Abstract
Runx2 has been identified as “a master gene” for the differentiation of osteoblasts and Runx2-deficient mice has demonstrated a complete absence of mature osteoblast and ossification. To further characterize the Runx2 responsive elements within the bone sialoprotein (BSP) promoter and further investigate into the role of Runx2 haploinsufficiency in osteoblast differentiation, mBSP9.0Luc mice and mBSP4.8Luc mice were crossed with Runx2-deficient mice respectively. Luciferase assay, micro CT scan, and histological analysis were performed using tissues isolated from mBSP9.0luc/Runx2+/− mice, mBSP4.8luc/Runx2+/− mice and their corresponding Runx2+/+ littermates. Alkaline phosphatase activity, mineralization assays and RT-PCR analysis using calvarial osteoblasts isolated from these transgenic mice were also performed. Luciferase assay demonstrated an early increase in luciferase expression in mBSP9.0luc/Runx2+/− mice before the expression level of luciferase dramatically decreased and turned lower than that in their control littermates in later stages. In contrast, luciferase expression in mBSP4.8luc/Runx2+/− failed to show such an early increase. Micro CT scan and histological analysis showed that BMD and trabecular bone volume were decreased and bone formation was delayed in Runx2+/− mice. Furthermore, mineralization assay and semi-quantitative RT-PCR assay demonstrated a gene-dose-dependent decrease in bone nodule formation and bone marker genes expression levels in cultured calvarial osteoblasts derived from Runx2 knockout mice. Reconstitution of Runx2-null cells with Runx2 vector partially rescued the osteoblast function defects. In conclusion, the 9.0 kb BSP promoter demonstrated a higher tissue-specific regulation of the BSP gene by Runx2 in vivo and full Runx2 gene dose is essential for osteoblast differentiation and normal bone formation.
Keywords: Runx2, Bone sialoprotein (BSP), Osteoblast differentiation, Transgenic mouse, Bone formation
INTRODUCTION
Bone sialoprotein (BSP) is a major non-collagenous extracellular matrix protein (Fisher et al., 1990) which has been shown to nucleate hydroxyapatite crystal formation in vitro and is a member of the SIBLING gene family of glycoproteins involved in regulating mineral crystal formation in bones and teeth. In mineralized tissues, BSP expression is associated with the onset of mineralization and is essentially restricted to differentiated osteoblasts, cementoblasts, odontoblasts and ameloblasts (Paz et al., 2005; Li et al., 2005). Together with alkaline phosphatase (ALP), type I collagen (Col1a1), and osteopontin (OPN), BSP has long served as an early marker of osteoblast differentiation (Huang et al., 2007). Based on the fact that BSP is a unique indicator for the formation of osteoblasts from preosteoblastic cells, which is a crucial step in osteogenesis, we believe that identifying molecular mechanisms that regulate the expression of this gene will provide important insights into the molecular control of osteodifferentiation.
To determine more definitively the sites of cell- and tissue-specific regulation of the BSP gene in vivo, we have generated transgenic mice harboring the 9.0 kb (mBSP9.0Luc) and 4.8 kb (mBSP4.8Luc) upstream mouse BSP promoter linked to a luciferase reporter gene respectively (Paz et al., 2005). Bone tissues isolated from mBSP9.0Luc mice such as calvaria, tibia and mandible, demonstrated an extremely high level of luciferase expression, whereas soft tissues, such as muscle, skin, intestine, spleen, liver and kidney showed negligible expression of the reporter gene. The transgenic mouse line mBSP4.8Luc also demonstrated selective expression of the reporter gene but the luciferase expression in mineralized tissues was significantly lower when compared with that in mBSP9.0Luc mice (Paz et al., 2005). Considering that BSP expression is an early marker for osteoblastic differentiation, these transgenic mice provide us a useful tool which enables us to further investigate BSP gene regulation as well as molecular mechanisms of osteogenic differentiation in vivo using a more convenient and more accurate method.
Among those genes which play important roles in osteogenic differentiation, Runx2 has been identified as “a master gene” for the differentiation of osteoblasts. Runx2, also known as Cbfa1, belongs to the runt-domain gene family. Analysis of Runx2-deficient mice has revealed a complete absence of mature osteoblast and ossification with only a few immature osteoblasts which expressed ALP weakly but not OPN and osteocalcin (OCN) (Otto et al., 1997; Komori et al., 1997). Runx2+/− mice demonstrated specific skeletal abnormalities that are characteristic of the human heritable skeletal disorder, cleidocranial dysplasia (CCD) (Otto et al., 1997). Moreover, Runx2 mutations have been identified as the etiological factor of CCD and heterozygous loss of Runx2 function is believed to be sufficient to produce the disorder (Mundlos et al., 1997). It was also reported that OSE2 (osteoblast-specific cis-acting element, which is the binding site of Runx2)-like elements, are present in the promoter of genes such as OCN, BSP and OPN and overexpression of exogenous Runx2 in MC3T3-E1 cells and C3H10T1/2 cells induced the expressions of OCN and BSP (Ducy et al., 1997).
To further characterize the Runx2 responsive elements within the BSP promoter and further investigate into the role of Runx2 haploinsufficiency in osteoblast differentiation, mBSP9.0luc mice and mBSP4.8luc mice were crossed with Runx2-deficient mice respectively to determine the extent of Runx2 regulation on BSP expression. We also performed in vitro experiments using Runx2-deficient cells to delineate the molecular mechanisms of BSP gene regulation by this essential transcription factor for osteogenic differentiation.
MATERIALS AND METHODS
Generation of mBSP9.0luc/Runx2+/− and mBSP4.8luc/Runx2+/− transgenic mouse line
Runx2 knock-out mice (all heterozygotes) were a generous gift from Dr. Mike Owen (London, UK). Our established transgenic mouse lines mBSP9.0luc and mBSP4.8luc (Paz et al., 2005) were crossed with Runx2+/− mouse line. The genotype of the offspring was confirmed by luciferase expression and Southern blotting of tail DNA with probes specific for Runx2 provided by Dr. Owen (Otto et al., 1997). All the embryos (E17.5) and neonatal offspring were harvested for analyses. mBSP9.0luc/Runx2+/+ and mBSP4.8luc/Runx2+/+ mice served as controls respectively.
Luciferase assays in tissues of transgenic mice
After euthanasia organs (calvaria, mandible, tibia and soft tissues) were excised from 1-, 7-, 14-, 42-, and 70-day-old mBSP9.0luc/Runx2+/− and mBSP4.8luc/Runx2+/− transgenic mice as well as their corresponding control mice. The isolated tissues were homogenized in buffer purchased from Analytical Luminescence (San Diego, CA). Luciferase assays were performed using a luminometer (Lumat LB9501, EG&G Berthold, Bad Wildbach, Germany) as described previously (Paz et al., 2005). Briefly, light emission was integrated for the initial 2 sec at 25 °C and recorded as a relative light unit (RLU). All tissue luciferase assay recordings were normalized for protein concentration determined spectrophotometrically (Du640 spectrophotometer, Beckman, Fullerton, CA) using the bicinchoninic acid (BCA) protein procedure (Pierce, Rockford, IL).
MicroCT measurement
The femurs isolated from 1-day-, 7-day-, and 8-week-old mBSP9.0luc/Runx2+/− mice, mBSP4.8luc/Runx2+/− mice, and their corresponding control mice were scanned by micro computed tomography (microCT) at 27 μm voxel resolution on a GE eXplore Locus Micro CT scanner. The following parameters were measured: bone volume, bone surface, and surface-to-volume ratio. At a 3D level the following calculations were made as previously published (Lane et al., 2005; Xiao et al., 2005): relative bone volume over total bone volume (BV/TV), trabecular number (Tb.N), trabecular thickness (Tb.Th), and trabecular separation (Tb.Sp). The diameter of spheres filling the structure was taken as Tb.Th, the thickness of the marrow spaces as Tb.Sp, and the inverse of the mean distances of the skeletal structure was calculated as Tb.N.
Tissue preparation and histological analysis
After euthanasia, tibias were isolated from 8-week-old mBSP9.0luc/Runx2+/− and mBSP4.8luc/Runx2+/− mice and their corresponding control mice. The tissues were fixed immediately in periodate-lysine-paraformaldehyde for 24 hours. The tissues were then demineralized in 0.2N HCL for 2 days with constant stirring. Demineralized tissues were washed in 0.1M phosphate buffer (pH 7.2) overnight. The tissues were then dehydrated in an ascending series of ethanol, cleared in xylene, and embedded in paraffin. Tissue sections, 6 μm thick, were mounted on glass slides and hematoxylin & eosin (H&E) staining was performed.
Calvarial osteoblast cells culture
Murine osteoblast precursor cells in the form of calvarial cells were prepared by enzymatic digestion as described by others (Komori T et al., 1997; Bellows et al., 1998) and routinely cultured in α-MEM (Life Technologies, Rockville, MD) supplemented with 10% HIFBS (Life Technologies, Rockville, MD), 2 mM L-glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin. Calvarial cultures were used up to passage 10 and medium was replaced every three days.
Alkaline phosphatase activity assay
An alkaline phosphatase (ALP) assay was performed in cell layers by colorimetric assay of enzyme activity using an alkaline phosphatase kit from BioAssay system (Hayward, CA) as recommended by the manufacturer. To prepare the cell lysates, cell layers were washed three times with TBS buffer (50 mM Tris, pH 7.4, and 0.15 M NaCl) and then scraped into TBS/Triton buffer (0.1% Triton X-100) followed by sonication and centrifugation to remove cellular debris. One hundred microliter of lysate was then mixed with 100 μl of the freshly prepared colorimetric substrate para-nitrophenyl phosphate, and incubated at 37 °C for 30 min. The enzymatic reaction was stopped by adding 100 μl of 0.2 N NaOH solution. The optical density of the yellow product para-nitrophenol was determined spectrophotometrically at 405 nm. Protein concentration of the cell lysates was measured with a BCA Protein Assay Kit (Pierce, Rockford, IL), and ALP activity was then expressed as para-nitrophenol produced in nmol/min/mg of protein.
Mineralization assays
In vitro alizarin red stainings were performed essentially as described (Tu et al., 2006) after calvarial cells were maintained in culture medium supplemented with 5 mM β-glycerophosphate and 50 μg/ml of ascorbic acid for 14 days. For quantification of mineralization, Alizarin Red-S was extracted with 10% cetylpyridinium chloride and assessed at 562 nm. Total protein content was measured with a BCA Protein Assay Kit (Pierce, Rockford, IL).
RNA isolation and RT-PCR Analysis
Total RNA were extracted by using an RNeasy Kit (Qiagen) following the manufacturer’s instructions. One microgram of the RNA were used to perform the RT-PCR using SuperScript™ one-step RT-PCR with platinum Taq (Invitrogen). The sequences of the primers for amplification of mouse BSP were: 5′-GGAGGGGGCTTCACTGAT-3′ and 5′-AACAATCCGTGCCACCA-3′ (product size: 1048 bp); for mouse glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were: 5′-ACCACAGTCCATGCCATCAC-3′ and 5′-TCCACCACCCTGTTGCTGTA-3′ (product size: 450 bp); for ALP were: 5′-GAAGACGTGGCGGTCTTTGC-3′ and 5′-GGGAATCTGTGCAGTCTGTG-3′ (product size: 457 bp); for OPN were: 5′-CTCCCGGTGAAAGTGACTGA-3′ and 5′-GACCTCAGAAGATGAACTCT-3′ (product size: 831 bp); for OCN were: 5′-CTCCCGGTGAAAGTGACTGA-3′ and 5′-GACCTCAGAAGATGAACTCT-3′ (product size: 371 bp); for Osx were: 5′-ATTCTCCCATTCTCCCTCCCT-3′ and 5′-GGAAGGGTGGGTAGTCATTTGC-3′ (product size: 376 bp), and for Runx2/Cbfa1 were: 5′-GAGGCCGCCGCACGACAACCG-3′ and 5′-CTCCGGCCCACAAATCTCAGA-3′ (product size: 294 bp). PCR products were electrophoresed on 1.5% agarose gel and the gel photographed and quantitated using UVP Image software. GAPDH amplification was performed for normalization purposes.
Reconstitution of calvarial cells by transfection of Runx2 vector
The Runx2 −/− calvarial cells were transfected with the expression plasmid for Runx2-Osf2/Cbfa1 (pCMV-Osf2/Cbfa1) or empty vector pCMV5 plasmid (a gift from Dr. Karsenty’s laboratory, Department of Genetics & Development, College of Physicians and Surgeons, Columbia University, New York, NY) using the liposome reagent (Qiagen, Valencia, CA) as previously described (Tu et al., 2004). Positive clones were then selected in G418-containing media for 14 days. Similarly, calvarial cells isolated from 1-day-old mBSP9.0luc/Runx2+/− and mBSP4.8luc/Runx2+/− mice were also transiently transfected with pCMV-Osf2/Cbfa1 or pCMV5 plasmid.
Statistical Analysis
All results are expressed as means ± SEM of 3 or more independent experiments. One-way ANOVA was used to test significance using the Statgraphic statistical graphics system software package (STSC, Rockville, MD). Values of p lower than 0.05 were considered statistically significant.
RESULTS
Identification of mBSP9.0luc/Runx2+/− and mBSP4.8luc/Runx2+/− transgenic mouse line
Luciferase assays of all the animals involved in the following studies demonstrated strong expression of luciferase and Southern blotting of tail DNA successfully identified mBSPluc/Runx2+/− mice from mBSPluc/Runx2+/+ mice. Southern blot analysis of E18 embryos identified mBSPluc/Runx2−/− embryos, which served as cell donors for in vitro experiments.
Luciferase expression patterns in tissues isolated from different transgenic mouse lines
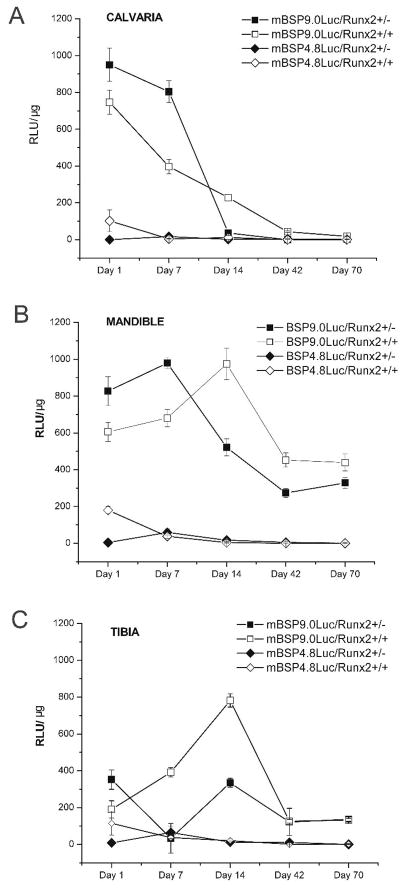
Luciferase assays of mineralized tissues (calvaria, mandible and tibia) isolated from mBSP9.0luc/Runx2+/− mice and mBSP9.0luc/Runx2+/+ mice demonstrated that in these mineralized tissues haploinsufficiency of Runx2 resulted in similar luciferase expression pattern changes which were characterized with an early increase in luciferase level in mBSP9.0luc/Runx2+/− mice when compared with that in mBSP9.0luc/Runx2+/+ mice. However, there were also some minor differences observed among different bone types which may be in part due to their different developmental schedules. Briefly, the calvaria and mandibules derived from 1-d- and 7-d-old mBSP9.0luc/Runx2+/− mice showed higher luciferase expression level than those from mBSP9.0luc/Runx2+/+ mice. However, luciferase expression in calvaria and mandibles isolated from older mBSP9.0luc/Runx2+/− mice (14-d-, 42-d-, and 70-d-old) was dramatically decreased compared with those at earlier stages and was lower than that in control mice. The difference in luciferase expression levels between older mBSP9.0luc/Runx2+/− mice and mBSP9.0luc/Runx2+/+ mice (14-d-, 42-d-, and 70-d-old) was more prominent in mandibles than in calvaria. The tibiae isolated from 1-d-old mBSP9.0luc/Runx2+/− mice also showed higher luciferase expression level than those from mBSP9.0luc/Runx2+/+ mice. However, in 7-d- and 14-d-old animals luciferase expression was observed to be lower in tibiae from mBSP9.0luc/Runx2+/− mice than those from mBSP9.0luc/Runx2+/+ mice. In tibiae from 42-d- and 70-d-old animals, no statistically significant difference was observed between those two mouse lines. In contrast, luciferase level in bone tissues of 1-d-old mBSP4.8luc/Runx2+/− mice was lower than that in the mBSP4.8luc/Runx2+/+ mice while in older mice the difference between these two mouse lines was not apparent or of no statistical significance (Figure 1A, B and C). Consistent with our previously published studies (Paz et al., 2005), there was no luciferase expression in intestine and liver tissues isolated from mBSP9.0luc/Runx2+/− mice, mBSP4.8luc/Runx2+/− mice, and the corresponding control mice. Luciferase also showed no expression in skeletal muscles isolated from mBSP9.0luc/Runx2+/− mice and the control mice. In contrast, both mBSP4.8luc/Runx2+/− mice and the control mice demonstrated a slight luciferase expression in their skeletal muscles which again confirmed our previous findings that a longer promoter is necessary for specific expression of BSP. Furthermore, consistent with previous findings that BSP expression can be detected in brain and kidney tissues, all the mouse lines demonstrated luciferase expression in the brain and kidney tissues. However, there is no statistically significant difference between luciferase expression levels in the soft tissues (brain, kidney, or muscle) isolated from mBSP9.0luc/Runx2+/− mice (or mBSP4.8luc/Runx2+/− mice) and the corresponding control mouse line (Data not shown).
Figure 1.
Luciferase expression patterns in tissues isolated from different transgenic mouse lines. Mineralized tissues (calvaria, mandible, tibia) were excised from 1-, 7-, 14-, 42-, and 70-day-old mBSP9.0luc/Runx2+/− and mBSP4.8luc/Runx2+/− transgenic mice as well as their corresponding control mice. Luciferase assays were performed using a luminometer (Lumat LB9501, EG&G Berthold, Bad Wildbach, Germany). Data is presented as mean ± SE. (A) The calvaria derived from 1-d- and 7-d-old mBSP9.0luc/Runx2+/− mice showed higher luciferase expression level than those from mBSP9.0luc/Runx2+/+ mice. However, luciferase expression in 14-d-old mBSP9.0luc/Runx2+/− mice was lower than that in control mice. There was no apparent difference in luciferase expression levels between 42-d- and 70-d-old mBSP9.0luc/Runx2+/− mice and mBSP9.0luc/Runx2+/+ mice. (B) The mandibules derived from 1-d- and 7-d-old mBSP9.0luc/Runx2+/− mice showed higher luciferase expression level than those from mBSP9.0luc/Runx2+/+ mice. However, luciferase expression in mandibles isolated from older mBSP9.0luc/Runx2+/− mice (14-d-, 42-d-, and 70-d-old) was dramatically decreased compared with those at earlier stages and was lower than that in control mice. (C) The tibiae isolated from 1-d-old mBSP9.0luc/Runx2+/− mice showed higher luciferase expression level than those from mBSP9.0luc/Runx2+/+ mice. However, in 7-d- and 14-d-old animals luciferase expression was observed to be lower in tibiae from mBSP9.0luc/Runx2+/− mice than those from mBSP9.0luc/Runx2+/+ mice. In tibiae from 42-d- and 70-d-old animals, no statistically significant difference was observed between those two mouse lines. (A, B, C) Luciferase level in bone tissues of 1-d-old mBSP4.8luc/Runx2+/− mice was lower than that in the mBSP4.8luc/Runx2+/+ mice while in older mice the difference between these two mouse lines was not apparent or of no statistical significance.
Runx2 haploinsufficiency resulted in decreased bone formation
To determine the effect of Runx2 haploinsufficiency on bone formation, femurs isolated from mBSPLuc/Runx2+/− mice and mBSPLuc/Runx2+/+ mice underwent quantitative microCT scan. In Runx2 deficient mice, we observed a gene-dose-dependent reduction in bone mineral density (BMD) and trabecular bone volume at all time points examined (1-day, 7-day and 8-week old mice, Table 1). Compared with the mBSPluc/Runx2+/+ mice, mBSPluc/Runx2+/− mice demonstrated decreased BMD (15.1–25.5%). The total trabecular volume (BV/TV) in mBSPluc/Runx2+/− mice decreased by 20.1–34.9%, and the trabecular number (Tb.N) decreased by 9.2–54.2 % when compared with the control mice. Trabecular thickness (Tb.Th) was 22.8–30.9% lower in mBSPluc/Runx2+/− mice than in control mice, whereas trabecular separation (Tb.Sp) was increased by 22.0–26.5% in mBSPluc/Runx2+/− mice compared with control mice.
Table 1.
Micro-CT analysis of femurs in 1-day, 7-day and 8-week-old wild type and Runx2 haploinsufficiency mice
| Genotype | Runx2+/+ 1 day | Runx2+/− 1 day | Runx2+/+ 7 day | Runx2+/− 7day | Runx2+/+ 8w | Runx2+/− 8w |
|---|---|---|---|---|---|---|
| BMD (mg/cc) | 9.56 ± 0.32 | 7.12 ± 0.29* | 11.69 ± 0.46 | 9.71 ± 0.23* | 24.12 ± 1.12 | 20.45 ± 1.23* |
| BV/TV (%) | 26.54 ± 1.43 | 18.26 ± 0.96* | 36.87 ± 1.84 | 23.98 ± 1.36* | 72.98 ± 3.18 | 58.33 ± 3.76* |
| Tb.Th (μm) | 45.65 ± 2.89 | 31.56 ± 1.59* | 78.12 ± 4.23 | 57.64 ± 4.56* | 189 ± 8.66 | 146 ± 5.6* |
| Tb.N (mm−1) | 5.12 ± 0.32 | 4.65 ± 0.26 | 4.42 ± 0.46 | 3.12 ± 0.21 | 3.67 ± 0.013 | 1.68 ± 0.006* |
| Tb.Sp (μm) | 134 ± 10.51 | 165 ± 15.6 | 150 ± 8.12 | 183 ± 11.65 | 181 ± 6.12 | 229 ± 11.23* |
Data are mean±SEM from 5 individual mice.
Significant difference from wild–type Runx2 +/+ mice.
Histological analysis
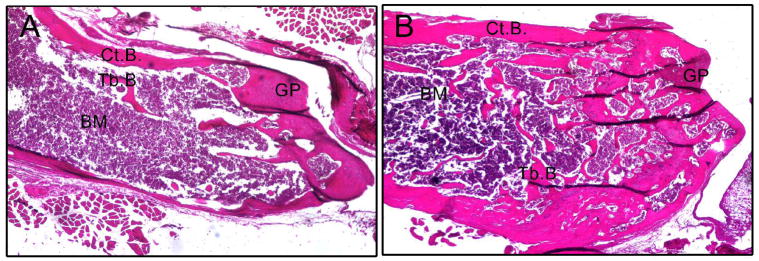
To understand the cellular basis for these structural changes and to identify potential mechanisms responsible for the reduced bone mass in Runx2 haploinsufficiency mice, we examined histological sections of the epiphyseal bone, growth plate, and metaphyseal bone of femurs isolated from 8-week-old wild-type and heterozygous mice (Figure 2). Heterozygous Runx2+/− mice displayed reduced trabecular bone volume, decreased cortical bone thickness and normal appearing growth plate as well as normal bone shape and length, which are consistent with Micro-CT structural analysis.
Figure 2.
Hematoxylin & Eosin staining sections of femur bones isolated from 8-week-old mBSP9.0luc/Runx2+/− mice (A) and mBSP9.0luc/Runx2+/+ mice (B). Heterozygous mBSP9.0luc/Runx2+/− mice showed decreased trabecular bone volume and thinner cortical bone compared with mBSP9.0luc/Runx2+/+ mice, while the growth plate appeared normal in mBSP9.0luc/Runx2+/− mice. Tb.B., trabecular bone; Ct.B., cortical bone; BM, bone marrow; GP, growth plate. Photographs were taken at 40x.
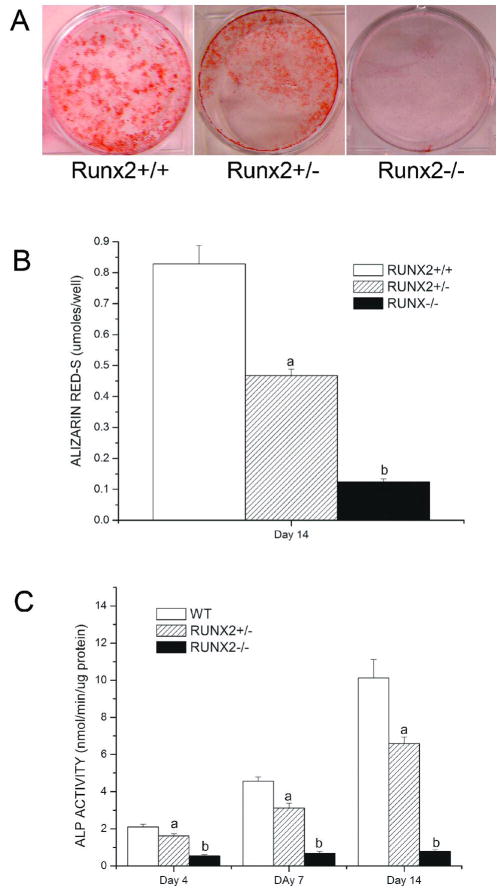
Alkaline phosphatase activity and mineralization assays in calvarial osteoblast cells
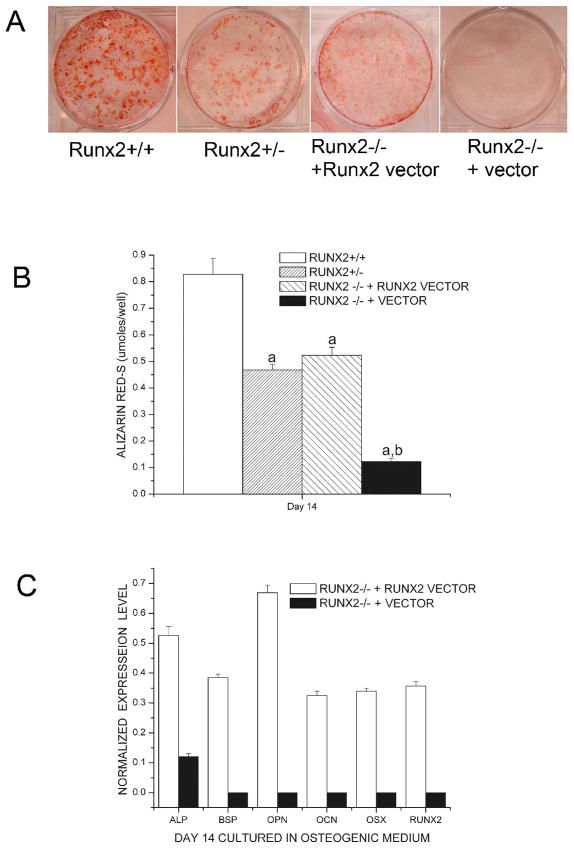
To further confirm the osteoblast function defects in Runx2 deficient mice, we performed calvarial cell cultures derived from Runx2+/+, Runx2+/− and Runx2−/− mice grown in osteoblastic differentiation media for up to 14 days. Compared with wild-type calvarial cells, Runx2 +/− culture showed less abundant mineralized nodules and had significantly lower Alizarin Red-S accumulation at day 14 of culture (Figure 3A, B). The Runx2+/− calvarial cultures also displayed significantly lower ALP activity during 14 days of culture compared with wild type cultures (Figure 3C). Furthermore, Runx2−/− cultures showed no mineralization and only background levels of ALP activity during the overall experimental period.
Figure 3.
Alkaline phosphatase activity and mineralization assays in calvarial osteoblast cells isolated from Runx2 deficient mice and wild type mice. Calvarial cells were isolated from Runx2+/+, Runx2+/− and Runx2−/− mice and were grown in osteoblastic differentiation media for up to 14 days. Compared with wild-type calvarial cells, Runx2 +/− culture showed less abundant mineralized nodules and had significantly lower Alizarin Red-S accumulation at day 14 of culture while Runx2−/− cultures showed no mineralization (A, B). Furthermore, the Runx2+/− calvarial cultures displayed significantly lower ALP activity during 14 days of culture compared with wild type mice and Runx2−/− cultures only showed background levels of ALP activity (C).
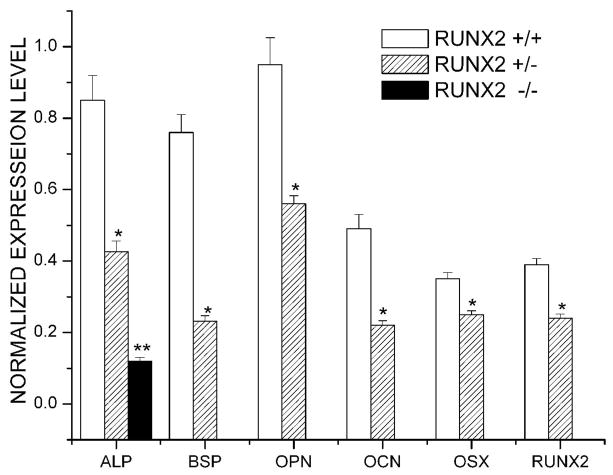
Insufficient Runx2 gene dosage resulted in decreased expressions of bone marker genes and downstream transcription factor
Using semiquantitative RT-PCR analysis, we measured the expression levels of ALP, BSP, OPN, OCN, Runx2 and osterix (Osx) mRNA in cultured calvarial osteoblasts 14 days after osteogenic induction. We found decreased expression levels of all the bone marker genes, Runx2, and Osx in osteoblast cultures derived from mBSPLuc/Runx2 +/− mice compared with those from mBSPLuc/Runx2 +/+ mice (Figure 4).
Figure 4.
Heterozygous Runx2 gene knockout resulted in decreased expressions of bone marker genes and downstream transcription factor. Calvarial cells were isolated from Runx2+/+, Runx2+/− and Runx2−/− mice and were grown in osteogenic media for up to 14 days. Semiquantitative RT-PCR analyses were carried out for early and late differentiation markers as described in methods. Levels of AP, BSP, OC, OPN, Cbfa1, and OSX were normalized with those of the loading control GAPDH in three independent experiments. *p < 0.05, ** P <0.01 vs. Runx2 +/+ control cells.
Overexpression of Runx2 induced osteoblast differentiation in Runx2−/− calvarial cells
To investigate whether exogenous overexpression of Runx2 could rescue the function defects of osteoblasts in Runx2-deficient calvarial cells, we transfected Runx2 −/− cells with Runx2 vector and compared the differentiation commitments with wild-type calvarial cells cultured in osteogenic medium. Alizarin-S red staining displayed that Runx2 deficient cells transfected with Runx2 vectors were capable of forming bone nodules and undergoing mineralization (Figure 5A and B). The results also showed that transfection with Runx2 vector induced the expression of bone markers, ALP, BSP, OCN, OPN and Osx, 14 days after osteogenic induction (Figure 5C).
Figure 5.
Reconstitution of Runx2-null cells partially rescued the phenotype of immortalized Runx2-deficient calvarial cells. Commitment of Runx2 null cells transfected with or without Runx2 vector to osteoblast lineages was tested in osteogenic media (a-MEM supplemented with 10% FBS, 10 mM of b-glycerol phosphate, and 50 mg/ml of ascorbic-acid) and cultured for 14 days (A, B). Total RNA was isolated from all treatment groups at indicated days and semiquantitative RT-PCR analyses were carried out for early and late differentiation markers as described in methods (C). Runx2-null cell cultured in osteogenic cocktail did not differentiate and induction of osteoblastic differentiation was noted only in Runx2-reconstituted cells. Wild-type (Runx2 +/+) and heterozygosity (Runx2+/−) calvarial cells served as controls. aP < 0.0.5 vs. Runx2 +/+ control cells; bP <0.05 vs. Runx2 −/− + Runx2 vector cells.
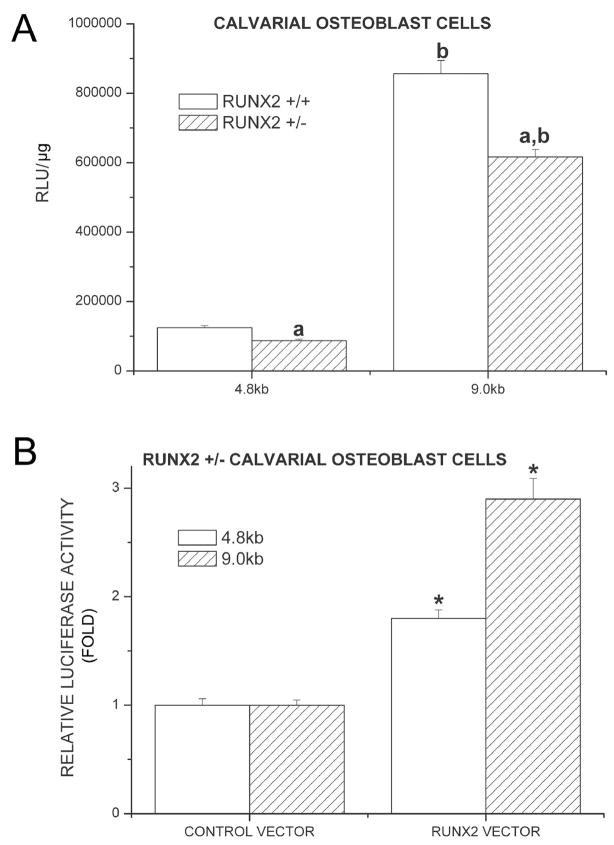
Runx2 enhanced luciferase expression in calvarial cells isolated from mBSP9.0luc/Runx2+/− mice and mBSP4.8luc/Runx2+/− mice in vitro
To further investigate the effect of Runx2 on BSP promoter, we isolated calvarial cells from 1-day-old mBSP9.0luc/Runx2+/− mice, mBSP4.8luc/Runx2+/− mice, and the corresponding control mice. Luciferase assays of these calvarial cells showed that although 1-day-old mBSP9.0luc/Runx2+/− mice demonstrated an increase in luciferase expression in vivo when compared with the control mice, the calvarial cells isolated from these mice demonstrated a lower luciferase activity compared with those from the control mice. Similarly, calvarial cells isolated from mBSP4.8luc/Runx2+/− mice also showed a decrease in luciferase expression level compared with those from the control mice, which was consistent with the results from the above in vivo studies (Figure 6A). To confirm that the direct effect of Runx2 on BSP promoter is to enhance its activity, we reconstituted the calvarial cells isolated from mBSP9.0luc/Runx2+/− mice and mBSP4.8luc/Runx2+/− mice with Runx2 vector. Luciferase assay demonstrated that Runx2 overexpression in calvarial cells isolated from mBSP9.0luc/Runx2+/− mice resulted in a 2.9-fold increase in luciferase level while Runx2 overexpression enhanced the luciferase expression in calvarial cells from mBSP4.8luc/Runx2+/− mice by 1.8-fold (Figure 6B).
Figure 6.
Overexpression of Runx2 enhanced luciferase expression in calvarial cells isolated from mBSP9.0luc/Runx2+/− mice and mBSP4.8luc/Runx2+/− mice in vitro. Luciferase assays of these calvarial cells showed that the calvarial cells isolated from these mice demonstrated a lower luciferase activity compared with those from the control mice (A). In the calvarial cells isolated from those mice, luciferase assay demonstrated that Runx2 overexpression in calvarial cells isolated from mBSP9.0luc/Runx2+/− mice resulted in a 2.9-fold increase in luciferase level while Runx2 overexpression enhanced the luciferase expression in calvarial cells from mBSP4.8luc/Runx2+/− mice by 1.8-fold (B). aP < 0.0.5 vs. Runx2 +/+ control cells; bP <0.05 vs. the cells from mBSP4.8luc/Runx2+/+ or +/− mice; *P < 0.05 vs. control vector transfected cells.
DISCUSSION
In one of our previous studies we first reported a high degree of tissue specificity using a 2.7 kb rat BSP promoter construct with a luciferase reporter gene in transgenic mouse lines. However, the specificity of the BSP promoter driven luciferase expression was not absolute (Chen et al., 1996). To investigate the role of Runx2 in BSP expression Benson et al (Benson et al., 1999) cloned a 2.5 kb fragment of mouse BSP promoter and discovered that although the -1335 site was able to bind with Runx2 in vitro, no significant enhancer activity in the -1335 element could be detected. However, recently the same group of researchers reported that two putative Runx2-binding sites (R1 and R2) were identified within the proximal region of the mouse, rat and human BSP genes which were demonstrated to bind Runx2 both in vitro and in vivo and act as osteoblast-specific transcriptional enhancers. It was also shown that Runx2 not only directly regulates BSP gene expression but also functions together with DLX5 or a related factor to activate BSP gene expression. As for their previous findings, the researchers believed that the -1335 site is not accessible to RUNX2 in vivo thus the site failed to demonstrate enhancer activity (Roca et al, 2005). However, these previous studies were mainly focused on the proximal 2.5 kb of the BSP promoter region and the researchers could not exclude the possibility that additional osteoblast-specific regulatory regions may reside in other parts of the BSP gene (Roca et al, 2005). Thus we have generated two transgenic mouse lines expressing reporter constructs driven by a BSP 9.0 kb promoter or a 4.8 kb promoter lacking two upstream Runx2 elements (Paz et al., 2005). It is revealed that the reporter gene demonstrated a more specific expression pattern in the transgenic mouse line in which the reporter construct is driven by a longer promoter.
To further investigate the transcriptional regulation of BSP by Runx2, our established transgenic mouse lines mBSP9.0Luc and mBSP4.8Luc (Paz et al., 2005) were crossed with Runx2+/− mouse line. Luciferase assays showed that there was no statistically significant difference between the luciferase expression levels in soft tissues isolated from mBSP9.0Luc/Runx2+/− mice (or mBSP4.8Luc/Runx2+/− mice) and the corresponding control mouse lines, which was consistent with the previous report that there is no Runx2 expression in the brain, heart, gut or liver (Huang et al., 2007). Luciferase assays of mineralized tissues demonstrated an early increase in luciferase expression in mBSP9.0Luc/Runx2+/− mice before the expression level of luciferase dramatically decreased and turned lower than that in the control mice at later developmental stages. In contrast, luciferase expression in mBSP4.8Luc/Runx2+/− mice failed to show such an early increase when compared with their control littermates. The early increase in luciferase level observed in mBSP9.0Luc/Runx2+/− mice due to Runx2 haploinsufficiency was unexpected considering that Runx2 has been shown to enhance BSP expression in vitro (Ducy et al., 1997). However, in a study of the gallus BSP gene promoter it was demonstrated that Runx2 mediated repression of the wild-type BSP promoter, although it is well known that Runx2 is able to activate multiple bone specific genes. The researchers suggested that the possible mechanisms could be a corepressor partner interacting with Runx2 or promoter architecture changes induced by Runx2 binding which allows binding by other potential transcription factors that suppress promoter activity (Javed et al., 2001). In a recent study on human BSP promoter, it was reported that Runx2 in pre-confluent Saos-2 human osteosarcoma cells suppressed BSP expression with histone deacetylase 3 (HDAC3) acting as a Runx2 co-repressor. When Saos-2 cells became increasingly confluent and differentiated towards mature osteoblasts, nuclear Runx2 protein level was dramatically down-regulated and BSP expression level was significantly increased (Lamour et al., 2007). In contrast to those of more closely related Gallus and human BSP, mouse BSP promoter organizational properties was believed to be different and the researchers suggested that down-regulation of BSP by Runx2 was more important for regulation in humans and Gallus than for that in rodent species (Javed et al., 2001). However, our results demonstrating elevated luciferase expression level induced by Runx2 haploinsufficiency in mBSP9.0Luc/Runx2+/− mice during the early stages of development also suggested a possible negative regulation of BSP promoter by Runx2. Moreover, our results also showed that mBSP4.8Luc/Runx2+/− failed to demonstrate such an early increase in luciferase expression and the luciferase level in these mice was significantly decreased due to the Runx2 haploinsufficiency at early developmental stages. These results are consistent with the previous findings that Runx2 binding sites in the proximal region of BSP promoter demonstrate enhancer activities (Roca et al., 2005). It was also suggest that the Runx2 binding sites deleted in mBSP4.8Luc/Runx2+/− mice may contribute to the suppression effect by Runx2 observed in mBSP9.0Luc/Runx2+/− mice during early developmental stages.
In order to investigate whether Runx2 suppress BSP promoter in a direct way, we performed several in vitro studies. Luciferase assays showed that although 1-day-old mBSP9.0luc/Runx2+/− mice demonstrated an increase in luciferase expression in vivo when compared with the control mice, the calvarial cells isolated from these mice showed a lower luciferase activity compared with those from the control mice. Similarly, calvarial cells isolated from mBSP4.8luc/Runx2+/− mice also showed a decrease in luciferase expression level compared with those from the control mice, which was consistent with the results from the above in vivo studies. Furthermore, Runx2 overexpression in calvarial cells isolated from mBSP9.0luc/Runx2+/− mice resulted in a 2.9-fold increase in luciferase level while Runx2 overexpression enhanced the luciferase expression in calvarial cells from mBSP4.8luc/Runx2+/− mice by 1.8-fold. All these results demonstrated that under simpler in vitro conditions, the effect of Runx2 on mouse BSP promoter is to enhance its activity. Moreover, Runx2 overexpression induced a higher increase in the activity of luciferase driven by a longer promoter. Taken together, our results and the previous findings suggested that the suppression effect of Runx2 on BSP promoter is in an indirect way. During early developmental stages a corepressor may interact with Runx2 and restrict excessive BSP expression. Furthermore, our results suggested that in vivo expressions of bone matrix proteins are regulated in a subtle and complicated way which may be very important for the accomplishment of normal bone development. Other studies have also shown that although Runx2 is a “master” gene during osteogenic differentiation, overexpression of Runx2 in a transgenic mouse line resulted in osteopenia, multiple bone fracture, and a significant decrease in terminally differentiated osteoblasts, which also demonstrated suppressive effect of Runx2 on osteogenic differentiation in vivo (Liu et al., 2001). For the first time our results demonstrated the dual effects of Runx2 on BSP promoter in vivo and these results further indicated that the complexity of in vivo environment could not be completely replicated by in vitro experiments.
Runx2 is a positive regulator of bone development, mineralization and osteoblast differentiation and its mutation and haploinsufficiency has been related to the human hereditary skeletal disorder, cleidocranial dysplasia (CCD) which is a disorder exhibiting defective endochondral and intra-membranous bone formation. Typical features include hypoplasia/aplasia of clavicles, patent fontanelles, Wormian bones (additional cranial plates caused by abnormal ossification of the calvaria), supernumerary teeth, short stature, and other skeletal changes (Mundlos et al., 1997). Tsuji et al. reported that after bone marrow ablation in the operated bone marrow cavity bone formation was significantly reduced in Runx2 heterozygous knockout mice while new bone formation was observed in WT mice. Furthermore, this effect was observed specifically in aged mice (Tsuji et al., 2004). It has also been reported that full Runx2 gene dosage is required for maintaining normal function of osteoblasts in mechanical unloading or nonphysiological condition (Salingcarnboriboon et al., 2006). In Runx2 heterozygous knockout mice, Yoda et al. observed delayed tooth eruption and decreased osteoclast number in the eruption pathway and concluded that although Runx2 haploinsufficiency does not retard the remodeling in femurs, full Runx2 gene dosage is required for the active alveolar bone resorption essential for the tooth eruption (Yoda et al., 2004). In a recent study immortalized Runx2−/− calvarial cells were shown to have no osteoblastic gene expression while cells infected with wild-type Runx2 adenovirus showed a robust temporal increase in the expression of osteoblast marker genes and were competent to respond to BMP2 (Bae et al., 2007). However, this study was mainly focused on the investigation of the specific Runx2-functional domains and did not provide more information of the gene-dose-dependent regulation of bone matrix proteins expression, osteoblast differentiation and mineralization by Runx2. Chung et al. also reported that Runx2 haploinsufficiency resulted in shorter tooth root, irregularities in root morphology and thinner parietal bone (Chung et al., 2004). Using micro CT and histological analysis of the long bones (femurs) we observed that BMD and trabecular bone volume were decreased and bone formation was delayed in Runx2+/− mice. Furthermore, in vitro mineralization assay and semi-quantitative RT-PCR assay demonstrated a gene-dose-dependent decrease in bone nodules formation and bone marker genes expression levels in cultured calvarial osteoblasts derived from Runx2 knockout mice. Reconstitution of Runx2-Null Cells with Runx2 vector showed a robust temporal increase in the expression of osteoblast marker genes and the restoration of bone nodule formation and mineralization. These results, together with the previous findings by other researchers, further confirmed that full Runx2 gene dose is essential for normal bone formation, bone remodeling and bone regeneration.
Acknowledgments
The study was supported by NIH grants DE 11088 and DE16710 to JC.
Contract grant sponsor: NIH
Contract grant numbers: DE 11088 and DE16710
LITERATURE CITED
- Bae JS, Gutierrez S, Narla R, Pratap J, Devados R, van Wijnen AJ, Stein JL, Stein GS, Lian JB, Javed A. Reconstitution of Runx2/Cbfa1-null cells identifies a requirement for BMP2 signaling through a Runx2 functional domain during osteoblast differentiation. J Cell Biochem. 2007 Feb 1;100(2):434–49. doi: 10.1002/jcb.21039. [DOI] [PubMed] [Google Scholar]
- Bellows CG, Ciaccia A, Heersche JN. Osteoprogenitor cells in cell populations derived from mouse and rat calvaria differ in their response to corticosterone, cortisol, and cortisone. Bone. 1998;23:119–125. doi: 10.1016/s8756-3282(98)00084-2. [DOI] [PubMed] [Google Scholar]
- Benson MD, Aubin JE, Xiao G, Thomas PE, Franceschi RT. Cloning of a 2.5 kb murine bone sialoprotein promoter fragment and functional analysis of putative Osf2 binding sites. J Bone Miner Res. 1999;14:396–405. doi: 10.1359/jbmr.1999.14.3.396. [DOI] [PubMed] [Google Scholar]
- Chen J, Thomas HF, Jin H, Jiang H, Sodek J. Expression of rat bone sialoprotein promoter in transgenic mice. J Bone Miner Res. 1996;11:654–664. doi: 10.1002/jbmr.5650110513. [DOI] [PubMed] [Google Scholar]
- Chung CR, Tsuji K, Nifuji A, Komori T, Soma K, Noda M. Micro-CT evaluation of tooth, calvaria and mechanical stress-induced tooth movement in adult Runx2/Cbfa1 heterozygous knock-out mice. J Med Dent Sci. 2004 Mar;51(1):105–13. [PubMed] [Google Scholar]
- D’Alonzo RC, Selvamurugan N, Karsenty G, Partridge NC. Physical interaction of the activator protein-1 factors c-Fos and c-Jun with Cbfa1 for collagenase-3 promoter activation. J Biol Chem. 2002;277:816–822. doi: 10.1074/jbc.M107082200. [DOI] [PubMed] [Google Scholar]
- Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89:747–754. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- Fisher LW, McBride OW, Termine JD, Young MF. Human bone sialoprotein. Deduced protein sequence and chromosomal localization. J Biol Chem. 1990;265:2347–2351. [PubMed] [Google Scholar]
- Ganss B, Kim RH, Sodek J. Bone sialoprotein. Crit Rev Oral Biol Med. 1999;10:79–98. doi: 10.1177/10454411990100010401. [DOI] [PubMed] [Google Scholar]
- Huang W, Yang S, Shao J, Li YP. Signaling and transcriptional regulation in osteoblast commitment and differentiation. Front Biosci. 2007;12:3068–3092. doi: 10.2741/2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javed A, Barnes GL, Jasanya BO, Stein JL, Gerstenfeld L, Lian JB, Stein GS. Runt homology domain transcription factors (Runx, Cbfa, and AML) mediate repression of the bone sialoprotein promoter: evidence for promoter context-dependent activity of Cbfa proteins. Mol Cell Biol. 2001;21(8):2891–905. doi: 10.1128/MCB.21.8.2891-2905.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- Lamour V, Detry C, Sanchez C, Henrotin Y, Castronovo V, Bellahcene A. RUNX2and HDAC3 mediated-repression is relieved in differentiating human osteoblast cells to allow high BSP expression. J Biol Chem. 2007 Oct;22 doi: 10.1074/jbc.M705833200. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Lane NE, Yao W, Nakamura MC, Humphrey MB, Kimmel D, Huang X, Sheppard D, Ross FP, Teitelbaum SL. Mice lacking the integrin beta5 subunit have accelerated osteoclast maturation and increased activity in the estrogen-deficient state. J Bone Miner Res. 2005;20:58–66. doi: 10.1359/JBMR.041017. [DOI] [PubMed] [Google Scholar]
- Li L, Zhu J, Tu Q, Yamauchi M, Sodek J, Karsenty G, Tang J, Chen J. An in vivo model to study osteogenic gene regulation: targeting an avian retroviral receptor (TVA) to bone with the bone sialoprotein (BSP) promoter. J Bone Miner Res. 2005;20:1403–1413. doi: 10.1359/JBMR.050316. [DOI] [PubMed] [Google Scholar]
- Liu W, Toyosawa S, Furuichi T, Kanatani N, Yoshida C, Liu Y, Himeno M, Narai S, Yamaguchi A, Komori T. Overexpression of Cbfa1 in osteoblasts inhibits osteoblast maturation and causes osteopenia with multiple fractures. J Cell Biol. 2001;155(1):157–66. doi: 10.1083/jcb.200105052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundlos S, Otto F, Mundlos C, Mulliken JB, Aylsworth AS, Albright S, Lindhout D, Cole WG, Henn W, Knoll JH, Owen MJ, Mertelsmann R, Zabel BU, Olsen BR. Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell. 1997;89:773–779. doi: 10.1016/s0092-8674(00)80260-3. [DOI] [PubMed] [Google Scholar]
- Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen BR, Selby PB, Owen MJ. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- Paz J, Wade K, Kiyoshima T, Sodek J, Tang J, Tu Q, Yamauchi M, Chen J. Tissue- and bone cell-specific expression of bone sialoprotein is directed by a 9.0 kb promoter in transgenic mice. Matrix Biol. 2005;24:341–352. doi: 10.1016/j.matbio.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Roca H, Phimphilai M, Gopalakrishnan R, Xiao G, Franceschi RT. Cooperative interactions between RUNX2 and homeodomain protein-binding sites are critical for the osteoblast-specific expression of the bone sialoprotein gene. J Biol Chem. 2005;280(35):30845–55. doi: 10.1074/jbc.M503942200. [DOI] [PubMed] [Google Scholar]
- Salingcarnboriboon R, Tsuji K, Komori T, Nakashima K, Ezura Y, Noda M. Runx2 is a target of mechanical unloading to alter osteoblastic activity and bone formation in vivo. Endocrinology. 2006;147:2296–2305. doi: 10.1210/en.2005-1020. [DOI] [PubMed] [Google Scholar]
- Selvamurugan N, Chou WY, Pearman AT, Pulumati MR, Partridge NC. Parathyroid hormone regulates the rat collagenase-3 promoter in osteoblastic cells through the cooperative interaction of the activator protein-1 site and the runt domain binding sequence. J Biol Chem. 1998;273:10647–10657. doi: 10.1074/jbc.273.17.10647. [DOI] [PubMed] [Google Scholar]
- Schmitt CP, Homme M, Schaefer F. Structural organization and biological relevance of oscillatory parathyroid hormone secretion. Pediatr Nephrol. 2005;20:346–351. doi: 10.1007/s00467-004-1767-7. [DOI] [PubMed] [Google Scholar]
- Teitelbaum SL. Mice lacking the integrin β5 subunit have accelerated osteoclast maturation and increased activity in the estrogen-deficient state. J Bone Miner Res. 2005;20:58–66. doi: 10.1359/JBMR.041017. [DOI] [PubMed] [Google Scholar]
- Thirunavukkarasu K, Mahajan M, McLarren KW, Stifani S, Karsenty G. Two domains unique to osteoblast-specific transcription factor Osf2/Cbfa1 contribute to its transactivation function and its inability to heterodimerize with Cbfbeta. Mol Cell Biol. 1998;18:4197–4208. doi: 10.1128/mcb.18.7.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji K, Komori T, Noda M. Aged mice require full transcription factor, Runx2/Cbfa1, gene dosage for cancellous bone regeneration after bone marrow ablation. J Bone Miner Res. 2004;19:1481–1489. doi: 10.1359/JBMR.040601. [DOI] [PubMed] [Google Scholar]
- Tu Q, Yamauchi M, Pageau SC, Chen J. Autoregulation of bone sialoprotein gene in pre-osteoblastic and non-osteoblastic cells. Biochem Biophys Res Commun. 2004;316:461–467. doi: 10.1016/j.bbrc.2004.02.068. [DOI] [PubMed] [Google Scholar]
- Tu Q, Valverde P, Chen J. Osterix enhances proliferation and osteogenic potential of bone marrow stromal cells. Biochem Biophys Res Commun. 2006;341:1257–1265. doi: 10.1016/j.bbrc.2006.01.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z, Awad HA, Liu S, Mahlios J, Zhang S, Guilak F, Mayo MS, Quarles LD. Selective Runx2-II deficiency leads to low-turnover osteopenia in adult mice. Dev Biol. 2005;283:345–356. doi: 10.1016/j.ydbio.2005.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoda S, Suda N, Kitahara Y, Komori T, Ohyama K. Delayed tooth eruption and suppressed osteoclast number in the eruption pathway of heterozygous Runx2/Cbfa1 knockout mice. Arch Oral Biol. 2004;49:435–442. doi: 10.1016/j.archoralbio.2004.01.010. [DOI] [PubMed] [Google Scholar]
- Yoshida CA, Furuichi T, Fujita T, Fukuyama R, Kanatani N, Kobayashi S, Satake M, Takada K, Komori T. Core-binding factor beta interacts with Runx2 and is required for skeletal development. Nat Genet. 2002;32:633–638. doi: 10.1038/ng1015. [DOI] [PubMed] [Google Scholar]