Abstract
In the past decade, the primary fungal pathogen Cryptococcus gattii has evolved and adapted to the temperate climate of the Pacific Northwest region of North America. This pathogen is now endemic and an increasingly common cause of life-threatening pulmonary and central nervous system infections that are difficult to manage and, in some cases, fatal to humans and other mammals throughout the region. A series of recent reports provide evidence that evolutionary, climatic, and anthropogenic factors may be causing the expansion of the Vancouver Island outbreak genotype into the United States, with the concomitant emergence of a unique genotype in the state of Oregon. Ongoing studies address the molecular epidemiology, roles of mating and genetic exchange, and geographic origins of this unprecedented outbreak of fungal infection of considerable public health magnitude.
Introduction and context
During the past decade (1999 to 2009) Cryptococcus gattii has emerged as a primary pathogen in western North America, including both Canada and the United States [1-4]. This novel emergence was unexpected based on the previous evidence that this pathogen, unlike Cryptococcus neoformans, was geographically restricted to tropical and subtropical regions throughout the world [5-8]. C. gattii can be classified into four discrete molecular types (Table 1), which represent cryptic species as no nuclear allelic exchange between groups has been observed [9,10]. This molecular classification is of fundamental importance. Of the four molecular types (VGI to VGIV), only one (VGII) is responsible for approximately 95% of all human and animal infections associated with the Vancouver Island outbreak and the subsequent expansion into the United States [1,3,10-14].
Table 1. Cryptococcus gattii molecular types.
| Species | Serotype | Molecular type | Description |
|---|---|---|---|
| C. gattii | B | VGI | Most common clinically, highly clonal |
| VGII | Responsible for Pacific NW Vancouver Island outbreak and Northwestern United States outbreak | ||
| C | VGIII | Highly fertile, more common in HIV+ patients | |
| VGIV | More common in HIV+ patients, rarely found |
A central question from the analysis of the Vancouver Island outbreak relates to the origin of a novel genotype, VGIIa/major, which is responsible for the vast majority of all infections reported in British Columbia [1,15]. As in the sibling species C. neoformans, many C. gattii populations are predominantly composed of α mating type isolates, and, to date, all isolates related to the outbreak have been exclusively α [10]. A seminal finding by Lin and colleagues in 2005 was the discovery that α-α monokaryotic fruiting represents a novel mode of sexual reproduction (including meiosis) [16-18]. This finding, in combination with the discovery of an α/α VGIIa/major diploid isolate from Vancouver Island (RB59), and molecular comparisons between the VGIIa/major genotype and the less prevalent VGIIb/minor genotype that is also found in Australia, led to the hypothesis that same-sex mating may have produced the hypervirulent VGIIa/major genotype and may be responsible for ongoing production of infectious spores [10]. An alternative hypothesis is that opposite-sex mating, possibly in South America where a isolates similar in genotype have been discovered [19], gave rise to the outbreak isolate genotype [10]. Mating of C. neoformans and C. gattii can be stimulated by plants or plant materials under laboratory conditions and may represent environmental niches in which sexual reproduction may occur [20,21]. In addition, the VGIIa/major subgroup has been shown to be more fertile in comparison to the VGIIb/minor subgroup [22]. While the origins of C. gattii VGIIa/major in North America remain elusive, it is clear that this emerging pathogen has invaded the United States, and that, in addition, a new unique United States genotype has arisen [11,12].
Major recent advances
In 2007 and 2008, the first reports of C. gattii in the Pacific Northwest were published. The report of Upton and colleagues [23] illustrated the first confirmed case of the Vancouver Island outbreak VGIIa/major genotype in the United States (2006) from a patient in Puget Sound, Washington, and MacDougall and colleagues [3] discovered related C. gattii VGII genotypes in the United States in 2005, including one later recognized as a VGIIc isolate. These studies prompted an increased surveillance in Washington and Oregon, and retrospective studies concluded that these were most likely the sentinel cases in the region [23]. Subsequent to these reports, the community has witnessed dozens of cases in the United States, all occurring after 2006 [11,13,14], with mortality in humans reaching levels over 25% (KA Marr and S West, unpublished observations). Beyond mortality, these infections are difficult to manage, prolonged, and a cause of significant morbidity. In addition to human cases, there has also been mortality among terrestrial companion, agrarian, and wild animals in the United States, as well as cases in wild marine mammals [11,12].
The dynamics of emerging outbreaks are often multifaceted, particularly in sexual pathogens, requiring in-depth molecular typing methods, including multilocus sequence typing (MLST), variable number of tandem repeats typing, and epidemiological analysis [24-27]. From a molecular epidemiological perspective, there were two recent findings of import regarding the specific outbreak dynamics in the Pacific Northwest region of the United States. The first finding is clear evidence that the range of the hypervirulent VGIIa/major genotype has expanded from British Columbia, Canada into the United States [11]. The second development is the discovery of a novel VGII genotype, VGIIc, found thus far exclusively in Oregon, occurring in both humans and other animals [11]. In addition to being geographically restricted, recent evidence suggests that this genotype, similar to VGIIa/major, is hypervirulent (EJB and JH, unpublished observations).
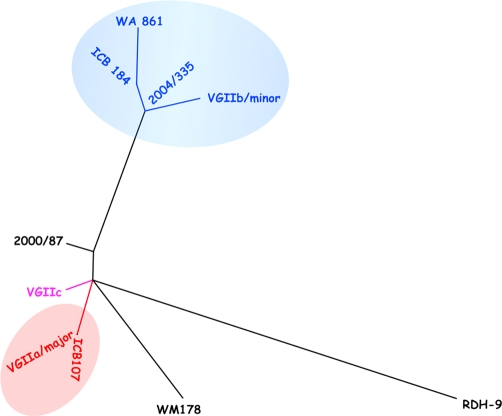
Analysis of the current molecular data suggests that the emergence in the United States may be more complicated than originally thought. The apparent increase in diversity seen in the US indicates that evolutionary forces may be causing an increase in diversity. Phylogenetic analysis using maximum likelihood analysis at seven MLST loci suggests that the novel VGIIc genotype is distinct but closely related to the VGIIa/major genotype when compared with several global VGII genotypes (Figure 1) [10,11]. While closely related, this novel genotype has never been found on Vancouver Island, even though this is one of the most sampled areas globally. This genotype may have originated elsewhere, or alternatively arose locally in Oregon via a genetic cross or a mutational process. This line of evidence leaves open the question of origin, and is a logical point of investigation for future studies.
Figure 1. Phylogeny of C. gattii VGII genotypes in the United States.
This unrooted maximum likelihood tree is based on concatenated sequences from the following seven genes that are commonly used in MLST analysis of C. gattii: IGS, TEF1, GPD1, LAC1, CAP10, PLB1, and MPD1. All corresponding sequences and associated GenBank accession numbers can be found in [10,11]. The blue shaded group includes the VGIIb/minor and closely related genotypes, while the red shaded group indicates the VGIIa/major genotype. Pink coloration indicates the VGIIc novel genotype, which is closely related to the VGIIa/major genotype based on this analysis. Sequence alignments were conducted using the Clustal W software package (http://www.ebi.ac.uk/Tools/clustalw2/index.html), and the maximum likelihood tree was constructed using the PhyML software package (http://www.atgc-montpellier.fr/phyml/).
Future directions
While aspects of the dynamics of the evolution of VGII in the United States remain unclear, recent population genetic and molecular studies of other C. gattii populations have yielded fundamental insights into hypotheses that should be explored in the US population. While isolates of C. gattii have been shown to undergo a-α mating under laboratory conditions [28], no a isolates have been found in the Pacific Northwest outbreak, making same-sex mating a more parsimonious explanation for the origins of novel isolates and the production of infectious spores, although no fruiting of C. gattii has been reported under defined laboratory conditions. Two recent studies from Australian populations, one clinical and one environmental, provide evidence that same-sex mating is occurring within these C. gattii and C. neoformans populations [25,26], and similar population-based approaches should be employed in the Washington and Oregon C. gattii populations to examine the possibility of ongoing recombination.
Another recent line of evidence suggests that mitochondrial inheritance and recombination could play significant roles in the evolution of C. gattii [29,30]. Mitochondrial recombination or exchange requires cell-cell fusion, and it is hypothesized that this type of event could also lead to other nuclear or plasmid genetic exchange. Detailed examination of mitochondrial genes in the outbreak population may help to illustrate levels of fusion and possible genetic recombination among this α VGII population. Overall, both population genetic and molecular studies focused on the United States C. gattii population will help establish the roles genetic exchange may have played in virulence acquisition and adaptive evolution to novel environments.
Acknowledgments
We thank Wenjun Li and Yonathan Lewit for critical reading. These studies were supported by the National Institutes of Health/National Institute of Allergy and Infectious Diseases (R01 grant AI39115).
Abbreviation
- MLST
multilocus sequence typing
Competing interests
The authors declare that they have no competing interests.
The electronic version of this article is the complete one and can be found at: http://F1000.com/Reports/Biology/content/1/62
References
- 1.Kidd SE, Hagen F, Tscharke RL, Huynh M, Bartlett KH, Fyfe M, Macdougall L, Boekhout T, Kwon-Chung KJ, Meyer W. A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada) Proc Natl Acad Sci U S A. 2004;101:17258–63. doi: 10.1073/pnas.0402981101. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 3.0 RecommendedEvaluated by Arturo Casadevall 08 Dec 2004
- 2.Stephen C, Lester S, Black W, Fyfe M, Raverty S. Multispecies outbreak of cryptococcosis on southern Vancouver Island, British Columbia. Can Vet J. 2002;43:792–4. [PMC free article] [PubMed] [Google Scholar]
- 3.MacDougall L, Kidd SE, Galanis E, Mak S, Leslie MJ, Cieslak PR, Kronstad JW, Morshed MG, Bartlett KH. Spread of Cryptococcus gattii in British Columbia, Canada, and detection in the Pacific Northwest, USA. Emerg Infect Dis. 2007;13:42–50. doi: 10.3201/eid1301.060827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duncan C, Schwantje H, Stephen C, Campbell J, Bartlett K. Cryptococcus gattii in wildlife of Vancouver Island, British Columbia, Canada. J Wildlife Dis. 2006;42:175–8. doi: 10.7589/0090-3558-42.1.175. [DOI] [PubMed] [Google Scholar]
- 5.Sorrell TC. Cryptococcus neoformans variety gattii. Med Mycol. 2001;39:155–68. [PubMed] [Google Scholar]
- 6.Kwon-Chung KJ, Bennett JE. Epidemiologic differences between the two varieties of Cryptococcus neoformans. Am J Epidemiol. 1984;120:123–30. doi: 10.1093/oxfordjournals.aje.a113861. [DOI] [PubMed] [Google Scholar]
- 7.Kwon-Chung KJ, Bennett JE. High prevalence of Cryptococcus neoformans var. gattii in tropical and subtropical regions. Zentralblatt Bakteriologie Mikrobiologie Hygiene. 1984;257:213–8. [PubMed] [Google Scholar]
- 8.Casadevall A, Perfect J. Cryptococcus neoformans. Washington DC: ASM Press; 1984. [Google Scholar]
- 9.Bovers M, Hagen F, Kuramae EE, Boekhout T. Six monophyletic lineages identified within Cryptococcus neoformans and Cryptococcus gattii by multi-locus sequence typing. Fungal Genet Biol. 2008;45:400–21. doi: 10.1016/j.fgb.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 10.Fraser JA, Giles SS, Wenink EC, Geunes-Boyer SG, Wright JR, Diezmann S, Allen A, Stajich JE, Dietrich FS, Perfect JR, Heitman J. Same-sex mating and the origin of the Vancouver Island Cryptococcus gattii outbreak. Nature. 2005;437:1360–4. doi: 10.1038/nature04220. [DOI] [PubMed] [Google Scholar]
- 11.Byrnes EJ, 3rd, Bildfell RJ, Frank SA, Mitchell TG, Marr KA, Heitman J. Molecular evidence that the range of the Vancouver Island outbreak of Cryptococcus gattii infection has expanded into the Pacific Northwest in the United States. J Infect Dis. 2009;199:1081–6. doi: 10.1086/597306. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6.0 Must ReadEvaluated by David Goldman 20 Mar 2009
- 12.Byrnes EJ, 3rd, Bildfell RJ, Dearing PL, Valentine BA, Heitman J. Cryptococcus gattii with bimorphic colony types in a dog in western Oregon: additional evidence for expansion of the Vancouver Island outbreak. J Vet Diagn Invest. 2009;21:133–6. doi: 10.1177/104063870902100122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Datta K, Bartlett K, Baer R, Byrnes EJ, 3rd, Galanis E, Heitman J, Hoang L, Leslie M, MacDougall L, Magill S, Morshed MG, Marr KA. Cryptococcus gattii: an emerging pathogenic fungus in Western North America. Emerg Infect Dis. 2009 [Epub ahead of print] [Google Scholar]
- 14.Bartlett KH, Kidd SE, Kronstad JW. The emergence of Cryptococcus gattii in British Columbia and the Pacific Northwest. Curr Infect Dis Rep. 2008;10:58–65. doi: 10.1007/s11908-008-0011-1. [DOI] [PubMed] [Google Scholar]
- 15.Kidd SE, Guo H, Bartlett KH, Xu J, Kronstad JW. Comparative gene genealogies indicate that two clonal lineages of Cryptococcus gattii in British Columbia resemble strains from other geographical areas. Eukaryot Cell. 2005;4:1629–38. doi: 10.1128/EC.4.10.1629-1638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin X, Patel S, Litvintseva AP, Floyd A, Mitchell TG, Heitman J. Diploids in the Cryptococcus neoformans serotype A population homozygous for the alpha mating type originate via unisexual mating. PLoS Pathog. 2009;5:e1000283. doi: 10.1371/journal.ppat.1000283. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 3.0 RecommendedEvaluated by Gerhard Braus 09 Feb 2009
- 17.Lin X, Hull CM, Heitman J. Sexual reproduction between partners of the same mating type in Cryptococcus neoformans. Nature. 2005;434:1017–21. doi: 10.1038/nature03448. [DOI] [PubMed] [Google Scholar]; F1000 Factor 4.8 Must ReadEvaluated by June Kwon-Chung 13 May 2005, John True 18 May 2005
- 18.Lin X, Litvintseva AP, Nielsen K, Patel S, Floyd A, Mitchell TG, Heitman J. alpha AD alpha hybrids of Cryptococcus neoformans: evidence of same-sex mating in nature and hybrid fitness. PLoS Genet. 2007;3:1975–90. doi: 10.1371/journal.pgen.0030186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Escandon P, Sanchez A, Martinez M, Meyer W, Castaneda E. Molecular epidemiology of clinical and environmental isolates of the Cryptococcus neoformans species complex reveals a high genetic diversity and the presence of the molecular type VGII mating type a in Colombia. FEMS Yeast Res. 2006;6:625–35. doi: 10.1111/j.1567-1364.2006.00055.x. [DOI] [PubMed] [Google Scholar]
- 20.Botes A, Boekhout T, Hagen F, Vismer H, Swart J, Botha A. Growth and mating of Cryptococcus neoformans var. grubii on woody debris. Microb Ecol. 2009;57:757–65. doi: 10.1007/s00248-008-9452-1. [DOI] [PubMed] [Google Scholar]
- 21.Xue C, Tada Y, Dong X, Heitman J. The human fungal pathogen Cryptococcus can complete its sexual cycle during a pathogenic association with plants. Cell Host Microbe. 2007;1:263–73. doi: 10.1016/j.chom.2007.05.005. [DOI] [PubMed] [Google Scholar]; F1000 Factor 3.0 RecommendedEvaluated by Guilhem Janbon 06 Jul 2007
- 22.Ngamskulrungroj P, Sorrell TC, Chindamporn A, Chaiprasert A, Poonwan N, Meyer W. Association between fertility and molecular sub-type of global isolates of Cryptococcus gattii molecular type VGII. Med Mycol. 2008;46:665–73. doi: 10.1080/13693780802210734. [DOI] [PubMed] [Google Scholar]
- 23.Upton A, Fraser JA, Kidd SE, Bretz C, Bartlett KH, Heitman J, Marr KA. First contemporary case of human infection with Cryptococcus gattii in Puget Sound: evidence for spread of the Vancouver Island outbreak. J Clin Microbiol. 2007;45:3086–8. doi: 10.1128/JCM.00593-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heitman J. Sexual reproduction and the evolution of microbial pathogens. Curr Biol. 2006;16:R711–25. doi: 10.1016/j.cub.2006.07.064. [DOI] [PubMed] [Google Scholar]
- 25.Saul N, Krockenberger M, Carter D. Evidence of recombination in mixed-mating-type and alpha-only populations of Cryptococcus gattii sourced from single eucalyptus tree hollows. Eukaryot Cell. 2008;7:727–34. doi: 10.1128/EC.00020-08. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 3.0 RecommendedEvaluated by Joe Heitman 22 May 2008
- 26.Bui T, Lin X, Malik R, Heitman J, Carter D. Isolates of Cryptococcus neoformans from infected animals reveal genetic exchange in unisexual, alpha mating type populations. Eukaryot Cell. 2008;7:1771–80. doi: 10.1128/EC.00097-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Byrnes EJ, 3rd, Li W, Lewit Y, Perfect JR, Carter D, Cox GM, Heitman J. First reported case of Cryptococcus gattii in the Southeastern USA: Implications for travel-associated acquisition of an emerging pathogen. PLoS ONE. 2009;4:e5851. doi: 10.1371/journal.pone.0005851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fraser JA, Subaran RL, Nichols CB, Heitman J. Recapitulation of the sexual cycle of the primary fungal pathogen Cryptococcus neoformans var. gattii: implications for an outbreak on Vancouver Island, Canada. Eukaryot Cell. 2003;2:1036–45. doi: 10.1128/EC.2.5.1036-1045.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bovers M, Hagen F, Kuramae EE, Boekhout T. Promiscuous mitochondria in Cryptococcus gattii. FEMS Yeast Res. 2009;9:489–503. doi: 10.1111/j.1567-1364.2009.00494.x. [DOI] [PubMed] [Google Scholar]
- 30.Xu J, Yan Z, Guo H. Divergence, hybridization, and recombination in the mitochondrial genome of the human pathogenic yeast Cryptococcus gattii. Mol Ecol. 2009 doi: 10.1111/j.1365-294X.2009.04227.x. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]