Abstract
The undisputed role of His64 in proton transfer during catalysis by carbonic anhydrases in the α class has raised questions concerning the details of its mechanism. The highly conserved residues Tyr7, Asn62, and Asn67 in the active-site cavity function to fine tune the properties of proton transfer by human carbonic anhydrase II (HCA II). For example, hydrophobic residues at these positions favor an inward orientation of His64 and a low pKa for its imidazole side chain. It appears that the predominant manner in which this fine tuning is achieved in rate constants for proton transfer is through the difference in pKa between His64 and the zinc-bound solvent molecule. Other properties of the active-site cavity, such as inward and outward conformers of His64, appear associated with the change in ΔpKa; however, there is no strong evidence to date that the inward and outward orientations of His64 are in themselves requirements for facile proton transfer in carbonic anhydrase.
Keywords: carbonic anhydrase, proton transfer, carbon dioxide, bicarbonate, hydration
An important advance in understanding catalysis by carbonic anhydrase came in the report in 1975 from Steiner, Jonsson, and Lindskog [1] who used hydrogen/deuterium isotope effects to deduce a rate-limiting, intramolecular proton transfer in the maximum velocity of catalysis by human carbonic anhydrase II (HCA II). They suggested that His64 was the likely shuttle group based on its position in the active site and its pKa near 7 that was consistent with the kinetic pKa of kcat. Direct evidence that this suggestion was correct came 14 years later; it was the reduction in maximal velocity by about a factor of 20 caused by the replacement of His64 in HCA II with Ala which cannot support proton transfer [2]. Moreover, maximal velocity catalyzed by the mutant H64A HCA II was rescued to levels close to that of the wild-type enzyme by imidazole buffer in solution [2].
Because of its rate-limiting, intramolecular proton transfer in catalysis, carbonic anhydrase has become a model for the examination of long range proton transfers and the role of hydrogen-bonded water networks [1, 3]. This is a step apparently shared by the numerous carbonic anhydrases classified into genetically distinct classes, such as α, β, and γ [4, 5], all of which carry out catalysis of CO2 hydration in a two-step mechanism. The first stage in the hydration direction is the reaction of zinc-bound hydroxide with carbon dioxide to form bicarbonate (eq 1). A water molecule subsequently displaces the bicarbonate at the metal shown in eq (1).
| (1) |
| (2) |
The second stage is the rate limiting proton transfer to regenerate the zinc-bound hydroxide (eq 2). The zinc-bound water transfers a proton to an exogenous proton acceptor or a residue of the extended active site represented by B in eq (2); the proton is ultimately removed to the bulk solvent in the hydration direction. Human carbonic anhydrase II, HCA II, the most studied of the enzymes of the α class, is emphasized in this review. Catalysis of the hydration of CO2 by HCA II is sustained at a maximal catalytic turnover of 1 µs−1, limited by proton transfer between the zinc-bound solvent and proton shuttle residue His64, represented as B in eq (2), when in the presence of excess external buffer.
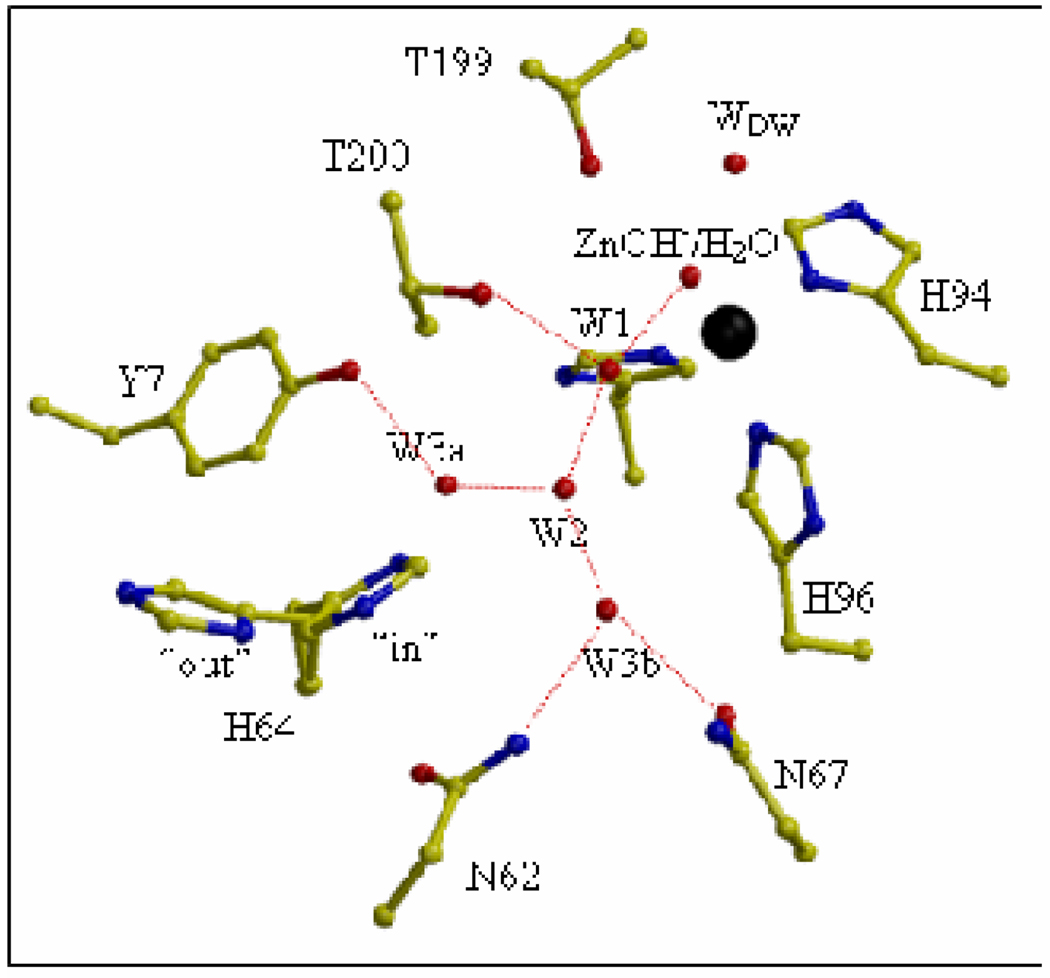
HCA II is monomeric, near 30 kDa molecular weight, and contains a catalytic zinc ion that is located at the bottom of a conical cavity about 15 Å deep, with tetrahedral coordination by three histidine residues (94, 96, and 119) and a highly polarized water molecule or hydroxyl group (Figure 1). In crystal structures, the active-site cavity is occupied with a hydrogen-bonded chain of ordered water molecules labeled W1, W2, W3a, and W3b leading out of the cavity toward the residue His64 (Figure 1). The cavity contains a region of hydrophobic residues (Val121, Val143, Leu198, and Trp209) which form a binding site for carbon dioxide that allows facile reaction with the zinc-bound hydroxide [6, 7]. Leading out of the cavity is a patch of more hydrophilic residues (Tyr7, Asn62, His64, Ans67, Thr199 and Thr200).
Figure 1.
The active site of HCA II in a ball-and-stick diagram taken from ref [15]. The zinc ion and the oxygen molecules of water molecules are shown as black and red spheres, respectively. The water network of the active-site is labeled W1, W2, etc. and WDW is the deep water. Hydrogen bonds involving solvent molecules are represented as dashed red lines. The dual conformation of the His64 side chain is shown in both inward and outward conformations.
Nair and Christianson [8] deduced from the crystal structure of HCA II that the side chain of His64 exists in two conformations related by a rotation about the Cα-Cβ side-chain angle χ1. These are referred to as the inward and outward conformations. In the inward orientation the imidazole of the side chain points toward the zinc (Figure 1)(χ1 and χ2 are 47° and 99° [9]); this is the predominant conformer at pH near 8.5 [8]. At pH 5.7 the outward conformer predominates (χ1 and χ2 are −55° and 111° [9]) in which the imidazole of the side chain points out of the active-site cavity near the hydrophobic residues Trp5, Tyr7, and Phe231. These inward and outward orientations of the side chain of His64 have been observed in the crystal structures of many variants of HCA II and, although the precise conformers differ somewhat in angles χ1 and χ2, for our purposes we will simplify by referring to the inward and outward conformational states only. Under crystallization conditions of pH 7.8 and at 1.05 Å resolution, the occupancy of the inward conformer is 80% and the outward 20% [9].
Proton shuttle residues have been identified in carbonic anhydrases of the β and γ class as well. His216 of the β carbonic anhydrase from Arabidopsis thaliana [10] and Glu84 of the γ carbonic anhydrase from Methanosarcina thermophila [11] have the properties of proton shuttles in the catalytic mechanisms. These examples of convergent evolution in the hydration of CO2 also demonstrate convergent evolution of proton transfer mechanisms. It is also useful to point out other carbonic anhydrases with dual conformations of proton shuttle residues. The proton shuttle residue Glu84 in the β carbonic anhydrase from M. thermophila is observed in crystal structures to have two distinct side chain conformations, one of which points toward the active site [12]. The activation of catalysis by reacting F65A-Y131C murine CA V with 4-chloromethylimidazole increased the rate of proton transfer in catalysis by three fold [13]. The electron density at 1.88Å resolution is interpreted as due to two different orientations of the modified side chain of Cys131 [14]. It is suggested that a solvent exposed and conformationally mobile side chain for the proton shuttle facilitates proton transfer between the active site and solvent in catalysis by carbonic anhydrase [14, 15]. These arguments appear straightforward. It would be an advantage in catalysis to have dual conformers of His64 so that the proton transferred to inward His64 in the hydration direction can flip to the outward orientation and deliver the proton easily to buffer in solution.
The energy barrier for the conformational change from inward to outward has not been approached by experimentation, but a ready answer is provided by molecular dynamics simulations. Maupin and Voth [16] considered the energy barrier for the conformational change from inward to outward in the EZnH2O2+-His system in which the rotation barrier is 5.6 kcal/mol with the inward orientation being more stable by 1.6 kcal/mol. For the EZnOH+-HisH+ system the rotation barrier from outward to inward is 6.2 kcal/mol with the outward conformer more stable by 3.2 kcal/mol. This means that the rotational mobility about χ1 is faster by several orders of magnitude than the proton transfer with an energy barrier near 10 kcal/mol. Because of the much slower rate of proton transfer with respect to conformer interconversion, an average or thermally equilibrated ratio of inward to outward occurs during the catalytic cycle. However, this ratio can change as catalysis proceeds.
Yet another consideration is the energy barrier to proton transfer between the zinc-bound solvent molecule and His64 that is entirely inward or entirely outward. We anticipate a larger barrier for His64 always outward because of the greater distance (by about 5 Å compared with inward) to the zinc-bound water and the need for more intervening solvent molecules for proton transfer in the outward orientation. Multistate empirical valence bond computations show the energy barrier for proton transfer is 11.4 kcal/mol for His64 always outward compared with 10.0 kal/mol for His64 always inward [17].
Distance between the proton donor and acceptor appears to be a significant factor for the efficiency of proton transfer within the active site of HCA II, according to a series of experiments given in Table 1. These experiments use histidine placed at various sites (positions 62, 64, 67, 200) in the active-site cavity and test their capacity to promote proton transfer, following the example of Liang et al. [18]. Other experiments use 4-methylimidazole (4-MI) in chemical rescue of mutants lacking a proton shuttle residue, with accompanying crystallographic studies to estimate the binding site of the 4-MI. These data show an optimal distance for the most efficient proton transfer near 7.5Å, which not surprisingly is the case for wild-type containing His64.
Table 1.
Rate constants for proton transfer from various sites in human carbonic anhydrase II and site-specific mutants.
However, it is not clear that distance is the feature that is optimized in the data of Table 1. In such a study, a combination of factors needs to be considered for each enzyme including structural and non-specific features of the active-site cavity, such as electrostatics. For example, in wild type and H64A-N67H HCA II the proton donor and acceptor are separated by at least two water molecules, and in H64A-T200H there are no water molecules intervening between donor and acceptor; in H64A-T200H the side chain His200 forms a direct hydrogen bond with the solvent molecule that coordinates the zinc directly. This close distance between proton donor and acceptor appears not to diminish proton transfer capacity in T200H by a very significant amount when compared with wild-type or H64A-N67H HCA II (Table 1). Other factors need to be considered, but it is not straightforward that there is a distance dependence on proton transfer in carbonic anhydrase, a point also discussed by Riccardi et al. [19].
In addition, experiments are not clear in supporting the hypothesis that an outward orientation for His64 would diminish catalysis. The first experiment to reflect on this hypothesis was the investigation of T200S HCA II in which the orientation of His64 appears predominantly outward in crystal structures (crystallization conditions at pH 8.0), within experimental uncertainties [20]. This outward orientation occurs even though the side chain of residue 200 is some 5Å away from His64 and there are no other conformational changes in T200S compared with wild type, nor is there a significant interaction gained in the outward His64. Catalysis by T200S HCA II shows no significant change in maximal velocity [20]. Certainly the conditions for crystallization of T200S HCA II and measurement of its catalysis are substantially different including apparent ionic strength and the presence of azide in the crystallization conditions [8]. That is, the preferred orientations of His64 could be different in the environment of the crystal and in the solution conditions under which catalytic activity was measured.
We now have additional crystal structures of variants of HCA II in which we can identify the orientation of His64 and for which we have rate constants for proton transfer in catalysis kB, all measured by the 18O-exchange method [21]. These enzymes are listed in Table 2. Except for wild type, they all contain one amino-acid replacement within the active-site cavity. The replacements are at Tyr7, Asn62, and Asn67. These are hydrophilic residues the side chains of which extend into the active site cavity and appear to form hydrogen bonds with the ordered, hydrogen-bonded water chain in the active-site cavity [9]. The side chains of residues 7, 62, and 67 are 7Å to 9Å from the zinc and 3Å to 6Å from the imidazole of His64 in its inward orientation. The replacement of these residues is shown to cause changes in the orientation of His64, changes in the pKa of the imidazole side chain of His64 (and through electrostatics in the pKa of the zinc-bound water), and changes in the structure of the ordered water network in the active-site cavity [22].
Table 2.
Rate constants kB for proton transfer in dehydration and pKa values for proton donor and acceptor in variants of HCA II.a
| Enzyme | Orientation of His64 |
pKa ZnH2O | pKa His64 | kB (µs−1) |
|---|---|---|---|---|
| 1 Y7F | in | 7.0 | 6.0 | 3.9 |
| 2 N62L | in | 6.0 | 6.0 | 0.2 |
| 3 N62A | in | 6.5 | 6.2 | 0.4 |
| 4 N62V | in | 5.9 | 5.9 | 0.35 |
| 5 Wild type | in/out | 6.8 | 7.2 | 0.8 |
| 6 N62T | in/out | 7.0 | 7.0 | 0.4 |
| 7 N67L | out | 6.0 | 7.5 | 0.2 |
| 8 N62D | out | 7.6 | 8.4 | 0.04 |
Replacement of Tyr7 and Asn62 by hydrophobic side chains (Y7F; N62L,A,V) is associated with a reduced pKa of His64 compared with wild type and an inward orientation of His64 (Table 2). These features are likely related. Introduction of hydrophobic residues near His64 will reduce the pKa, favoring the uncharged form of the imidazole side chain. Simulations show that the inward orientation of His64 in the EZnH2O2+-His system of HCA II is favored by 1.6 kcal/mol compared with the outward [16]. On the other hand, the outward orientation of His64 in N62D is understood by the opposite rationale. The presence of the charged carboxylate of Asp62 promotes an increase in the pKa of His64 (Table 2), and simulations confirm that in the system EZnH2O+-HisH+ the outward orientation is preferred [16]. Why N67L HCA II has His64 in the outward orientation is unexplained. For another mutant N62T, one assumes that the uncharged, hydrophilic Thr62 is sufficiently similar to Asn62 in the wild type that, like the wild type, this mutant promotes an orientation of His64 that is inward and outward (Table 2) [23]. Other than the differences in orientation of His64, there are no significant changes observed in the backbone or side-chain structures of the mutants of Table 2, with a notable exception (N62D) mentioned below.
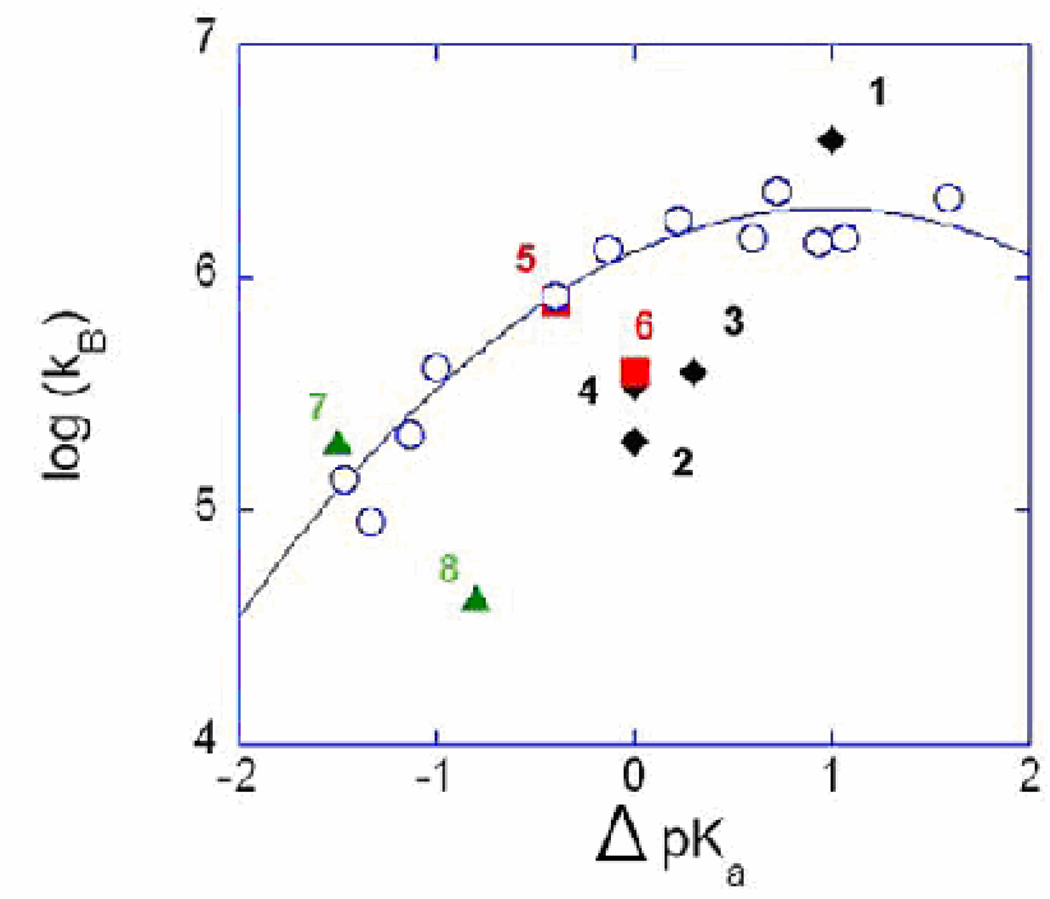
The values of the rate constants for proton transfer kB measured in the dehydration direction by 18O exchange [21] for each of the mutants of Table 2 are presented in a free energy plot in Figure 2 (filled symbols). Superimposed on this plot are the rate constants for proton transfer in catalysis by H64A HCA II caused by the presence of imidazole and pyridine derivatives which act as proton donors from solution (open circles) [24]. The usefulness of these superimposed data is that they represent proton transfer from a donor chemically similar to His64 but not tethered to the enzyme. Extensive study of this intermolecular proton transfer shows that the rate constants kB for a number of different exogenous proton donors can be fit to Marcus Rate Theory [25, 26], which is significant because it allows the properties of proton transfer in carbonic anhydrase to be compared with proton transfer in a number of model systems, both experimental and theoretical.
Figure 2.
Free energy plot of the logarithm of kB (s−1) versus ΔpKa (pKa ZnH2O – pKa His64) for (filled symbols) the wild type and the mutants of HCA II identified by number in Table 2; and (open circles) for H64H HCA II with proton transfer provided predominantly by derivatives of imidazole and pyridine acting as exogenous proton donors with the solid line a best fit of Marcus Rate Theory (Figure 5 from ref [24]). Data were obtained by 18O exchange at 25 °C.
In fact, the rate constant for proton transfer kB in catalysis by wild-type HCA II falls on a Marcus curve [24], an observation that is again made in Figure 2. The two enzymes (wild type and N62T) showing both the inward and outward orientation of His64 have high catalytic activity at ΔpKa near zero. This is a significant region of the free energy plot of Figure 2 since carbonic anhydrase must catalyze both hydration and dehydration reactions in its physiological function; this region of the curve at ΔpKa near zero provides the most facile intramolecular proton transfer when both the hydration and dehydration directions are considered. Although Table 2 is a very limited sample of enzymes, it gives little support to the suggestion that the conformational access to both the inward and outward orientations is an important contributor to efficiency of proton transfer. Two enzymes with predominantly inward orientations of His64, N62L and N62V, show nearly as great rate constants for proton transfer kB as wild type. In addition, the mutant N62T with dual orientations of His64 has about the same kB as N67L with an outward orientation of His64. Overall, accumulated data to date included in Table 2 do not show convincing evidence that the inward and outward conformations of His64 in crystal structures are relevant to proton transfer.
We observe from Figure 2 that the data for proton transfer from the exogenous proton donors lie closely along the Marcus curve, and the data for the site-specific mutants are much more scattered. What are the factors that account for the scatter of data points for intramolecular proton transfer in Figure 2? This is a difficult question; the difference in energy barrier between the least and most efficient enzyme of Figure 2 is only 2.4 kcal/mol. First we observe that the kinetic constants kB for the dehydration direction for the mutants of Table 2 generally increase with increasing ΔpKa (= pKa ZnH2O – pKa His64). This is to acknowledge that the more acidic the imidazole of His64, the better a proton donor it is. Other factors must be involved otherwise the data of Table 2 would not be so scattered in Figure 2. Perhaps the data can be interpreted more completely by a multi-state Marcus model as described for example by Warshel and colleagues [27].
A structural difference can be invoked to account for the low value of kB for N62D HCA II. In this mutant the side chain of Asn67 and the side chain of Gln92 are shifted closer to His64 by as much as 1.4Å compared with wild type [23]. Along with the presence of Asp62, this increases the hydrophilic character of the active site cavity and may be a significant contributor to the high pKa of His64 in N62D. As pointed out above, the side chains that are replaced in HCA II for the mutants of Table 2 (residues 7, 62, 67) are 3 – 6Ǻ from the imidazole ring of His64. In an earlier report, measurements of kB for mutants of K64H HCA III were fit to a Marcus plot with less scatter than we observe for HCA II in Figure 2 [25]. The difference is probably that in K64H HCA III a perturbing mutation was to replace Phe198 that is 12Ǻ from His64. This is much more distant than the side chains replaced in Table 2. However, another perturbing replacement in K64H HCA III was R67N which is 6Ǻ from His64, comparable to this study.
The role of the ordered water in the active-site cavity may be discerned in two mutants of Table 2, Y7F and N67L. For Y7F, the structure of ordered water is altered from a branched, hydrogen-bonded chain in wild type to an array of single water molecules, not branched, that extend from the zinc-bound solvent to His64 [22]. Computations show a more rapid proton transfer through a single, non-branched chain of water molecules compared with branched chains [28–30]. Yet experimental data show that the rapid rate constant for proton transfer observed for Y7F falls close to the Marcus curve (Figure 2) which is the activity curve predicted solely on the basis of ΔpKa. Correspondingly, a prominent feature of the crystal structure of N67L is that there is no ordered water observed in this mutant [22], yet the proton transfer activity of N67L also falls on the Marcus curve. Current concepts of the energetics of proton transfer drawn from experimental evidence certainly emphasize the formation of the water chain connecting donor and acceptor as a precondition for proton transfer [26, 31]; however, the relevance of the ordered water structure observed in the crystal structures of the enzymes of Table 2 is uncertain.
Finally, it is appropriate to point out results supporting the conclusion that a change in orientation of His64 does not contribute to the energy barrier of intramolecular proton transfer in catalysis by carbonic anhydrase. Shimahara et al. [32] have devised a mechanistic scheme in which the two neutral tautomers of the imidazole of His64 participate in a manner that does not require reorientation of the side chain. In another approach, Riccardi et al. [19] have presented computations pointing out that electrostatic interactions make a predominant contribution to the energy barrier of proton transfer in carbonic anhydrase. Under these computations, distance between donor and acceptor is not a factor, and both the inward and outward orientations of His64 support proton transfer with equivalent facility.
Acknowledgement
We thank our colleague of many years Dr. Chingkuang Tu for invaluable assistance. We are grateful to Dr. Robert McKenna and many students in his lab for their continuing collaboration.
Abbreviations
- HCA II
human carbonic anhydrase isozyme II
- 4-MI
4-methylimidazole
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Steiner H, Jonsson BH, Lindskog S. Catalytic Mechanism of Carbonic-Anhydrase -Hydrogen-Isotope Effects on Kinetic-Parameters of Human C Isoenzyme. European Journal of Biochemistry. 1975;59:253–259. doi: 10.1111/j.1432-1033.1975.tb02449.x. [DOI] [PubMed] [Google Scholar]
- 2.Tu CK, Silverman DN, Forsman C, Jonsson BH, Lindskog S. Role of histidine 64 in the catalytic mechanism of human carbonic anhydrase II studied with a site-specific mutant. Biochemistry. 1989;28:7913–7918. doi: 10.1021/bi00445a054. [DOI] [PubMed] [Google Scholar]
- 3.Silverman DN, Lindskog S. The Catalytic Mechanism of Carbonic-Anhydrase -Implications of a Rate-Limiting Protolysis of Water. Accounts of Chemical Research. 1988;21:30–36. [Google Scholar]
- 4.Chegwidden WR, Carter ND, Edwards YH. The Carbonic Anhydrases New Horizons. Basel: Birkhauser Verlag; 2000. [Google Scholar]
- 5.Supuran CT, Scozzafava A, Conway J. Carbonic Anhydrase - Its Inhibitors and Activators. Boca Raton: CRC Press; 2004. [Google Scholar]
- 6.Domsic JF, Avvaru BS, Kim CU, Gruner SM, Agbandje-McKenna M, Silverman DN, Mckenna R. Entrapment of Carbon Dioxide in the Active Site of Carbonic Anhydrase II. Journal of Biological Chemistry. 2008;283:30766–30771. doi: 10.1074/jbc.M805353200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sjoblom B, Polentarutti M, Djinovic-Carugo K. Structural study of X-ray induced activation of carbonic anhydrase. Proc. Natl. Acad. Sci. USA. 2009;106:10609–10613. doi: 10.1073/pnas.0904184106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nair SK, Christianson DW. Unexpected pH-Dependent Conformation of His-64, the Proton Shuttle of Carbonic Anhydrase-II. Journal of the American Chemical Society. 1991;113:9455–9458. [Google Scholar]
- 9.Fisher SZ, Maupin CM, Budayova-Spano M, Govindasamy L, Tu C, Agbandje-McKenna M, Silverman DN, Voth GA, McKenna R. Atomic crystal and molecular dynamics simulation structures of human carbonic anhydrase II: Insights into the proton transfer mechanism. Biochemistry. 2007;46:2930–2937. doi: 10.1021/bi062066y. [DOI] [PubMed] [Google Scholar]
- 10.Rowlett RS, Tu C, McKay MM, Preiss JR, Loomis RJ, Hicks KA, Marchione RJ, Strong JA, Donovan GS, Jr, Chamberlin JE. Kinetic characterization of wild-type and proton transfer-impaired variants of beta-carbonic anhydrase from Arabidopsis thaliana. Arch Biochem Biophys. 2002;404:197–209. doi: 10.1016/s0003-9861(02)00243-6. [DOI] [PubMed] [Google Scholar]
- 11.Tripp BC, Ferry JG. A structure-function study of a proton transport pathway in the gamma-class carbonic anhydrase from Methanosarcina thermophila. Biochemistry. 2000;39:9232–9240. doi: 10.1021/bi0001877. [DOI] [PubMed] [Google Scholar]
- 12.Iverson TM, Alber BE, Kisker C, Ferry JG, Rees DC. A closer look at the active site of gamma-class carbonic anhydrases: high-resolution crystallographic studies of the carbonic anhydrase from Methanosarcina thermophila. Biochemistry. 2000;39:9222–9231. doi: 10.1021/bi000204s. [DOI] [PubMed] [Google Scholar]
- 13.Earnhardt JN, Wright SK, Qian MZ, Tu CK, Laipis PJ, Viola RE, Silverman DN. Introduction of histidine analogs leads to enhanced proton transfer in carbonic anhydrase V. Archives of Biochemistry and Biophysics. 1999;361:264–270. doi: 10.1006/abbi.1998.0984. [DOI] [PubMed] [Google Scholar]
- 14.Jude KM, Wright SK, Tu C, Silverman DN, Viola RE, Christianson DW. Crystal structure of F65A/Y131C-methylimidazole carbonic anhydrase V reveals architectural features of an engineered proton shuttle. Biochemistry. 2002;41:2485–2491. doi: 10.1021/bi015808q. [DOI] [PubMed] [Google Scholar]
- 15.Silverman DN, McKenna R. Solvent-Mediated Proton Transfer in Catalysis by Carbonic Anhydrase. Acc Chem Res. 2007;40:669–675. doi: 10.1021/ar7000588. [DOI] [PubMed] [Google Scholar]
- 16.Maupin CM, Voth GA. Preferred orientations of His64 in human carbonic anhydrase II. Biochemistry. 2007;46:2938–2947. doi: 10.1021/bi062170f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maupin CM, McKenna R, Silverman DN, Voth GA. Elucidation of the Proton Transport Mechanism in Human Carbonic Anhydrase II. Journal of the American Chemical Society. 2009;131:7598–7608. doi: 10.1021/ja8091938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang ZW, Jonsson BH, Lindskog S. Proton-Transfer in the Catalytic Mechanism of Carbonic-Anhydrase.2. Effects of Placing Histidine-Residues at Various Positions in the Active-Site of Human Isoenzyme-II. Biochimica Et Biophysica Acta. 1993;1203:142–146. doi: 10.1016/0167-4838(93)90048-v. [DOI] [PubMed] [Google Scholar]
- 19.Riccardi D, Konig P, Guo H, Cui Q. Proton transfer in carbonic anhydrase is controlled by electrostatics rather than the orientation of the acceptor. Biochemistry. 2008;47:2369–2378. doi: 10.1021/bi701950j. [DOI] [PubMed] [Google Scholar]
- 20.Krebs JF, Fierke CA, Alexander RS, Christianson DW. Conformational Mobility of His-64 in the Thr200Ser Mutant of Human Carbonic Anhydrase-II. Biochemistry. 1991;30:9153–9160. doi: 10.1021/bi00102a005. [DOI] [PubMed] [Google Scholar]
- 21.Silverman DN. Carbonic anhydrase: oxygen-18 exchange catalyzed by an enzyme with rate-contributing proton-transfer steps. Methods Enzymol. 1982;87:732–752. doi: 10.1016/s0076-6879(82)87037-7. [DOI] [PubMed] [Google Scholar]
- 22.Fisher SZ, Tu CK, Bhatt D, Govindasamy L, Agbandje-McKenna M, McKenna R, Silverman DN. Speeding up proton transfer in a fast enzyme: kinetic and crystallographic studies on the effect of hydrophobic amino acid substitution in the active site of human carbonic anhydrase II. Biochemistry. 2007;42:3803–3813. doi: 10.1021/bi602620k. [DOI] [PubMed] [Google Scholar]
- 23.Zheng JY, Avvaru BS, Tu C, McKenna R, Silverman DN. Role of Hydrophilic Residues in Proton Transfer during Catalysis by Human Carbonic Anhydrase II. Biochemistry. 2008;47:12028–12036. doi: 10.1021/bi801473w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.An H, Tu C, Duda D, Montanez-Clemente I, Math K, Laipis PJ, McKenna R, Silverman DN. Chemical rescue in catalysis by human carbonic anhydrases II and III. Biochemistry. 2002;41:3235–3242. doi: 10.1021/bi0120695. [DOI] [PubMed] [Google Scholar]
- 25.Silverman DN, Tu C, Chen X, Tanhauser SM, Kresge AJ, Laipis PJ. Rate-equilibria relationships in intramolecular proton transfer in human carbonic anhydrase III. Biochemistry. 1993;32:10757–10762. doi: 10.1021/bi00091a029. [DOI] [PubMed] [Google Scholar]
- 26.Kresge AJ, Silverman DN. Application of Marcus rate theory to proton transfer in enzyme-catalyzed reactions. Enzyme Kinetics and Mechanism, Pt E. 1999;308:276–297. doi: 10.1016/s0076-6879(99)08014-3. [DOI] [PubMed] [Google Scholar]
- 27.Braun-Sand S, Strajbl M, Warshel A. Studies of proton translocations in biological systems: simulating proton transport in carbonic anhydrase by EVB-based models. Biophys J. 2004;87:2221–2239. doi: 10.1529/biophysj.104.043257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cui Q, Karplus M. Is a "proton wire" concerted or stepwise? A model study of proton transfer in carbonic anhydrase. Journal of Physical Chemistry B. 2003;107:1071–1078. [Google Scholar]
- 29.Wu Y, Voth GA. A computer simulation study of the hydrated proton in a synthetic proton channel. Biophys J. 2003;85:864–875. doi: 10.1016/S0006-3495(03)74526-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brewer ML, Schmitt UW, Voth GA. The formation and dynamics of proton wires in channel environments. Biophys J. 2001;80:1691–1702. doi: 10.1016/S0006-3495(01)76140-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peters KS. A Theory-Experiment Conundrum for Proton Transfer. Accounts of Chemical Research. 2009;42:89–96. doi: 10.1021/ar8001156. [DOI] [PubMed] [Google Scholar]
- 32.Shimahara H, Yoshida T, Shibata Y, Shimizu M, Kyogoku Y, Sakiyama F, Nakazawa T, Tate S, Ohki SY, Kato T, Moriyama H, Kishida K, Tano Y, Ohkubo T, Kobayashi Y. Tautomerism of histidine 64 associated with proton transfer in catalysis of carbonic anhydrase. J Biol Chem. 2007;282:9646–9656. doi: 10.1074/jbc.M609679200. [DOI] [PubMed] [Google Scholar]
- 33.Elder I, Tu CK, Ming LJ, McKenna R, Silverman DN. Proton transfer from exogenous donors in catalysis by human carbonic anhydrase II. Archives of Biochemistry and Biophysics. 2005;437:106–114. doi: 10.1016/j.abb.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 34.Bhatt D, Tu C, Fisher SZ, Hernandez Prada JA, McKenna R, Silverman DN. Proton transfer in a Thr200His mutant of human carbonic anhydrase II. Proteins. 2005;61:239–245. doi: 10.1002/prot.20615. [DOI] [PubMed] [Google Scholar]
- 35.Fisher Z, Prada JAH, Tu C, Duda D, Yoshioka C, An HQ, Govindasamy L, Silverman DN, McKenna R. Structural and kinetic characterization of active-site histidine as a proton shuttle in catalysis by human carbonic anhydrase II. Biochemistry. 2005;44:1097–1105. doi: 10.1021/bi0480279. [DOI] [PubMed] [Google Scholar]
- 36.Bhatt D, Fisher SZ, Tu C, McKenna R, Silverman DN. Location of binding sites in small molecule rescue of human carbonic anhydrase II. Biophys J. 2007;92:562–570. doi: 10.1529/biophysj.106.093203. [DOI] [PMC free article] [PubMed] [Google Scholar]