis of intact proteins and post-translational modifications to biomarker discovery and targeted multiple reaction monitoring, each with their own experimental specifics. Given the rising tide of interest in mass spectrometry based imaging, we focus here on LC-MS proteomics applied to voxel based tissue imaging with emphasis on the methodological aspects of this novel application.
2. Concept of Accurate Mass and Time tag LC-MS
The main bottleneck in voxelation based imaging studies is the sample throughput of the analytical platform. The accurate mass and time (AMT) tag approach is one of the top choices when there is a need for high-throughput proteomics. The concept of the AMT tag approach is that peptide identification is based not on MS/MS fragmentation patterns, like in a classical and widely used LC-MS/MS approach, but on directly matching LC-MS features to a pre-established AMT tag database using accurately measured mass and LC elution time information [11,12].
The problem with the more common LC-MS/MS approach is that it selects only a limited number of ions from the MS spectra for MS/MS fragmentation. The typical setting ranges from the top 3 to 10 most abundant ions. However, an MS spectrum, especially in the middle of the chromatogram, may contain a significantly larger (>100) number of peptides. Thus serial MS/MS fragmentation will likely not identify all peptides visible in the MS scan, resulting in significant undersampling. This occurs even with the dynamic exclusion possible on modern automated MS/MS instrumentation, where ions are not selected for fragmentation for a period of time to avoid redundant fragmentation of the same ion type. However, dynamic exclusion requires significantly extended separation times, 2-D LC separation techniques or running the sample multiple times so that at least some low abundance peptides not previously selected for fragmentation subsequently get analyzed. In all cases the time required for sample analysis is thus increased.
In the AMT tag approach there is no need for MS/MS fragmentation, since the information from an MS scan with high mass accuracy and high-resolution together with LC elution time are sufficient for unambiguous peptide identification by matching to a pre-existing extensive AMT tag database. All peptides from an MS scan can potentially be identified, providing that the tag database approaches comprehensive coverage. This results in less need for increased separation time and dimensions, making the AMT tag approach suitable for projects requiring higher throughput.
The strategy does require effort to build an AMT tag database (or look-up table) of theoretical monoisotopic masses and observed elution times from LC-MS/MS analyses done beforehand. Such a look-up table identifies peptides by matching mass and elution times of observed LC-MS features with tabulated values. Nevertheless, building the database is required only once for similar specimen types and does not slow throughput for the bulk of samples. Furthermore, restricting the database only to peptides that are actually present in the sample as opposed to all possible peptides in the genome substantially reduces the number of false peptide identifications, permitting high data confidence.
3. Voxel sample preparation for LC-MS
3.1. Harvesting voxels
Voxelation used a two dimensional array of blades in a criss-cross pattern [5]. The blades were fabricated by photoetching from stainless steel combined with precision stack lamination. As a simple and cost effective alternative, an array of interlocking razor blades can also be used. The pixels were square with 1 mm edges. The array consisted of a total cutting area of 400 voxels, 20/20 voxels. Voxels were harvested from 1 mm thick coronal sections of the mouse brain obtained using commercially available rodent brain matrices.
The sensitivity of the LC-MS is such that the 1 mm3 voxels provided enough material for analysis. This obviated the need for pooling of voxels and permits between-animal variance to be estimated. Harvested voxels were placed into 1 ml U-bottom shape 96 deep-well plates (Beckman Coulter, Fullerton, CA) [7]. Typically, a coronal slice from the mouse brain contains 10 to 70 voxels of 1 mm3 and the entire brain 650 to 670. Voxels were stored at −80°C before analysis.
3.2. Tissue processing and trypsin digestion
Voxel samples were managed through the processing and analysis steps in 96 deep-well plates [7]. All liquid handling was performed on a Biomek FX robotic station (Beckman Coulter, Fullerton, CA). The Biomek FX robot is equipped with Span-8 and AD96 multichannel pipette heads, orbital shaker, absorbance detector and FX Stacker 10 carousel.
Due to the large number of samples, we considered a protocol with as few steps as possible to facilitate more automatable sample processing. Noteworthy, a typical sample preparation protocol for LC-MS(/MS) analysis often involves C18 solid-phase extraction sample clean-up. Due to low sample sizes and potentially significant losses we decided to avoid this step, developing a method so that all reagents were volatile and could be removed by lyophilization. This negates the use of typical reagents such as urea for denaturation and iodoacetamide for protection of free cysteine residues by alkylation. We therefore turned to a protocol based on the organic solvent 2,2,2-trifluoroethanol (TFE) for sample processing, obviating the need for further clean-up [13].
Prior to processing, voxel samples were defrosted for 15–30 min at +4°C and spun down at +4°C for 3 min at 3,000 rpm in an Eppendorf Centrifuge 5810R (Eppendorf, Hamburg, Germany) equipped with A-4–81-MTP swing bucker rotor. Samples were resuspended in 120 µl of 50% 2,2,2-trifluoroethanol (TFE) (Sigma-Aldrich) in 50 mM NH4HCO3 (pH 7.8) with 5 mM tributilphosphine (TBP) (Sigma-Aldrich) for protein reduction.
The initial choice of TBP as a reducing agent was due to its volatility; however, recently we have observed dimerized (MW of the most abundant isotope 436.3599 Da) and trimerized (MW of the most abundant isotope 654.5398 Da) oxidation products of TBP. These by-products are not volatile and co-elute with peptides during reverse phase chromatography. This partially suppresses peptide ionization at the electrospray, inhibiting detection. The other option is to use β-mercaptoethanol, which is volatile; however, it has weaker reducing power. Alternatively, DTT can be used as a reducing agent for this protocol.
Tissue homogenization was achieved by performing sonication in 50% TFE solution. For high-throughput ultrasonic homogenization we used a probe-based system, SonicMan (Matrical, Spokane, WA), with dispensable 96-pin lids designed for 96 deep-well plates. The solution volume of 120 µl was dictated by the length of the pins. A lower volume would result in the pins not being submerged deeply enough into the sample, decreasing sonication efficiency. The sonication cycle was 10 times 1 sec at 100% with 3 sec cooling intervals. We used 4 sonication cycles per plate, alternating front and back positions by turning the plate 180° between cycles.
To ensure that all voxel samples were homogenized, the plates were visually inspected and samples with non-homogenized tissue taken out, sonicated individually in ultrasonic water bath 5510 Branson (Branson Ultrasonics, Danbury, CT) and put back into the well. The same approach can be used for low-throughput homogenization, when the specimen number is relatively low. After homogenization, samples can be frozen and stored at −80°C until further processing.
For protein denaturation and reduction, samples were thawed, shaken on a Biomek FX orbital shaker for 12 sec at 600 rpm and incubated at 60°C for 2 hours following the addition of 5 mM TBP. Samples were allowed to cool down to room temperature and the foil cover switched to the air permeable membrane AirPore™ (Qiagen, Venlo, Netherlands). To reduce the amount of TFE, samples were dried down to a lowest volume of about 60 µl in a SpeedVac (ThermoFisher, Waltham, MA) equipped with a swing-bucket rotor. In our experience, TFE at high concentration inhibits trypsin activity.
After reducing the sample volume, we diluted samples to 300 µl with 50 mM NH4HCO3 (pH 7.8) supplemented with 1 mM CaCl2 and added 30 µl of 0.1 µg/µl trypsin (Promega, Madison, WI). After each step, samples were mixed by pipetting the liquid up and down. Samples were subjected to tryptic digestion by incubating overnight at 37°C with gentle shaking at 300 rpm in an Innova 4320 incubator shaker (New Brunswick Scientific, Edison, NJ). After digestion, samples can be frozen and stored at −80°C for further processing.
Samples were resuspended after lyophilization in 60 µl of 25 mM NH4HCO3. To ensure peptide solubilization, we applied three cycles of shaking on the orbital shaker for 12 sec at 1000 rpm and mixed by pipetting 10 times by 50 µl. If the samples will later be supplemented with 18O-labeled internal standard for quantitation, it is important to deactivate the trypsin. This can be achieved, for example, by incubating the samples in the boiling water bath for 10 min and then immediately cooling in ice for 10 min. Samples can be frozen at this point and stored at −80°C for further processing.
It is necessary to remove any remaining unsolubilized particles in the samples as they may clog the LC capillary columns. Particles were filtered away using 0.22 µm hydrophilic PVDF filters (Millipore, Billerica, MA). A sandwich was constructed consisting of a 96-well PVDF filter plate between two deep well plates. The sandwich was spun 3 times for 5 min at 3,900 rpm on the Eppendorf Centrifuge 5810R (Eppendorf, Hamburg, Germany) equipped with A-4–81-MTP swing bucker rotor. After this step, the cleaned samples can be frozen and stored at −80°C.
Peptide concentrations in the samples were measured with the BCA assay (Pierce, Rockford, IL). On average the peptide yield was 30 µg per 1 mm3 voxel. Concentrations were properly adjusted with 25 mM NH4HCO3 (pH 7.8) to equal levels.
Samples can optionally be supplied with equal amounts of a universal 18O-labeled internal standard for peptide quantitation based on 16O/18O ratios. The universal internal standard is a pooled voxel sample and can be obtained simply by digestion of the entire region of the brain under study. In this way virtually every peptide in any voxel sample will have an isotopically labeled counterpart. Preparation of the internal standard involved essentially the same protocol described above, followed by an 18O-labeling procedure described elsewhere [14].
4. LC-MS analysis
As is evident from its name, the Accurate Mass and Time (AMT) tag approach relies on accurate measurements of a peptide's elution time and mass. It is thus crucial to have both high-resolution LC separation and mass spectrometry instrumentation.
4.1. LC system
The high performance capillary LC separation systems with peak capacities about 500 we used are described in detail in [15]. Briefly, for mouse brain voxelation studies in one case we employed a 100 min long gradient on a 150 µm inner diameter (i.d.) × 65 cm long LC column packed with 3 µm Jupiter C18 particles (Phenomenex, Torrance, CA). The mobile phase solvents consisted of (A) 0.2% acetic acid and 0.05% TFA in water and (B) 0.1% TFA in 90% acetonitrile. An exponential gradient was used for the separation, starting with 100% A and gradually increasing to 60% B over 100 min.
With larger sample numbers, as in 3D-whole brain mapping, we used an LC system with a shorter gradient time of 35 minutes at the expense of peak capacity, which was decreased to about 200. For the faster system, we used a 75 µm i.d. × 15 cm long capillary column using the same type of packing material as above. The mobile phase solvents consisted of (A) 0.1% formic acid in water and (B) 0.1% formic acid in 90% acetonitrile. An exponential gradient was used for the separation, starting with 100% A and gradually increasing to 60% B over 35 min.
The systems were run under constant pressure of 10,000 psi in the case of the 100 min set-up and 5,000 psi in the case of the 35 min set-up. To maximize throughput, the LC carts were fully automated two- or four-column systems capable of handling the samples from the 96 deep-well plates. A detailed description of the LC system can be found elsewhere [16].
4.2. Mass spectrometry instrumentation
Typical high-resolution mass spectrometer instrumentation in the AMT tag approach is either a time-of-flight, Fourier transform (FT) ion cyclotron resonance (ICR) mass analyzer or the recently developed Orbitrap™ mass analyzer [17] based on the Kingdon trap. In particular, for mouse brain voxel samples we coupled the LC systems with either an in-house built 11-Tesla FTICR mass spectrometer [18] in the case of the 100 min gradient set-up or the commercially available LTQ-Orbitrap™ (ThermoFisher, Waltham, MA) in the case of the faster 35 min gradient set-up.
The 11-Tesla mass spectrometer is not equipped with automated gain control, which allows regulation of the number of ions in the ICR cell. Nevertheless the typical resolution for MS spectra was about 100,000. The MS spectra on the LTQ-Orbitrap™ were acquired with a resolution setting of 60,000 and automated gain control set to 5×105 charges.
Typical duty cycles for the 11-Tesla FTICR and LTQ-Orbitrap™ were 1.8 and 1.7 seconds on average, respectively. Thus, peptide elution peaks on average span 6 scans for the 100 min LC set-up coupled with the 11 Tesla instrument and 3 scans for the 35 min LC set-up coupled with the LTQ-Orbitrap™. The amount of peptide loaded on the LC column was 1.75 µg (0.875 µg sample plus 0.875 µg 18O-labeled internal standard) for 150 µm i.d. LC columns coupled with 11 Tesla instruments or 0.25 µg for 75 µm i.d. LC columns coupled with LTQ-Orbitrap™.
The throughput of the systems allowed us to analyze a single coronal slice consisting of 71 voxels on the 100 min LC gradient system coupled with the 11 Tesla ICR mass spectrometer in a one week timeframe and a whole mouse brain consisting of 664 voxels on the faster 35 min LC gradient system coupled with the LTQ-Orbitrap™ in a three week timeframe in non-stop fashion.
5. LC-MS data analysis
5.1. Picking LC-MS features
To identify peptides, we first need to detect and extract items observed in LC-MS space (Fig. 2). A single spectrum may contain hundreds of species and a single LC-MS experiment will typically result in 70,000 to 100,000 isotopic distributions for the 35 min gradient LC system coupled with the LTQ-Orbitrap™ and 25,000 to 35,000 distributions after applying the detection threshold cut-off for the 100 min gradient system coupled to the 11 Tesla FTICR.
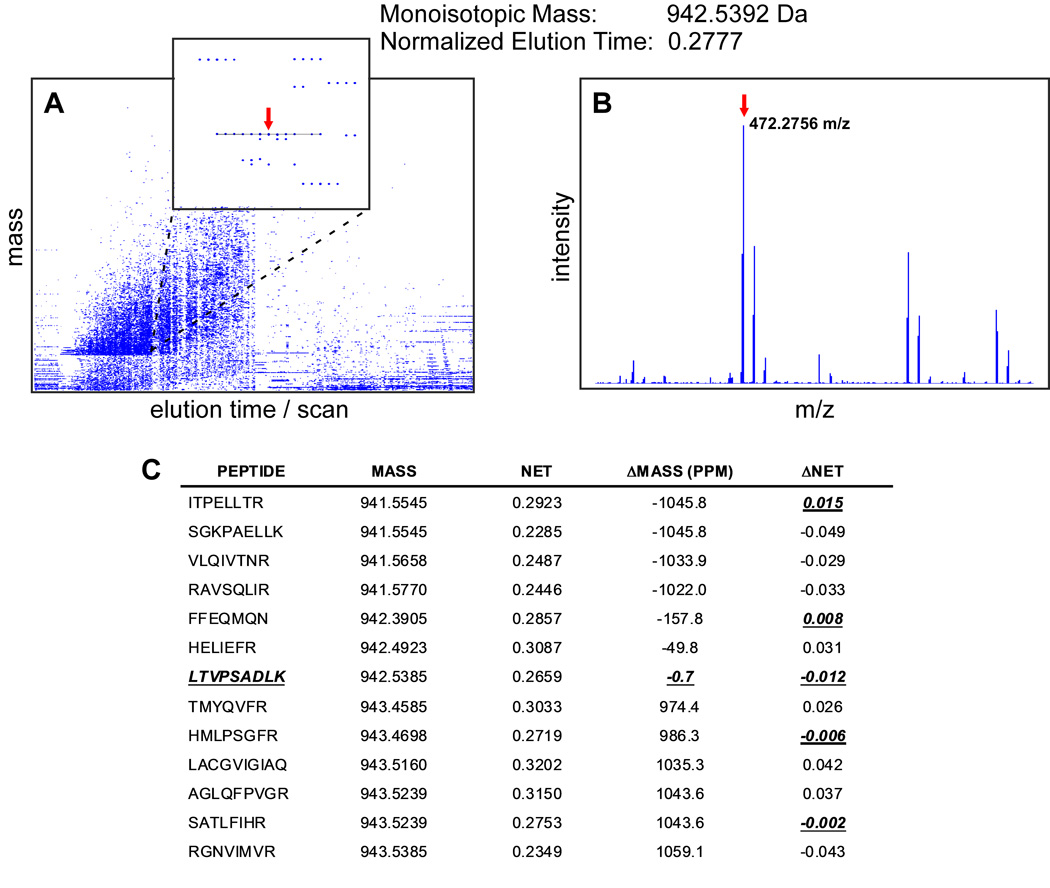
Fig. 2.
Peptide identification using the AMT tag approach. (A) Plot of mass vs. LC elution time, deisotoped spectra employed. Zoom box shows an example of an LC-MS feature highlighted with line. Estimated monoisotopic mass and normalized elution time (NET) of the feature are 942.5392 Da and 0.2777, respectively. Red arrow points to the monoisotopic mass in the middle of the feature. (B) Monoisotopic mass in the corresponding MS spectrum. (C) All peptides in the “look-up” table within 1 Da and 0.05 NET tolerance windows. There is only one peptide, LTVPSADLK, which passes both thresholds on maximum mass and NET deviations of 2 ppm and 0.02 NET, respectively. The peptide sequence can be unambiguously assigned to the protein product of the Slc6a11 gene, GABA transporter 4 protein (see Fig. 3).
Raw spectra are processed to detect isotopic distributions and determine the monoisotopic masses of the species present. The conversion of isotopic distributions to text reports including both monoisotopic masses and corresponding intensities for all detected species in each spectrum is called deisotoping. This procedure is performed by in-house developed software ICR-2LS or Decon2LS (http://ncrr.pnl.gov/software) utilizing an approach based on the THRASH algorithm [19]. More details on spectra deisotoping can be found in [11,12].
Typically, LC-MS analyses are set up so that the elution peak width of the peptide measured in seconds is at least a few-fold larger than the instrument duty cycle, the time between MS scans. In such an arrangement the peptides and other eluting species are observed over multiple consecutive scans and their intensities form the shape of an elution peak. To further reduce data dimensionality, sets of monoisotopic masses are grouped together if they are observed in sequential spectra and do not deviate from the mean cluster monoisotopic mass by greater than the user defined threshold (Fig. 2).
Each cluster, termed an LC-MS feature, has a mass estimated as a median of the monoisotopic masses in the feature, an elution time corresponding to the time of the scan with the most intense monoisotopic peak, and abundance computed as the sum of monoisotopic peak intensities. If the sample contains an 18O internal standard, 16O/18O pairs are identified using an algorithm seeking co-eluting peptides with 4.0085-Da mass difference. Feature and pair finding, as well as a number of the following steps, are performed with VIPER software [20] and are described in detail elsewhere [12].
5.2. Description of AMT database
Recognizing peptides from LC-MS feature mass and elution times employs an AMT tag database. This database or “look-up” table contains theoretical peptide monoisotopic masses and observed elution times confidently identified by previous extensive LC-MS/MS surveys.
Peptide identification is achieved by matching MS/MS spectra against peptides from a protein sequence database. We typically use the International Protein Index databases provided by the European Bioinformatics Institute (http://www.ebi.ac.uk/IPI/IPImouse.html). SEQUEST is the most widely used MS/MS search engine and performs peptide identification by matching and scoring the observed MS/MS spectra against the theoretical peptide spectra [21]. Additional software tools include Mascot [22], X!Tandem [23], OMMSA [24] and a number of others.
To assess the confidence of peptide identification or control the false discovery rate, that is the rate of incorrect peptide identifications, the same searches are done against the protein database using reversed protein sequences [25,26]. The typical false discovery rate for the peptides being included in the AMT tag databases is less than 1%.
For peptide elution time, we either used the time of the scan with the highest observed theoretical spectra matching score or extracted the peptide’s elution profiles and derived the time of the maximum elution peak with MASIC software (http://ncrr.pnl.gov/software). In the case of LC systems operated under constant pressure there is a possibility of variation of the flow rate, which may result in some systematic variations in elution times. To account for such variations, elution times from separate runs were aligned with predicted relative elution time values [27], so they scaled from 0 to 1, and termed normalized elution times (NET).
The compiled AMT tag database consists of a list of unique, confidently identified peptides with their sequences, theoretical masses and NET values averaged across the multiple runs. Typically, a database contains 10,000–40,000 unique confidently identified peptides corresponding to 3,000–10,000 proteins. A description of the AMT tag database used for the mouse brain studies can be found elsewhere [7].
5.3. Peptide identification by peak matching
As previously mentioned, peptide identification in the AMT tag approach is based on goodness of match between the experimentally observed and tabulated values of the monoisotopic mass and elution time in the AMT tag database. Thus, it is important to have very high accuracy in the measurement of both values. Measurement errors can be reduced by modeling systematic error followed by subtraction, thus leaving only random error. For example, variations in elution time caused by flow rate variation can be corrected by a warping procedure in which the elution times of the observed features are aligned to the tabulated elution times from the AMT tag database [28].
Systematic mass measurement errors can be detected and eliminated by regression analysis of the error residuals against a set of parameters like scan acquisition time, mass to charge ratio, ion intensity, total ion current and other factors [29]. Those approaches significantly reduce mass and elution time error spreads, enhancing match stringencies. The maximum allowable deviation is typically based on 2 standard deviations, thus retaining about 95% of correct matches assuming a normal distribution of error residuals. For complex samples such as mouse brain, routinely achievable maximum allowable deviations for mass and elution time measurements are about 2 ppm for FTICR or LTQ-Orbitrap™ instruments and 1–2% NET for 35 or 100 min LC system set ups.
After applying maximum allowable deviation criteria for mass and NET measurements, most LC-MS features have only one unambiguous match within the “look-up” table (Fig. 2). For example, 86.8% of LC-MS features from 664 voxel samples corresponding to the entire mouse brain had only one AMT tag matching within ±2 ppm and ±0.02 NET tolerances. From the opposite side, 97.1% of the AMT tags had only one matching LC-MS feature. Thus for the most part, LC-MS feature to peptide matching is unambiguous.
To select the most probable Mass and Time tag match for the 13.2% ambiguously matching features, we score goodness of match as a distance function. The probabilistic or Mahalanobis distance D between the observed LC-MS feature and a matching peptide from AMT tag “look-up” table is calculated based on observed differences DMass and DNET as in equation (1)
| (1) |
, where σmass and σnet are standard deviations of the error residual distributions of mass and NET measurements, respectively.
The probability density function of a true match to be at distance Di assuming a bivariate normal distribution of mass and NET measurement error residuals with no mutual correlation is given by equation (2)
| (2) |
Assuming equal a priori probability p of all peptide matches, the probability of the peptide match at distance Di to be true P(+|Di) according to Bayes' law is
| (3) |
Equal a priori probability is certainly an assumption, albeit one significantly simplifying computation. However, there are approaches for probability estimation of SEQUEST search results using empirical statistical models [30]. In addition, proposed modifications allow statistical mixture modeling to be used in other search engines such as MASCOT and X!Tandem [31].
Applying selection criteria to the maximum allowable probabilistic distance D and its rank as well as the spatially localized confidence scoring (SLiC) yields highly confident identifications from the LC-MS features. Highly confident identification typically means <5% false discovery rate, that is the estimated percentage of falsely identified LC-MS features within all identified LC-MS features after imposing the selection criteria. The false discovery rate can be estimated empirically by matching features to a database containing only false answers or to a normal database concatenated with a database containing false answers. This approach is akin to using reverse protein sequences to estimate false discovery rates for peptide identification based on MS/MS fragmentation [25,26]. To construct the AMT tag database containing only false answers we shifted all the peptide masses by an integer value. For example, peptides shifted by 11 Dalton quite closely approximate the behavior of false matches (Fig. 2 of [7]). However, such an estimate is quite similar for all tested integer shifting values from −20 Da to +20 Da (Supplementary Fig. 7 in [7]).
Recently a more sophisticated approach has been developed to estimate the probability of correct identification for individual LC-MS/peptide matches [32]. It relies on modeling distributions of true and false matches along the DMass and DNET dimensions and applying Bayesian formula to calculate the probability of correct assignment assuming equal a priori probabilities. However unlike the SLiC score this statistical model does not explicitly take into account the local density of LC-MS feature to peptide matches.
For current LC-MS platforms a typical maximum probabilistic distance criterion is about D = 2.8, that is no more than a 2 ppm mass difference and a 2% NET difference assuming a typical 1 ppm standard deviation of mass error distribution on the LTQ Orbitrap or FTICR instrument and a 1% standard deviation of NET error distribution. To avoid ambiguity in the matching, further criteria are applied such as rank of distance D (lowest distance has rank = 1) or SLiC score > 0.5, that is selecting the closest peptide match to the LC-MS feature. These criteria are quite typical for AMT tag based studies, giving reasonable confidence in LC-MS feature to peptide assignments. However, the false discovery rate in voxelation can be further reduced by imposing the requirement for a match to be present in multiple neighboring voxel samples. The latter criterion quite effectively filters out false matches due to random, but not systematic, errors that primarily appear in one voxel.
5.4. Peptide-to-protein assignment
Peptide-to-protein assignment for mammalian organisms is not straightforward. Most peptides have a sequence match to multiple proteins. For example, within 16,875 peptides identified in 664 voxel samples, only 6,618 or 39% matched a unique protein. Most (61%) peptides had multiple protein matches within the IPI database. For instance one extreme peptide, IFVGGIK, matched 58 items in the IPI database, corresponding to multiple homologs and isoforms of the heterogeneous nuclear ribonucleoprotein.
A conservative strategy would be to retain only those peptides unambiguously matching protein sequences. However such an approach results in most identified peptides being ignored in follow up quantitation. A more sophisticated strategy is to group highly homologous proteins, indistinguishable by the bottom-up proteomic approach, into one entity or protein group. It is then possible to try and derive the most likely protein set based on the number of peptides matching a protein, the presence of unambiguous matches and the combination of each. Both the latter tasks, that is grouping of indistinguishable proteins and assigning probability values, can be performed by adopting the Protein Prophet software [33].
5.5. Quantitating protein relative abundances
5.5.1. Sample-to-sample normalization
For peptide quantitation we used either LC-MS feature abundances directly or their ratios to the corresponding paired features in the 18O-labeled internal standard. To remove systematic errors arising from technical issues, it is necessary to perform a peptide intensity or 16O/18O ratio normalization procedure between LC-MS runs. We believe that most systematic biases arise from differences in ESI tip-orifice alignment, inaccuracies during liquid handling and imprecision originating from sample concentration measurement. For the most part these biases can be corrected by multiplying the observed peptide intensities Pij (j = 1,…, J) for a given LC-MS run i (i = 1,…, I) by a constant factor Fi.
To estimate the factor F we assumed that most peptides do not change in abundance and only a minority change from sample to sample. In computing the factor, we used only peptides observed across > 95% of runs. First, we calculated the grand median peptide abundance Mj = mediani(Pij) for all selected peptides across all LC-MS datasets. Missing values were simply ignored in this calculation. For each LC-MS dataset we then calculated the factor Fi = median(Pij/Mi), that is, the magnitude by which all peptide intensities in a given dataset are above or below their median values. All peptide abundance values were then normalized by dividing by the derived factor Fi.
5.5.2. Adjustment of peptide intensities and 16O/18O ratios
It is known that peptides originating from the same protein may be observed with different intensities due to variations in ionization efficiencies. It is also true that divergent efficiency of 18O trypsin catalyzed labeling may cause systematic biases in 16O/18O ratios. In the next step we applied a simple procedure in an attempt to correct these errors.
Peptide abundance profiles originating from the same protein were adjusted to an approximately equal level using multiplication by a peptide-specific factor. A peptide observed across the most voxels was picked as a reference peptide for a given protein.
To adjust abundances of another peptide, we first calculated the ratios of abundances of the given peptide to the reference peptide and then divided its abundances across all datasets by the median ratio value. After this adjustment, different peptides corresponding to the same protein follow not only the same abundance pattern but also have approximately equal abundance values. An arbitrary protein abundance was calculated as the median of its adjusted peptide abundances. Such an approach based on a reference peptide along with others has been recently implemented in DAnTE software (http://ncrr.pnl.gov/software).
5.5.3. Deriving protein abundance patterns
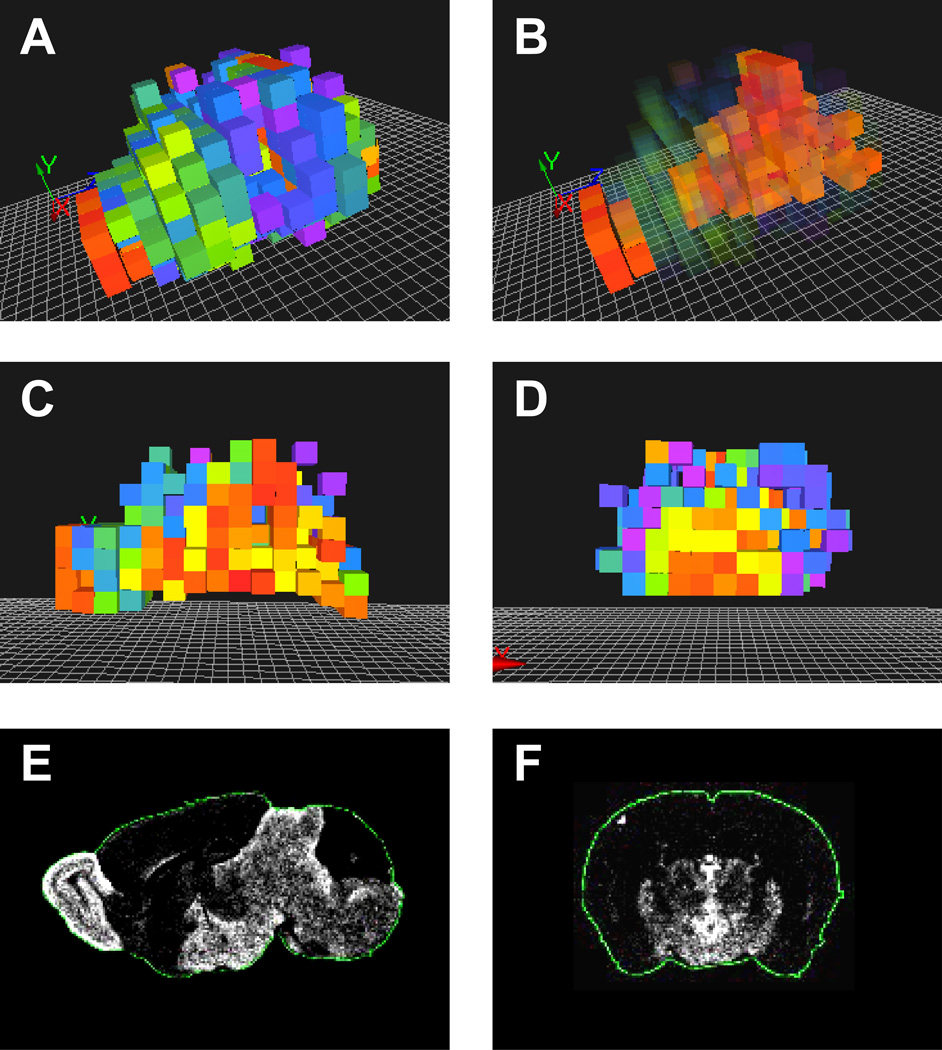
An abundance pattern for a given protein was finally generated by dividing the arbitrary protein abundances by its median protein abundance across voxels. Missing peptide and protein abundance values were simply ignored. For convenience of visualization, the derived patterns can be log2 transformed. An example of the 3D protein abundance pattern for the GABA transporter 4 protein, encoded by the Slc6a11 gene, is shown in Fig. 3.
Fig. 3.
Spatial distribution of GABA transporter 4 protein (Slc6a11 gene) depicted as (A) general 3D view, (B) view with increased transparency for voxels with lower protein abundances, (C) midline sagittal section view, (D) coronal section view through hypothalamus. Voxelated protein abundance patterns were visualized with custom software written in Python language and using OpenGL library for 3D rendering. The protein abundance agrees well with the mRNA distribution retrieved from the Allen Brain Atlas (http://www.brain-map.org) shown as (E) sagittal and (F) coronal sections views.
6. Concluding remarks
Voxelation coupled with LC-MS provides a convenient discovery-driven approach to image the brain proteome in the adult and in development and to understand the etiology of neurodegenerative and other brain disorders. We expect the greatest benefits in applying the approach when little is know about the disease, particularly the relevant molecular pathways and affected brain regions. Voxelation followed by LC-MS analysis offers an approach to interrogating both domains at the same time.
Brain structures can certainly be non-destructively imaged by CT, PET or MRI. However, even if there are no detectable alterations in brain structure at the morphological or metabolic level, there may be considerable changes detectable at the molecular or protein level. There are also advantages in studying the protein content of individual voxels rather than in the entire brain or dissected brain regions. For example, if a change was restricted to only a small part of a brain area, it would be more difficult to detect using the entire brain or region. The ability to quantify relative abundance in spatially localized regions also gives additional improvements in dynamic range.
There are many brain disorders about which our knowledge is rudimentary, ranging from schizophrenia to autism. The reported incidence rates of brain disorders affecting children and especially seniors have been increasing over the last few decades and are now approaching epidemic levels. In this situation it is important to continue developing novel approaches to studying the normal and malfunctioning brain at the molecular level.
Acknowledgements
We thank The Allen Institute for Brain Science for permission to use their data in this publication. Portions of the research were supported by the NIH National Center for Research Resources (RR18522 to R.D.S.) and NIH grant R01 NS050148 to D.J.S. Proteomic analyses were performed in the Environmental Molecular Sciences Laboratory, a US Department of Energy (DOE) national scientific user facility located at the Pacific Northwest National Laboratory (PNNL) in Richland, Washington. PNNL is a multi-program national laboratory operated by Battelle Memorial Institute for the DOE under Contract DE-AC05-76RL01830.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lein ES, Hawrylycz MJ, Ao N, et al. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 2.Magdaleno S, Jensen P, Brumwell CL, et al. PLoS Biol. 2006;4:e86. doi: 10.1371/journal.pbio.0040086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sunkin SM, Hohmann JG. Hum Mol Genet. 2007;16:R209–R219. doi: 10.1093/hmg/ddm183. Spec No. 2. [DOI] [PubMed] [Google Scholar]
- 4.Gong S, Zheng C, Doughty ML, et al. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- 5.Liu D, Smith DJ. Methods. 2003;31:317–325. doi: 10.1016/s1046-2023(03)00162-2. [DOI] [PubMed] [Google Scholar]
- 6.Singh RP, Smith DJ. Biol Psychiatry. 2003;53:1069–1074. doi: 10.1016/s0006-3223(03)00238-5. [DOI] [PubMed] [Google Scholar]
- 7.Petyuk VA, Qian WJ, Chin MH, et al. Genome Res. 2007;17:328–336. doi: 10.1101/gr.5799207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McDonnell LA, Heeren RM. Mass Spectrom Rev. 2007;26:606–643. doi: 10.1002/mas.20124. [DOI] [PubMed] [Google Scholar]
- 9.Reyzer ML, Caprioli RM. Curr Opin Chem Biol. 2007;11:29–35. doi: 10.1016/j.cbpa.2006.11.035. [DOI] [PubMed] [Google Scholar]
- 10.Aebersold R, Mann M. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 11.Liu T, Belov ME, Jaitly N, et al. Chem Rev. 2007;107:3621–3653. doi: 10.1021/cr068288j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zimmer JS, Monroe ME, Qian WJ, et al. Mass Spectrom Rev. 2006;25:450–482. doi: 10.1002/mas.20071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang H, Qian WJ, Mottaz HM, et al. J Proteome Res. 2005;4:2397–2403. doi: 10.1021/pr050160f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qian WJ, Monroe ME, Liu T, et al. Mol Cell Proteomics. 2005;4:700–709. doi: 10.1074/mcp.M500045-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen Y, Smith RD. Electrophoresis. 2002;23:3106–3124. doi: 10.1002/1522-2683(200209)23:18<3106::AID-ELPS3106>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 16.Livesay EA, Tang K, Taylor BK, et al. Anal Chem. 2008;80:294–302. doi: 10.1021/ac701727r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scigelova M, Makarov A. Proteomics. 2006;6 Suppl 2:16–21. doi: 10.1002/pmic.200600528. [DOI] [PubMed] [Google Scholar]
- 18.Gorshkov MV, Pasa Tolic L, Udseth HR, et al. J Am Soc Mass Spectrom. 1998;9:692–700. doi: 10.1016/s1044-0305(98)00037-3. [DOI] [PubMed] [Google Scholar]
- 19.Horn DM, Zubarev RA, McLafferty FW. J Am Soc Mass Spectrom. 2000;11:320–332. doi: 10.1016/s1044-0305(99)00157-9. [DOI] [PubMed] [Google Scholar]
- 20.Monroe ME, Tolic N, Jaitly N, et al. Bioinformatics. 2007;23:2021–2023. doi: 10.1093/bioinformatics/btm281. [DOI] [PubMed] [Google Scholar]
- 21.Yates JR, 3rd, Eng JK, McCormack AL, et al. Anal Chem. 1995;67:1426–1436. doi: 10.1021/ac00104a020. [DOI] [PubMed] [Google Scholar]
- 22.Perkins DN, Pappin DJ, Creasy DM, et al. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 23.Fenyo D, Beavis RC. Anal Chem. 2003;75:768–774. doi: 10.1021/ac0258709. [DOI] [PubMed] [Google Scholar]
- 24.Geer LY, Markey SP, Kowalak JA, et al. J Proteome Res. 2004;3:958–964. doi: 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
- 25.Qian WJ, Liu T, Monroe ME, et al. J Proteome Res. 2005;4:53–62. doi: 10.1021/pr0498638. [DOI] [PubMed] [Google Scholar]
- 26.Elias JE, Gygi SP. Nat Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 27.Petritis K, Kangas LJ, Yan B, et al. Anal Chem. 2006;78:5026–5039. doi: 10.1021/ac060143p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jaitly N, Monroe ME, Petyuk VA, et al. Anal Chem. 2006;78:7397–7409. doi: 10.1021/ac052197p. [DOI] [PubMed] [Google Scholar]
- 29.Petyuk VA, Jaitly N, Moore RJ, et al. Anal Chem. 2008;80:693–706. doi: 10.1021/ac701863d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keller A, Nesvizhskii AI, Kolker E, et al. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 31.Choi H, Ghosh D, Nesvizhskii AI. J Proteome Res. 2008;7:286–292. doi: 10.1021/pr7006818. [DOI] [PubMed] [Google Scholar]
- 32.May D, Liu Y, Law W, et al. J Proteome Res. 2008;7:5148–5156. doi: 10.1021/pr8004502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nesvizhskii AI, Keller A, Kolker E, et al. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]