Abstract
The in vitro selection of antimicrobial resistance in important pathogens can provide critical information on the genetic basis of drug resistance, and such information can be used to predict, anticipate and even contain the spread of resistance in clinical practice. For instance, the discovery of the role of pfmdr1 in mefloquine resistance in malaria parasites resulted from in vitro studies. However, the in vitro selection of resistance is difficult, challenging and time consuming. In this review, we discuss the key parameters that impact on the efficiency of the in vitro selection of resistance, and propose strategies to improve and streamline this process.
Keywords: ‘accelerated resistance to multidrug’, clones, per-parasite resistant frequency, mutation rate, resistance indexes
Introduction
The control of malaria rests on the use of mosquito vector control and antimalarial chemotherapy. However, the latter strategy is hampered by the emergence of antimalarial resistance. Currently, there is no antimalarial drug that malaria parasites have not developed resistance against. Worryingly, there is evidence that malaria parasites in Cambodia are becoming resistant to artemisinin derivatives,1 providing a potential threat to artemisinin-based combination therapy (ACT), the cornerstone of current treatment. This emphasizes the need to reduce drug resistance selection pressure by withdrawing monotherapies,2 to develop new and affordable drug combinations, and to understand the mechanisms that underlie resistance to existing antimalarials. This could lead to: (i) the development of simple means of tracking the selection and spread of resistance; and (ii) the design and discovery of new and more potent antimalarials.
To study mechanisms of resistance, one needs to obtain well-characterized drug-resistant strains. However, such strains are not generally available for most antimalarials. Murine malaria—Plasmodium berghei, Plasmodium chabaudi, Plasmodium yeolii and Plasmodium vinckei—have been used as surrogates for Plasmodium falciparum to study the mechanisms of drug resistance by inducing resistance in vivo. This approach has led to the selection of drug-resistant parasite lines and subsequent studies on mechanisms of drug resistance.3 However, for drugs such as chloroquine and probably artemisinin, mechanisms of resistance in murine and P. falciparum malaria are different, highlighting the limitations of the murine malaria model.4–8
The development of the in vitro system for the routine culture of P. falciparum in 1976 provided a tool to induce resistance in vitro.9 In 1978, Nguyen-Dinh and Trager10 reported for the first time the selection of chloroquine resistance in vitro. This study heralded a new era and showed that selection of antimalarial resistance could be achieved in vitro in P. falciparum, thus permitting the study of the mechanisms of antimalarial resistance even before drugs were used in clinical practice. While this approach has been used to select some drug-resistant strains (see Table 1), it has not yet been used to study most of the antimalarials that are either in clinical use, such as the artemisinin derivatives, lumefantrine and piperaquine, or drugs in the pipeline and likely to be used in the near future, such as tafenoquine and pyronaridine.1,11–13
Table 1.
Summary of main findings of in vitro selection of resistance to antimalarials in P. falciparum strains
| Drugs | Strains (drug resistance profilea) | Ric (IC50) | Ratio of HDCd per IC50 after drug pressure | Time required to select resistance (months) | Stability: period of drug-free culture (months) | Work carried out on parasite lines: main finding | Ref. |
|---|---|---|---|---|---|---|---|
| CQ | FCR3 (CQ and CG resistant) | NM | NM | 4 | 1 | NM | 10 |
| FAC8 (CQ resistant) | 2.34 (83 ng/mL) | 1.23 | NM | 1 | deamplification of pfmdr1 in association with CQ resistance | 34 | |
| HB3 (PM resistant) | 1.64 (28 ng/mL) | 1.74 | 30 | NM | DNA amplification in chromosomes 3 and 12; deamplification in chromosome 3 after drug removal | 39 | |
| 106/01 (CQ susceptible)b | 12 (37 nM) | 0.23 | 2 | NM | evidence that the presence of pre-existing mutations in pfcrt lead to a rapid selection of the key 76 mutation | 38 | |
| MFQ | FCK (CQ resistant) | 16 (8 nmol/L) | 0.5 | 3 | NM | NM | 27 |
| Smith (CQ, PM and SD resistant) | 3.4 (3.5 µg/L) | inverse relationship between MFQ and CQ | 28 | ||||
| Camp (CQ susceptible) | 2.4 (4.9–12 µg/L) | 1.66 | >1e | 6 and cryopreservation | |||
| W2 (CQ, PM and SD resistant) | 4.6 (4.5 nM) | 1.93 | 22.4 | 12 | 1. inverse relationship between CQ and MFQ activity | 21 | |
| 2. identification of pfmdr1 as MFQ resistance marker | |||||||
| K1 (CQ, PM and SD resistant) | 4.07 (22.4 ng/mL) | 0.8 | NM | NM | 1. MFQ resistance associated with pfmdr1 overamplification | 29 | |
| W2mef (CQ, PM, SD and MFQ resistant) | 1.41 (58.88 ng/mL) | 1.08 | NM | NM | 2. evidence of inverse relationship with CQ | ||
| W2mef (CQ, PM, SD and MFQ resistant) | 1.07 (15.2 ng/mL) | 148.1 | 18 | NM | evidence of cross-resistance with HFT and QN and inverse relationship with CQ | 26 | |
| HFT | T9.96 (CQ susceptible) | 3.3 (6.6–22 nM) | 0.45 | 6 | 6 and cryopreservation | cross-resistance with QN but inverse relationship with CQ | 71 |
| K1 (CQ and SD resistant) | 9 (2.2 nM) | 0.4 | 2 | ||||
| PM | FCR3 | NM (15 nM) | NM | 7 | DNA amplification (chromosome containing dhfrf) | 40 | |
| 5FO | W2 | 100 (2 nM) | 1 | 2 | NM | evidence that resistance emerges quicker in already resistant strains | 63 |
| FCR3 | NM | 1 | 2 | NM | |||
| ATV | W2 | 30 (3 nM) | 1.1 | 2 | NM | evidence that resistance emerges quicker in already resistant strains | 63 |
| K1 | 837 (13.6 nM) | 1.6 | NM | <3 | evidence that mutations in cytochrome b are associated with ATV resistance | 46 | |
| BMS-3888891 | Dd2 (CQ, QN, PM and SD resistant) | 12 (10 nM) | NM | 2.66 | NM | evidence that resistance is associated with point mutation in protein farnesyl transferase | 51 |
| N-89 | FCR3 | 10 (25 nM) | NM | 24 | NM | no cross-resistance between the endoperoxides N-89 and artemisinin | 70 |
| AZ | Dd2 | 15.3 (124 nM) | NM | 0.7 | NM | AZ resistance is associated with point mutation in ribosomal protein L4 (pfRpL4) | 59 |
| 7G8 (CQ and PM resistant) | 17.5 (228 nM) | NM | 0.7 | NM |
CQ, chloroquine; CG, cycloguanil; SD, sulfadoxine; PM, pyrimethamine; HFT, halofantrine; MFQ, mefloquine; ATV, atovaquone; 5FO, 5-fluoro-orotate; AZ, azithromycin; QN, quinine; NM, not mentioned.
aInformation on resistance phenotype was presented in the references listed in the table and in Nkrumah et al.71
bThis strain is CQ susceptible; it has mutations in five codons of pfcrt, but not at codon 76. After drug pressure, two parasite lines were obtained: one with pfcrt-76 asparagine and a second with pfcrt-76 isoleucine, with IC50 values of 302.2 and 443.1 nM, respectively. We used the highest IC50 (443.1 nM) in the table. The normal mutant in CQ resistance is pfcrt-76 threonine.
cRi, resistance index: the ratio of the IC50 of the selected parasite line to the IC50 of the parent strain (before drug pressure).
dHDC, highest drug concentration tested, in which the parasite lines could grow after drug pressure.
eExact time period was not given.
fdhfr, dihydrofolate reductase gene.
Among these drugs, artemisinins are the most important. Indeed, this drug family is currently the backbone of malaria therapy as they form the basis of ACTs, and artesunate is replacing quinine in the treatment of severe malaria.14–16 A major concern is that resistance to this drug family is now emerging in South East Asia. Strategies are being put in place to predict, control and contain the emergence of artemisinin resistance.1,17 These strategies would be enhanced if the mechanism of resistance was identified and a simple marker developed for epidemiological studies. PfATPase6 has been proposed as the molecular target and polymorphisms have been associated with artemether resistance,18 but this has not been confirmed elsewhere.1 Consequently, in the absence of well-characterized drug-resistant strains, the mechanism of artemisinin resistance still awaits characterization. Could the in vitro culture system permit the selection of stable resistant parasites to these important antimalarials?
The selection of in vitro resistance is very slow; it can take months or even years for a stable drug-resistant line to emerge. This limitation combined with the difficulties of the in vitro culture technique (risk of contamination of cultures and slow parasite growth) might explain why for many antimalarials no stable resistant strains have been reported up to now. A recent review has presented the methods and application of the in vitro and in vivo selection of resistance.19 In the current paper, we have reviewed work carried out on the in vitro selection of resistance for the past 30 years. This review has identified the key parameters that are critical in the success of this process. We propose strategies to streamline the in vitro selection of antimalarial drug resistance.
First and benchmark work
The first work on the selection of P. falciparum in vitro was reported in 1978 by Nguyen-Dinh and Trager10 using the then newly developed method of malaria culture, the Petri dish method.9 A Gambian (West Africa) strain (FCR3), whose growth was completely inhibited at a concentration of 100 ng/mL chloroquine, was grown in the presence of increasing chloroquine concentrations, starting from 10 ng/mL. After 15 cycles (2 months), a parasite line that could grow in the presence of 100 ng/mL chloroquine was selected. This resistant phenotype was stable, since the parasite could grow in drug-free medium for several weeks without losing the selected phenotype. This work was perceived as a landmark, as it opened up the possibility that resistance could be selected against antimalarials in vitro.
How to interpret the data on the in vitro selection of resistance
We focused our review on publications that provided sufficient information on the in vitro selection experiment, including IC50 values (concentration that inhibits 50% of parasite growth), the protocol used and stability information. We reviewed 16 publications and summarize the results in Table 1.
Right shift of the dose–response curve
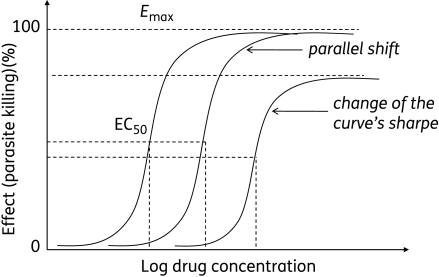
In vitro, drug activity is assessed by growing the parasite in the presence of various drug concentrations (serial dilution). The equation for the parasite growth rate as a function of the drug concentration generates a sigmoid dose–response curve, from which are deduced IC50/IC90 values (inhibitory concentrations that inhibit 50% or 90% of parasite growth). Resistance is defined by a right shift of the curve (Figure 1). The shape of the shift can be parallel, with the same maximum parasite killing effect (Emax) as the parent strain, or this shape could change, with a reduced maximum parasite killing effect (Figure 1).20
Figure 1.
Drug–response as a function of the concentration. Any right shift of the curve denotes an increase in the IC50 and, thus, the resistance. The shift can be parallel or the shape of the curve and the maximum effect (Emax) could change. Reproduced from Trends in Parasitology, 18(10), White NJ, The assessment of antimalarial drug efficacy, 458–64, 2002, with permission from Elsevier.20
We define the resistance index (Ri) as the ratio of the IC50 of the resistant line to that of the parent strain. Thus, the higher the Ri, the higher the level of resistance. However, a low Ri does not necessarily mean a low level of resistance. As shown in Table 1, Ri values can vary from 1 to >800. It is clear that an Ri of 100 indicates high-level resistance (found on strain W2 after drug pressure of atovaquone and 5-fluoro-orotate) (Table 1). An Ri of <10 may indicate an intermediate resistance level. However, it is interesting to note that the mefloquine-resistant parasite line (from W2), which was used to identify the mechanism of mefloquine resistance, had an Ri of only 4.6 (Table 1), implying that even a small right shift of the curve could be associated with resistance.21,22 In vivo, the relationship between the degree of resistance and the resulting therapeutic responses is complex, and depends on parameters such as the host immune response and the pharmacokinetic and pharmacodynamic properties of the drug.68
Variation of in vitro assay data
There is interlaboratory (interassay) variation of IC50/IC90. The culture conditions, mainly the initial parasitaemia, haematocrit, the time of incubation, the timepoint when the label is added (hypoxanthine for instance), the use of normal or substitute serum and the gas mixture composition, have been singled out as influencing the test.23,24 A right shift as a result of interassay variation could be wrongly interpreted as reflecting resistance. Therefore, any right shift is considered valid only if all in vitro culture parasite parameters are maintained constant.
Instability of the resistant phenotype
The resistant phenotype is stable when it remains unchanged after growing parasites in drug-free medium. In Table 1, in studies in which stability was reported, parasite lines remained resistant when grown in the absence of the drug during time periods varying from 2 weeks to 1 year and, in two studies, resistance remained unchanged after parasite cryopreservation. The unstable phenotypes could be associated with reduced parasite fitness, explaining why, once drug pressure is removed, the phenotype reverts to normal. For instance, a recent study has shown that parasites grown under mefloquine pressure expressed multiple pfmdr1 copy numbers and that these parasites had a significantly decreased survival fitness compared with parasites with a single pfmdr1 copy number.25
Existence of more than one parasite population during drug pressure
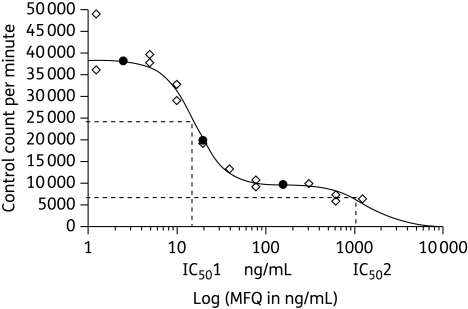
We computed the ratio of the highest drug concentration at which the parasite grew normally during the selection process with the parasite drug IC50 after drug selection (highest drug concentration tested/IC50, Table 1). This ratio is expected to be <1, since it represents the ratio of a subinhibitory concentration (thus, <IC50) divided by the IC50. In 7/15 strains, this ratio was ≥1.1. The most striking data were observed with W2mef on mefloquine,26 showing a ratio >148. This indicates that this parasite line could grow in the presence of a drug concentration 148 times higher than its own IC50. This led to the discovery that, in fact, there were two parasite populations with different IC50 values—one with a lower value, which corresponds to the dominant population, and another minor population, with a higher IC50 value. The second population is the one that persists when parasites are cultured in the presence of high drug concentrations. When determining IC50 values, drug pressure is removed and, because of the fitness disadvantage conferred by the drug-resistant phenotype, the minor population is replaced by the dominant and drug-susceptible one, reflecting low IC50 values. Since the determination of IC50 values is based on the use of a ‘monophasic’ drug–response curve, only one IC50 is obtained, which represents the dominant population. However, the presence of two populations in a mixture can be identified using a ‘biphasic’ drug–response curve.26 Figure 2 summarizes the graphic representation of the use of a biphasic drug–response curve leading to the identification of two populations.26 Thus, a ratio of >1 indicates the existence of at least two parasite populations and selection emerges on the backdrop of the minor population, which gradually becomes dominant as the drug-pressure experiment continues.
Figure 2.
Dose concentration–response of an in vitro-selected mefloquine (MFQ)-resistant line that could grow in the presence of >1200 ng/mL mefloquine. The dose–response was analysed using a modification of the logistic logarithmic function that permits evaluation of a biphasic concentration–response relationship. Two parasite populations with different IC50s can be defined from this graph, yet the use of a monophasic concentration–response would have generated one single IC50 (inhibitory concentration that kills 50% of parasites). Reproduced, with permission, from Peel et al. (Figure 4b).26
How useful has in vitro resistance been?
The main objective of inducing in vitro resistance is to generate parasite lines that can be used to study the mechanism of resistance. How much has already been done with the P. falciparum cell lines that were selected in vitro?
Mefloquine and chloroquine
The first selection of in vitro mefloquine resistance was carried out in the early 1980s.27,28 In 1988, Oduola et al.21 selected the mefloquine-resistant strain W2mef, from the parent strain W2. The analysis of P-glycoprotein pfmdr1 between W2 and W2mef led to the discovery of the association between amplification of this gene and mefloquine resistance.22 Thereafter, the WEHI group (Walter and Eliza Hall Institute) induced mefloquine resistance against the K1 strain and further against the W2mef line, and confirmed the involvement of pfmdr1 in mefloquine resistance.29 Since then, these findings have been supported in field isolates30–33 and in rodent malaria.5
The aforementioned studies and the one by Barnes et al.34 on the selection of chloroquine resistance (Table 1) showed the existence of an inverse relationship between the pfmdr1 copy number (mefloquine resistance) and chloroquine susceptibility. This prompted further investigations into pfmdr1 polymorphism in relation to chloroquine susceptibility. Several single nucleotide polymorphisms in this gene have been associated with chloroquine resistance and it has now been established that this gene plays an ancillary role in chloroquine resistance, the key gene being pfcrt.30,35–37
The use of in vitro-selected chloroquine-resistant parasite lines has also led to a better understanding of the mechanisms of chloroquine resistance. Chloroquine resistance was induced in the Sudanese strain 106/1.38 This strain contains seven mutations associated with chloroquine resistance in pfcrt (at codons 74, 75, 220, 271 and 371), but the key one, at codon 76, is wild type (lysine), while chloroquine-resistant isolates have threonine at this codon. This strain was chloroquine susceptible. By subjecting it to chloroquine pressure for 2 months, a chloroquine-resistant line was selected. Subsequent genetic analyses demonstrated that this selected chloroquine-resistant phenotype was associated with the presence of a mutation at pfcrt codon 76 (from lysine to isoleucine or asparagine). Though this mutated amino acid was not threonine, this study confirmed the critical role of the mutation at codon 76 in chloroquine resistance.
The HB3 strain was used to select chloroquine resistance and further analysis demonstrated the existence of DNA amplification in chromosomes 3 and 12 in this parasite line.39 It is interesting to note that neither pfmdr1 nor pfcrt are located on chromosomes 3 or 12. Since the mechanism of chloroquine resistance is known to be multifactorial, other genes might be involved; however, no further studies have been carried out on these cell lines.
Pyrimethamine
Banyal and Inselburg40 selected FCR3 parasite lines resistant to pyrimethamine. This phenotype was found to be associated with DNA amplification on chromosome 4 in a region corresponding to the dihydrofolate reductase–thymidine synthase gene (dhfr-ts).41,42 It was also found that these parasite lines harbour a point mutation at codon 223 of the dhfr domain,42 in line with the role of this gene in pyrimethamine resistance.43–45
Atovaquone
This drug has been combined with proguanil to form Malarone®, one of the most potent antimalarial prophylactic agents. Atovaquone binds to cytochrome b (Cyt b), leading to the inhibition of mitochondrial electron transfer, and mutation in Cyt b has been associated with atovaquone resistance in vivo.46 Using drug pressure on the K1 strain, Korsinczky et al.46 selected parasite lines resistant to atovaquone, with Ri varying from 76- to 837-fold. Sequencing of the Cyt b gene of these parasites showed point mutations at codons 275, 272, 280, 281, 284 and 280, and these codons are located in the binding pocket of atovaquone to Cyt b.46 Interestingly, the mutation at 268, which is commonly found in atovaquone-resistant field isolates, is also located in this binding pocket.47–49
BMS-388891
This compound is an inhibitor of protein farnesyl transferase (PFT), which catalyses the transfer of the farnesyl group to the C-terminus of a specific set of proteins, including those that are essential in cell multiplication, and the blockade of this farnesylation is associated with cell death.50 Inhibition of the PFT enzyme is a good drug target against malaria, and many inhibitors have been synthesized and evaluated as potential antimalarials.50–52 Eastman et al.51 induced resistance to an inhibitor of PFT, BMS-388891, by culturing the Dd2 strain in the presence of continuous drug pressure. Within 80 days, a line with a 12-fold decreased activity was selected and genetic analyses have shown the existence of a point mutation at codon 837 of PFT. Thus, polymorphism in the PFT enzyme could be associated with in vivo resistance to inhibitors of PFT in the clinical setting.
Azithromycin
The macrolide azithromycin, along with erythromycin, has proved to be active against P. falciparum, and several clinical trials of azithromycin and erythromycin alone or in combination with chloroquine, quinine or artesunate have been conducted. These macrolide agents are promising antimalarials, especially in the treatment of malaria in pregnancy.53–58 An azithromycin-pressure experiment on Dd2 and 7G8 strains led to the selection of resistant lines within 21 days, and these lines showed cross-resistance to erythromycin.59 This phenotype was associated with point mutations in P. falciparum apicoplast-encoded ribosomal protein L4 (pfRpL4), one of the proteins that form the 50S ribosomal subunit,59 in line with the macrolide resistance mechanism in bacteria.59 Though azithromycin and erythromycin are still experimental drugs, the use of laboratory-induced resistant lines has led to the identification of drug resistance genes before these drugs enter clinical use. The ease with which in vitro resistance to macrolides has been selected in vitro may indicate that the selection and spread of in vivo resistance may be rapid. This drug is still in clinical evaluation against malaria and it remains to be seen whether this prediction will be confirmed in vivo.
This information clearly demonstrates the value of drug-resistant parasite lines selected in vitro. For instance, pfmdr1, one of the genes central to resistance to quinoline or quinoline-related drugs, was identified using in vitro-selected drug resistance parasite lines. The use of the same approach has led to the confirmation of pyrimethamine and chloroquine mechanisms of resistance. Work on the macrolide azithromycin and an inhibitor of PFT has demonstrated that it is possible to understand the mechanism of drug resistance before the drug is used in the clinical setting, and before resistant parasites are found in the field.51,59 In theory, the same could be achieved with any other antimalarial drugs.
Are there strains more prone to becoming drug resistant?
Early reports using murine malaria indicate that a strain resistant to one drug is more amenable to give rise to resistant lines to another drug, compared with strains that are fully drug susceptible. Pyrimethamine- and chloroquine-resistant parasites were more easily generated from drug-resistant strains than from drug-susceptible ones in P. chabaudi and P. vinckei.60–62 Evidence that the same phenomenon prevails in P. falciparum has been provided by Rathod's group.63 They induced resistance to atovaquone and 5-fluoro-orotate against the multidrug-resistant strain W2 (cycloguanil, pyrimethamine and sulfadoxine resistant), FCR3 (pyrimethamine and cycloguanil resistant), HB3 (pyrimethamine resistant), 3D7 (sulfadoxine resistant) and the fully drug-susceptible D6. They demonstrated that W2 was more prone to generate drug-resistant parasite lines than FCR3 and that the latter was more prone than 3D7. Not a single resistant line was obtained from the fully drug-susceptible D6. The W2 strain acquired resistance to these drugs with a frequency 10- to 1000-fold higher than the frequency in other strains. This phenomenon was named ‘accelerated resistance to multidrug (ARMD)’ and is different to the known multidrug-resistant phenotype.63 ARMD is characterized by the ability of a strain to generate a drug-resistant clone when put under drug pressure. This results from the high mutation rate during parasite multiplication. The low efficiency of the mechanisms of DNA repair has been proffered as one of the factors that contribute to this ARMD phenotype.19,64
For instance, HB3 is susceptible to almost all antimalarial drugs, except pyrimethamine. It has one single point mutation at codon 108 in dhfr65 and, thus, has a low level of pyrimethamine resistance. As shown in Table 1, chloroquine drug pressure for 30 months gave rise to a line with an Ri of only 1.64, a value that is among the lowest in Table 1. On the other hand, high level of drug-resistant phenotype does not necessarily imply a higher ability to generate drug-resistant lines. This is the case for W2mef. This parasite line is resistant to many antimalarials and is, thus, more prone to generate resistant clones (as discussed earlier). However, it could not generate resistant clones when subjected to mefloquine drug pressure (Ri < 2, see Table 1).26,29 The reason could be that it was already mefloquine resistant, as a result of pfmdr1 being overexpressed. W2mef, therefore, needed to develop new and additional mechanisms to cope with the higher mefloquine concentration, explaining, at least partially, the difficulties in generating stable clones with higher mefloquine resistance. This is supported by the fact that, in vivo, no new mechanisms of mefloquine resistance have been reported other than pfmdr1 amplification. Thus, the ability of a parasite to generate resistant lines will vary from one strain to another.
What should be considered when designing an ‘in vitro drug selection’ study?
The following are critical parameters that have to be considered when setting up the experiment.
Initial strains
Strains are different; there are those that are more prone to generate resistant parasite lines than others, and these are called ARMD strains (see previous section). Thus, in vitro selection for resistance should focus on drug-resistant strains/isolates.
Initial parasitaemia
One of the key parameters in the success of these experiments is the initial parasitaemia. Work in animals has shown that the higher the initial parasitaemias, the higher is the chance of selecting drug-resistant mutants. For instance, Ramakrishnan et al.66 could select sulfadiazine resistance in P. berghei in mice at 10% parasitaemia, but failed to do so at 1%. The most interesting evidence was provided in humans, where it was shown that the chance of developing pyrimethamine resistance after a single-dose treatment was associated with the size of the parasite population.67
Genetic events that confer de novo drug resistance (gene point mutations or copy number variations) are spontaneous and rare, and are drug independent. In vitro, parasites undergo mitotic nuclear division exclusively. Thus, the chance of obtaining a mutant strain that will confer drug resistance will be a function of the number of mitotic events and, thus, the size of the parasite population.
The generation of de novo resistance is also dependent upon the rate at which parasites generate drug-resistant clones. Since this rate, the per-parasite resistance frequency (PRF), varies depending on the mutation (or the gene), different drugs will have different rates; if resistance involves one single gene, the PRF is likely to be higher than when resistance is multifactorial. For instance, in vivo, the PRF for pyrimethamine, atovaquone and mefloquine has been estimated to be 1011, 1012 and 1014, respectively; chloroquine has been proposed to be ∼1019; and values <1018 have been proposed for artemisinin.68 Interestingly, in general, PRF rates are found to be higher in vivo, e.g. atovaquone-resistant mutants arise at a frequency of 1 in 105,63 and the rate with mefloquine is 1 in 108 and 1 in 1013 when resistance is associated with one and two/three pfmdr1 copy numbers, respectively.25 Thus, different drugs will have a different chance to encounter a drug-resistant clone in vitro.
There are ∼5 × 1016–5 × 1017 circulating parasites in the human population.2 If such numbers of parasites could be put into a flask to induce in vitro resistance, one could expect that resistance strains would emerge against any given drug, since, in theory, resistant strains would already be present in the population. Generally, cultures are carried out in small flasks (15 mL) containing 5 mL of the culture suspension [2% haematocrit, equivalent to 100 µL of red blood cells (RBCs)]. Assuming that 10% parasitaemia could be reached after culture, only 108 infected RBCs could be obtained using such flasks.
For drugs such as atovaquone with a higher PRF, chances are high that a resistant clone will emerge after some mitotic events with a starting parasitaemia of 108; thus, under these conditions, atovaquone resistance would be relatively easy to select, as shown in Table 1. On the other hand, for drugs with a low PRF, such as artemisinin, a starting parasitaemia of 108 will not be favourable to the selection of resistance. Thus, current experimental conditions do not favour the emergence of resistant mutants against drugs with a low PRF. Thus, experiments should be carried out under conditions where a higher parasite number could be used. There should not be a limitation of parasite number, apart from that imposed by the experimental conditions.
Use of current multidrug-resistant strains
One of the characteristics of ARMD strains is that they are multidrug resistant. The more drugs that parasites have been exposed to, the higher their chance of harbouring an ARMD phenotype. The current strains used in selection of resistance studies were collected 20–30 years ago. For instance, W2, the most commonly used strains to induce resistance, were collected in the 1970s. Since then, parasites have been exposed to new antimalarials. It is reasonable to state that the current multidrug strains are more prone to generate resistance than strains collected 30 years ago. Consequently, the selection of resistance should also include contemporary parasite strains to maximize the chance of obtaining drug-resistant clones. Parasites from malaria-endemic areas where drug resistance is high, for instance South East Asia, should be considered. Another approach would be to use genetically modified parasites that are more prone to generate new mutants. Such parasites could be those with impaired DNA repair mechanisms;64 however, they are not available yet.
Time of experiment
The success of the selection of resistance depends on the length of the experimental time period. The longer the selection of resistance takes, the higher the chance of obtaining resistant lines. In theory, for any drug, resistance will eventually emerge when given sufficient time. The question is therefore how long the process should take or when to stop.
The process can stop for two reasons. The first could be that parasites have reached the limit of acceptable concentration beyond which they can no longer grow. Alternative drug-free and short exposures to high drug concentrations could overcome this limitation.26 The second reason, which is more common, is that the process is stopped by the experimenter. For instance, two experiments in which the authors failed to select artesunate (an artemisinin derivative) and ferroquine resistance were stopped within 1 month of drug pressure.6,69 As we discussed, for a drug such as artemisinin, which has the lowest PRF, it would be surprising that resistant parasites would emerge in vitro within 1–2 months. Thus, it is reasonable to expect that more time, even in terms of years, would be required to induce in vitro resistance against this drug. It is therefore critical that the experimenter bears these considerations in mind when setting up drug resistance studies. For instance, Aly et al.70 selected a parasite line resistant to the endoperoxide N-89 after 2 years of continuous drug pressure. However, this N-89-resistant strain was susceptible to the endoperoxide artemisinin, indicating that the mechanisms of resistance between these drugs are different.
Conclusions
Thirty years ago, the first report that chloroquine resistance could be induced in vitro heralded a new era with the expectation that antimalarial-resistant parasite lines could be obtained, allowing study of the mechanism of resistance before drugs reached clinical use. Although the approach has been used to discover and/or confirm the mechanism of resistance to various antimalarials, including mefloquine, chloroquine and pyrimethamine, it is surprising to note that for important drugs such as artemisinin or lumefantrine, which now form the backbone of malaria treatment, no well-characterized resistant parasite lines exist, limiting investigations on their mechanism of resistance. Yet the use of an in vitro system could allow the selection of resistant parasites against any antimalarials. Attempts have been made to select resistance to artemisinin in vitro, but in vain. The selection of resistance is challenging and time consuming. Our review of the literature shows that strategies exist to streamline this process. With the development of new molecular technologies, such as Solexa Illumina and 454-sequencing, which allow high-throughput DNA sequencing and whole transcriptome analysis, it is now possible to study drug resistance mechanisms by exploring the parasite whole genome variation at a relatively low cost and in a short time period. However, the challenge remains the generation of drug-resistant parasite lines.
Funding
This study was supported by the European Developing Countries Clinical Trials Partnership (EDCTP) and The Wellcome Trust WT077092 and WT084538.
Transparency declarations
None to declare.
Acknowledgements
We thank the Director of the Kenya Medical Research Institute for permission to publish this review. We also thank Professor Nick White for helpful comments.
References
- 1.White NJ. Qinghaosu (artemisinin): the price of success. Science. 2008;320:330–4. doi: 10.1126/science.1155165. [DOI] [PubMed] [Google Scholar]
- 2.White NJ. Antimalarial drug resistance. J Clin Invest. 2004;113:1084–92. doi: 10.1172/JCI21682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peters W. Chemotherapy and Drug Resistance in Malaria. London: Academic Press; 1987. [Google Scholar]
- 4.Afonso A, Hunt P, Cheesman S, et al. Malaria parasites can develop stable resistance to artemisinin but lack mutations in candidate genes atp6 (encoding the sarcoplasmic and endoplasmic reticulum Ca2+ ATPase), tctp, mdr1, and cg10. Antimicrob Agents Chemother. 2006;50:480–9. doi: 10.1128/AAC.50.2.480-489.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carlton JM, Hayton K, Cravo PV, et al. Of mice and malaria mutants: unravelling the genetics of drug resistance using rodent malaria models. Trends Parasitol. 2001;17:236–42. doi: 10.1016/s1471-4922(01)01899-2. [DOI] [PubMed] [Google Scholar]
- 6.Hunt P, Afonso A, Creasey A, et al. Gene encoding a deubiquitinating enzyme is mutated in artesunate- and chloroquine-resistant rodent malaria parasites. Mol Microbiol. 2007;65:27–40. doi: 10.1111/j.1365-2958.2007.05753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hunt P, Cravo PV, Donleavy P, et al. Chloroquine resistance in Plasmodium chabaudi: are chloroquine-resistance transporter (crt) and multi-drug resistance (mdr1) orthologues involved? Mol Biochem Parasitol. 2004;133:27–35. doi: 10.1016/j.molbiopara.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 8.Hunt P, Martinelli A, Fawcett R, et al. Gene synteny and chloroquine resistance in Plasmodium chabaudi. Mol Biochem Parasitol. 2004;136:157–64. doi: 10.1016/j.molbiopara.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 9.Trager W, Jensen JB. Human malaria parasites in continuous culture. Science. 1976;193:673–5. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- 10.Nguyen-Dinh P, Trager W. Chloroquine resistance produced in vitro in an African strain of human malaria. Science. 1978;200:1397–8. doi: 10.1126/science.351801. [DOI] [PubMed] [Google Scholar]
- 11.Kokwaro G, Mwai L, Nzila A. Artemether/lumefantrine in the treatment of uncomplicated falciparum malaria. Expert Opin Pharmacother. 2007;8:75–94. doi: 10.1517/14656566.8.1.75. [DOI] [PubMed] [Google Scholar]
- 12.Myint HY, Ashley EA, Day NP, et al. Efficacy and safety of dihydroartemisinin–piperaquine. Trans R Soc Trop Med Hyg. 2007;101:858–66. doi: 10.1016/j.trstmh.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 13.Ramharter M, Kurth F, Schreier AC, et al. Fixed-dose pyronaridine–artesunate combination for treatment of uncomplicated falciparum malaria in pediatric patients in Gabon. J Infect Dis. 2008;198:911–9. doi: 10.1086/591096. [DOI] [PubMed] [Google Scholar]
- 14.Checkley AM, Whitty CJ. Artesunate, artemether or quinine in severe Plasmodium falciparum malaria? Expert Rev Anti Infect Ther. 2007;5:199–204. doi: 10.1586/14787210.5.2.199. [DOI] [PubMed] [Google Scholar]
- 15.Dondorp A, Nosten F, Stepniewska K, et al. Artesunate versus quinine for treatment of severe falciparum malaria: a randomised trial. Lancet. 2005;366:717–25. doi: 10.1016/S0140-6736(05)67176-0. [DOI] [PubMed] [Google Scholar]
- 16.Jones KL, Donegan S, Lalloo DG. Artesunate versus quinine for treating severe malaria. Cochrane Database Syst Rev. 2007:CD005967. doi: 10.1002/14651858.CD005967.pub2. issue 4. [DOI] [PubMed] [Google Scholar]
- 17.Dondorp AM, Nosten F, Yi P, et al. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2009;361:455–67. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jambou R, Legrand E, Niang M, et al. Resistance of Plasmodium falciparum field isolates to in-vitro artemether and point mutations of the SERCA-type PfATPase6. Lancet. 2005;366:1960–3. doi: 10.1016/S0140-6736(05)67787-2. [DOI] [PubMed] [Google Scholar]
- 19.Witkowski B, Berry A, Benoit-Vical F. Resistance to antimalarial compounds: methods and applications. Drug Resist Updat. 2009;12:42–50. doi: 10.1016/j.drup.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 20.White NJ. The assessment of antimalarial drug efficacy. Trends Parasitol. 2002;18:458–64. doi: 10.1016/s1471-4922(02)02373-5. [DOI] [PubMed] [Google Scholar]
- 21.Oduola AM, Milhous WK, Weatherly NF, et al. Plasmodium falciparum: induction of resistance to mefloquine in cloned strains by continuous drug exposure in vitro. Exp Parasitol. 1988;67:354–60. doi: 10.1016/0014-4894(88)90082-3. [DOI] [PubMed] [Google Scholar]
- 22.Wilson CM, Serrano AE, Wasley A, et al. Amplification of a gene related to mammalian mdr genes in drug-resistant Plasmodium falciparum. Science. 1989;244:1184–6. doi: 10.1126/science.2658061. [DOI] [PubMed] [Google Scholar]
- 23.Basco LK. Molecular epidemiology of malaria in Cameroon. XX. Experimental studies on various factors of in vitro drug sensitivity assays using fresh isolates of Plasmodium falciparum. Am J Trop Med Hyg. 2004;70:474–80. [PubMed] [Google Scholar]
- 24.Briolant S, Parola P, Fusai T, et al. Influence of oxygen on asexual blood cycle and susceptibility of Plasmodium falciparum to chloroquine: requirement of a standardized in vitro assay. Malar J. 2007;6:44. doi: 10.1186/1475-2875-6-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Preechapornkul P, Imwong M, Chotivanich K, et al. Plasmodium falciparum pfmdr1 amplification, mefloquine resistance, and parasite fitness. Antimicrob Agents Chemother. 2009;53:1509–15. doi: 10.1128/AAC.00241-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peel SA, Merritt SC, Handy J, et al. Derivation of highly mefloquine-resistant lines from Plasmodium falciparum in vitro. Am J Trop Med Hyg. 1993;48:385–97. doi: 10.4269/ajtmh.1993.48.385. [DOI] [PubMed] [Google Scholar]
- 27.Brockelman CR, Monkolkeha S, Tanariya P. Decrease in susceptibility of Plasmodium falciparum to mefloquine in continuous culture. Bull World Health Organ. 1981;59:249–52. [PMC free article] [PubMed] [Google Scholar]
- 28.Lambros C, Notsch JD. Plasmodium falciparum: mefloquine resistance produced in vitro. Bull World Health Organ. 1984;62:433–8. [PMC free article] [PubMed] [Google Scholar]
- 29.Cowman AF, Galatis D, Thompson JK. Selection for mefloquine resistance in Plasmodium falciparum is linked to amplification of the pfmdr1 gene and cross-resistance to halofantrine and quinine. Proc Natl Acad Sci USA. 1994;91:1143–7. doi: 10.1073/pnas.91.3.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duraisingh MT, Cowman AF. Contribution of the pfmdr1 gene to antimalarial drug-resistance. Acta Trop. 2005;94:181–90. doi: 10.1016/j.actatropica.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 31.Price RN, Cassar C, Brockman A, et al. The pfmdr1 gene is associated with a multidrug-resistant phenotype in Plasmodium falciparum from the western border of Thailand. Antimicrob Agents Chemother. 1999;43:2943–9. doi: 10.1128/aac.43.12.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Price RN, Uhlemann AC, Brockman A, et al. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet. 2004;364:438–47. doi: 10.1016/S0140-6736(04)16767-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uhlemann AC, McGready R, Ashley EA, et al. Intrahost selection of Plasmodium falciparum pfmdr1 alleles after antimalarial treatment on the northwestern border of Thailand. J Infect Dis. 2007;195:134–41. doi: 10.1086/509809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barnes DA, Foote SJ, Galatis D, et al. Selection for high-level chloroquine resistance results in deamplification of the pfmdr1 gene and increased sensitivity to mefloquine in Plasmodium falciparum. EMBO J. 1992;11:3067–75. doi: 10.1002/j.1460-2075.1992.tb05378.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanchez CP, Stein WD, Lanzer M. Is PfCRT a channel or a carrier? Two competing models explaining chloroquine resistance in Plasmodium falciparum. Trends Parasitol. 2007;23:332–9. doi: 10.1016/j.pt.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 36.Valderramos SG, Fidock DA. Transporters involved in resistance to antimalarial drugs. Trends Pharmacol Sci. 2006;27:594–601. doi: 10.1016/j.tips.2006.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ekland EH, Fidock DA. Advances in understanding the genetic basis of antimalarial drug resistance. Curr Opin Microbiol. 2007;10:363–70. doi: 10.1016/j.mib.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cooper RA, Ferdig MT, Su XZ, et al. Alternative mutations at position 76 of the vacuolar transmembrane protein PfCRT are associated with chloroquine resistance and unique stereospecific quinine and quinidine responses in Plasmodium falciparum. Mol Pharmacol. 2002;61:35–42. doi: 10.1124/mol.61.1.35. [DOI] [PubMed] [Google Scholar]
- 39.Lim AS, Cowman AF. Plasmodium falciparum: chloroquine selection of a cloned line and DNA rearrangements. Exp Parasitol. 1996;83:283–94. doi: 10.1006/expr.1996.0076. [DOI] [PubMed] [Google Scholar]
- 40.Banyal HS, Inselburg J. Plasmodium falciparum: induction, selection, and characterization of pyrimethamine-resistant mutants. Exp Parasitol. 1986;62:61–70. doi: 10.1016/0014-4894(86)90008-1. [DOI] [PubMed] [Google Scholar]
- 41.Inselburg J, Bzik DJ, Horii T. Pyrimethamine resistant Plasmodium falciparum: overproduction of dihydrofolate reductase by a gene duplication. Mol Biochem Parasitol. 1987;26:121–34. doi: 10.1016/0166-6851(87)90136-8. [DOI] [PubMed] [Google Scholar]
- 42.Tanaka M, Gu HM, Bzik DJ, et al. Mutant dihydrofolate reductase–thymidylate synthase genes in pyrimethamine-resistant Plasmodium falciparum with polymorphic chromosome duplications. Mol Biochem Parasitol. 1990;42:83–91. doi: 10.1016/0166-6851(90)90115-3. [DOI] [PubMed] [Google Scholar]
- 43.Gregson A, Plowe CV. Mechanisms of resistance of malaria parasites to antifolates. Pharmacol Rev. 2005;57:117–45. doi: 10.1124/pr.57.1.4. [DOI] [PubMed] [Google Scholar]
- 44.Nzila A. The past, present and future of antifolates in the treatment of Plasmodium falciparum infection. J Antimicrob Chemother. 2006;57:1043–54. doi: 10.1093/jac/dkl104. [DOI] [PubMed] [Google Scholar]
- 45.Hawkins VN, Joshi H, Rungsihirunrat K, et al. Antifolates can have a role in the treatment of Plasmodium vivax. Trends Parasitol. 2007;23:213–22. doi: 10.1016/j.pt.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 46.Korsinczky M, Chen N, Kotecka B, et al. Mutations in Plasmodium falciparum cytochrome b that are associated with atovaquone resistance are located at a putative drug-binding site. Antimicrob Agents Chemother. 2000;44:2100–8. doi: 10.1128/aac.44.8.2100-2108.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Berry A, Senescau A, Lelievre J, et al. Prevalence of Plasmodium falciparum cytochrome b gene mutations in isolates imported from Africa, and implications for atovaquone resistance. Trans R Soc Trop Med Hyg. 2006;100:986–8. doi: 10.1016/j.trstmh.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 48.Musset L, Bouchaud O, Matheron S, et al. Clinical atovaquone–proguanil resistance of Plasmodium falciparum associated with cytochrome b codon 268 mutations. Microbes Infect. 2006;8:2599–604. doi: 10.1016/j.micinf.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 49.Pimentel S, Nogueira F, Benchimol C, et al. Detection of atovaquone–proguanil resistance conferring mutations in Plasmodium falciparum cytochrome b gene in Luanda, Angola. Malar J. 2006;5:30. doi: 10.1186/1475-2875-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Buckner FS, Eastman RT, Yokoyama K, et al. Protein farnesyl transferase inhibitors for the treatment of malaria and African trypanosomiasis. Curr Opin Investig Drugs. 2005;6:791–7. [PubMed] [Google Scholar]
- 51.Eastman RT, White J, Hucke O, et al. Resistance to a protein farnesyltransferase inhibitor in Plasmodium falciparum. J Biol Chem. 2005;280:13554–9. doi: 10.1074/jbc.M413556200. [DOI] [PubMed] [Google Scholar]
- 52.Olepu S, Suryadevara PK, Rivas K, et al. 2-Oxo-tetrahydro-1,8-naphthyridines as selective inhibitors of malarial protein farnesyltransferase and as anti-malarials. Bioorg Med Chem Lett. 2008;18:494–7. doi: 10.1016/j.bmcl.2007.11.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Noedl H, Krudsood S, Leowattana W, et al. In vitro antimalarial activity of azithromycin, artesunate, and quinine in combination and correlation with clinical outcome. Antimicrob Agents Chemother. 2007;51:651–6. doi: 10.1128/AAC.01023-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kalilani L, Mofolo I, Chaponda M, et al. A randomized controlled pilot trial of azithromycin or artesunate added to sulfadoxine–pyrimethamine as treatment for malaria in pregnant women. PLoS ONE. 2007;2:e1166. doi: 10.1371/journal.pone.0001166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Noedl H, Krudsood S, Chalermratana K, et al. Azithromycin combination therapy with artesunate or quinine for the treatment of uncomplicated Plasmodium falciparum malaria in adults: a randomized, phase 2 clinical trial in Thailand. Clin Infect Dis. 2006;43:1264–71. doi: 10.1086/508175. [DOI] [PubMed] [Google Scholar]
- 56.Miller RS, Wongsrichanalai C, Buathong N, et al. Effective treatment of uncomplicated Plasmodium falciparum malaria with azithromycin–quinine combinations: a randomized, dose-ranging study. Am J Trop Med Hyg. 2006;74:401–6. [PubMed] [Google Scholar]
- 57.Dunne MW, Singh N, Shukla M, et al. A double-blind, randomized study of azithromycin compared to chloroquine for the treatment of Plasmodium vivax malaria in India. Am J Trop Med Hyg. 2005;73:1108–11. [PubMed] [Google Scholar]
- 58.Dunne MW, Singh N, Shukla M, et al. A multicenter study of azithromycin, alone and in combination with chloroquine, for the treatment of acute uncomplicated Plasmodium falciparum malaria in India. J Infect Dis. 2005;191:1582–8. doi: 10.1086/429343. [DOI] [PubMed] [Google Scholar]
- 59.Sidhu AB, Sun Q, Nkrumah LJ, et al. In vitro efficacy, resistance selection, and structural modeling studies implicate the malarial parasite apicoplast as the target of azithromycin. J Biol Chem. 2007;282:2494–504. doi: 10.1074/jbc.M608615200. [DOI] [PubMed] [Google Scholar]
- 60.Peters W, Porter M. The chemotherapy of rodent malaria, XXVI. The potential value of WR 122,455 (a 9-phenanthrenemethanol) against drug-resistant malaria parasites. Ann Trop Med Parasitol. 1976;70:271–81. doi: 10.1080/00034983.1976.11687123. [DOI] [PubMed] [Google Scholar]
- 61.Powers KG, Jacobs RL, Good WC, et al. Plasmodium vinckei: production of chloroquine-resistant strain. Exp Parasitol. 1969;26:193–202. doi: 10.1016/0014-4894(69)90112-x. [DOI] [PubMed] [Google Scholar]
- 62.Rosario VE. Genetics of chloroquine resistance in malaria parasites. Nature. 1976;261:585–6. doi: 10.1038/261585a0. [DOI] [PubMed] [Google Scholar]
- 63.Rathod PK, McErlean T, Lee PC. Variations in frequencies of drug resistance in Plasmodium falciparum. Proc Natl Acad Sci USA. 1997;94:9389–93. doi: 10.1073/pnas.94.17.9389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Trotta RF, Brown ML, Terrell JC, et al. Defective DNA repair as a potential mechanism for the rapid development of drug resistance in Plasmodium falciparum. Biochemistry. 2004;43:4885–91. doi: 10.1021/bi0499258. [DOI] [PubMed] [Google Scholar]
- 65.Peterson DS, Walliker D, Wellems TE. Evidence that a point mutation in dihydrofolate reductase–thymidylate synthase confers resistance to pyrimethamine in falciparum malaria. Proc Natl Acad Sci USA. 1988;85:9114–8. doi: 10.1073/pnas.85.23.9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ramakrishnan SP, Prakash S, Chowdhury DS, et al. Studies on Plasmodium berghei Vincke and Lips, 1948. XXIX. The size of parasite population and its relation to the selection of a strain resistant to sulphadiazine. Indian J Malariol. 1961;15:95–106. [PubMed] [Google Scholar]
- 67.Martin DC, Arnold JD. The effect of parasite populations on the curative action of pyrimethamine. Trans R Soc Trop Med Hyg. 1968;62:379–84. doi: 10.1016/0035-9203(68)90089-8. [DOI] [PubMed] [Google Scholar]
- 68.White NJ, Pongtavornpinyo W. The de novo selection of drug-resistant malaria parasites. Proc Biol Sci. 2003;270:545–54. doi: 10.1098/rspb.2002.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Daher W, Biot C, Fandeur T, et al. Assessment of Plasmodium falciparum resistance to ferroquine (SSR97193) in field isolates and in W2 strain under pressure. Malar J. 2006;5:11. doi: 10.1186/1475-2875-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Aly NS, Hiramoto A, Sanai H, et al. Proteome analysis of new antimalarial endoperoxide against Plasmodium falciparum. Parasitol Res. 2007;100:1119–24. doi: 10.1007/s00436-007-0460-8. [DOI] [PubMed] [Google Scholar]
- 71.Nkrumah LJ, Riegelhaupt PM, Moura P, et al. Probing the multifactorial basis of Plasmodium falciparum quinine resistance: evidence for a strain-specific contribution of the sodium–proton exchanger PfNHE. Mol Biochem Parasitol. 2009;165:122–31. doi: 10.1016/j.molbiopara.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]