Abstract
Glutaredoxins, proteins traditionally involved in redox reactions, are also required for iron–sulphur cluster assembly and haem biosynthesis. These new roles are likely related to the ability of some glutaredoxins to bind labile [2Fe–2S] clusters and to transfer them rapidly and efficiently to acceptor proteins. Recent results point to putative roles for glutaredoxins in the sensing of cellular iron and in iron–sulphur cluster biogenesis, either as scaffold proteins for the de novo synthesis of iron–sulphur clusters or as carrier proteins for the transfer of preformed iron–sulphur clusters. Based on prokaryote genome analysis and in vivo studies of iron regulation in yeast, we propose putative new roles and binding partners for glutaredoxins in the assembly of metalloproteins.
Pathways for iron–sulphur cluster assembly
Iron is required for the function of many metalloproteins including those containing iron–sulphur (Fe–S) clusters, haems, mono- or bi-nuclear non-haem iron centres, and the more complex homometallic or heterometallic active sites of nitrogenases, hydrogenases, sulphite and nitrite reductases, CO dehydrogenases and acetyl-CoA synthases. In both eukaryotes and prokaryotes, metalloproteins are first synthesized in an apoform via the cellular translational machinery, and then the prosthetic group is inserted into the polypeptide. Mitochondria and chloroplasts have a high iron demand as many processes including photosynthesis, respiration, sulphur and nitrogen assimilation and chlorophyll catabolism require the presence of numerous Fe–S cluster-containing proteins. Similarly, other processes such as DNA repair, ribosome biogenesis or tRNA thiomodification also rely on the functionality of Fe–S cluster-containing proteins [1,2]. In general, Fe–S clusters perform a wide diversity of functions ranging from electron transfer, (de)hydration reactions, radical-generation, disulphide cleavage and small molecule redox reactions, regulation of biological processes in response to external stimuli; they can also play purely structural roles [3,4].
The components for Fe–S cluster assembly belong to several systems called NIF (nitrogen fixation), SUF (sulphur mobilization), ISC (iron–sulphur cluster) and CIA (cytosolic iron–sulphur cluster assembly) machineries [1,5]. Prokaryotes utilize three systems: the NIF system is specific for proteins associated with nitrogen fixation, the SUF system functions in archaea and under oxidative-stress and iron-limitation conditions in many bacteria, and the ISC system is the primary Fe–S cluster assembly machinery in many bacteria. In eukaryotes, the ISC assembly system is present in mitochondria whereas the SUF machinery functions in plastids of photosynthetic organisms [1,2,6]. The CIA machinery is devoted to the maturation of cytosolic and nuclear proteins [1]. Because the bacterial components of these Fe–S cluster assembly machineries are organized into operons, a minimal set of proteins required for this process has been initially identified. The primary function for these assembly machineries consist in the building of an Fe–S cluster on so-called scaffold proteins (in general classified as U or A-type scaffold proteins) from iron- and sulphur-delivery proteins for its subsequent transfer to recipient apoproteins (Figure 1). The nature of the iron donor remains a matter of debate, whereas the sulphur donor is a pyridoxal phosphate-dependent cysteine desulphurase (NifS, SufS, IscS) [1,7,8]. Additional proteins such as electron donors or chaperones are required for this reaction. As an example, recent work showed that cysteine desulphurases are associated with specific proteins: SufE for the bacterial and plant SUF systems and Isd11 for the yeast ISC system. SufE most likely serves to liberate sulphur atoms for a transfer to scaffold proteins [1,8,9]. Although generally not present in the bacterial nif, suf or isc operons, some glutaredoxins (Grxs), small proteins traditionally involved in redox reactions, are also implicated in Fe–S cluster biogenesis as accessory proteins. Here, we outline recent progress explaining how Grxs might be necessary for Fe–S cluster biogenesis and more generally for the regulation of iron homeostasis. We also discuss the use of genome analysis to identify new Grx-binding partners which participate in metal homeostasis.
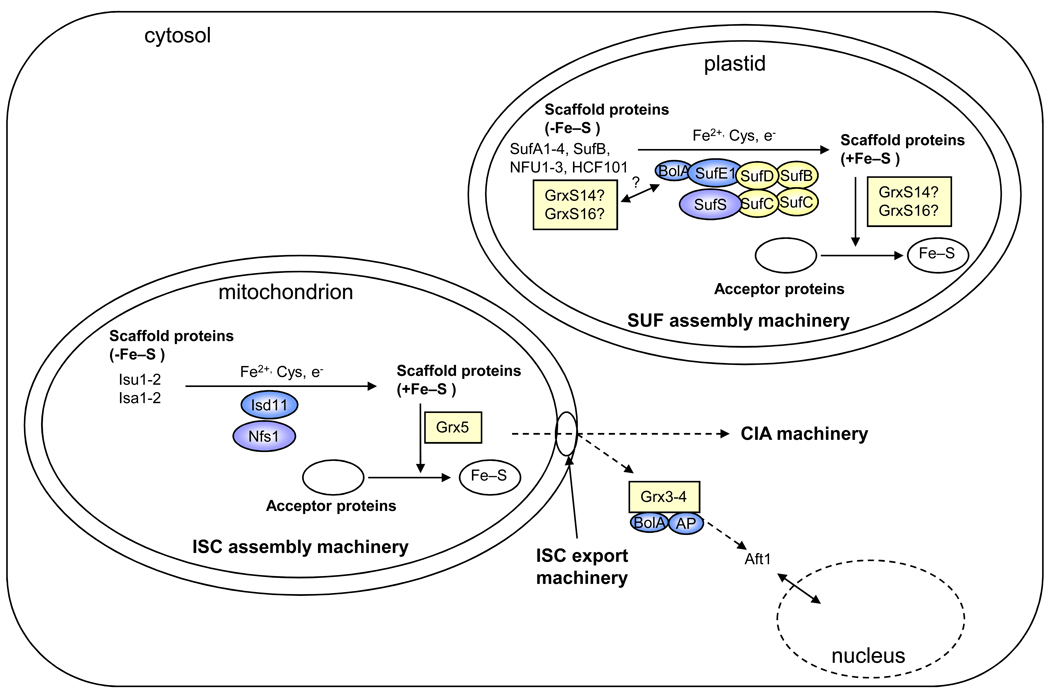
Figure 1. [sc1] Simplified view of the putative Grx roles in Fe–S cluster assembly machineries in eukaryote organelles.
This scheme has been drawn based on the current models of ISC assembly in yeast mitochondria and of the plant plastidial and bacterial SUF assembly machineries [1,6,8,64]. The SUF system is composed of a SufBCD (color) and SufSE (blue and purple) complexes. The nomenclature of Arabidopsis thaliana members has been used for the plastidial model. In mitochondria, Grx5p (color) is most likely involved in Fe–S cluster (color) transfer from Isu1p to acceptor proteins. As orthologs exist in most living organisms and as they generally complement the yeast mutant strain, it is tempting to speculate that this function is conserved among kingdoms. Isu and Isa correspond to U- and A-type scaffold proteins, respectively, Nfs1p (purple) to the cysteine desulphurase and Isd11p (blue) to a protein required for Nfs1p activity. In plastids, it is hypothesized that GrxS14 and GrxS16 (color) can perform scaffolding functions, participate in cluster trafficking between other identified scaffolds (SufA1-4, SufB, NFU1-3, HCF101) and recipient proteins in a manner analogous to the mitochondrial Grx5p, or regulate SUF function through an interaction with the BolA domain of SufE1. Notably, these roles are not mutually exclusive. To complete this view, the CIA (cytosolic iron–sulphur cluster assembly) and ISC (iron–sulphur cluster) export machineries are shown, and the current model for the role of cytosolic Grx3p and Grx4p (color) in iron sensing is depicted. AP: aminopeptidase P-like protein.
Many Grxs for distinct functions?
Traditionally, Grxs have been defined as thiol-disulphide oxidoreductases which exhibit a thioredoxin (Trx) fold architecture and a Cxx[C/S] active site. Grx was initially identified as a glutathione (GSH)-dependent electron donor for ribonucleotide reductase in Escherichia coli [10]. Since then, many additional functions have been proposed for Grxs both in prokaryotes and eukaryotes, in particular, in relation to glutathionylation, a protein regulatory or cysteine protective mechanism in which a glutathione is covalently attached as a mixed disulphide on a cysteine residue [11–13]. Grxs also appear to be efficient catalysts for the opposite reaction, i.e. deglutathionylation, and therefore the number of known Grx target proteins continues to increase.
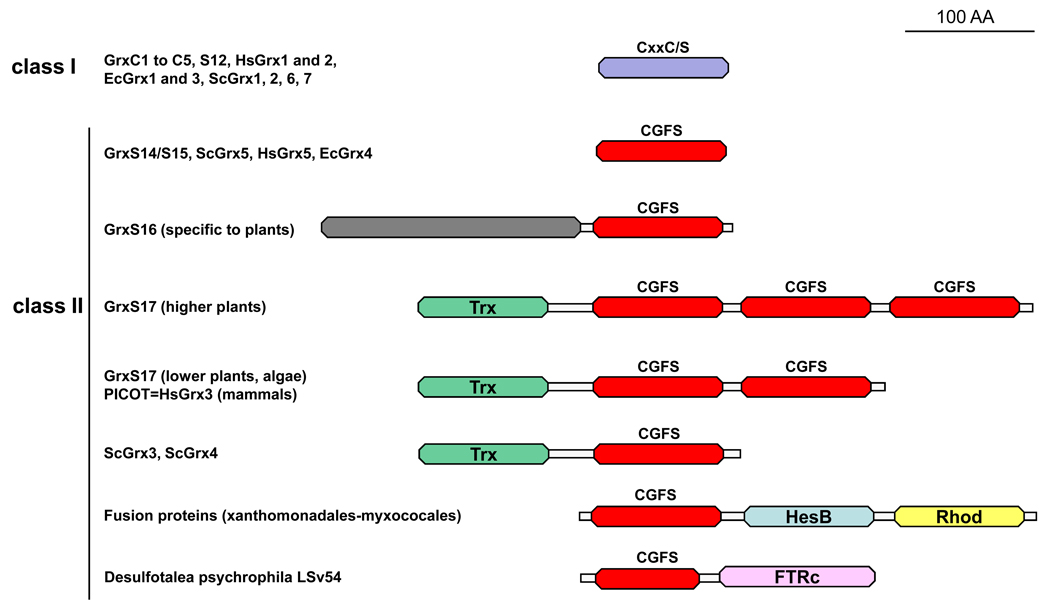
In most organisms possessing GSH and Grx proteins (some archaea and bacteria phyla lack GSH or Grxs or both), Grxs are encoded by multigenic families [14,15]. The largest number of grx genes is found in the sequenced model plant Populus trichocarpa (38 genes), thus pointing toward considerable functional diversity for Grxs in plants [14]. Grxs were initially classified as monothiol or dithiol proteins, based on the nature of the active sites (either CxxC or CGFS). However, comparative genomic analyses led to the identification of several Grx isoforms with diverse active site sequences as well as fusion proteins comprising a Grx domain. Hence, a refined phylogenetic classification in photosynthetic organisms led to organizing these sequences into six classes, based not only on the active site sequence, but also on conserved motifs involved in glutathione binding [14,16]. This classification can be extended to non-photosynthetic organisms. Classes I and II are the most widespread as, with a few exceptions, they are found in most prokaryotic and eukaryotic organisms (Figure 2). Class I generally contains Grxs comprising a single domain and exhibiting monothiol or dithiol active sites such as CPY[C/S], CGYC, CPFC or CSY[C/S]. It includes human Grx1 and 2, Saccharomyces cerevisiae Grx1 and 2, E. coli Grx1 to 3. In higher plants, these Grxs are named GrxC1 to C5 and GrxS12 (Figure 2). Class II includes all Grxs which contain a CGFS active site (hereafter referred to as CGFS Grxs). In general, this class contains two groups of proteins: those with a single Grx domain (ScGrx5, HsGrx5, EcGrx4 and plant GrxS14 and S15) and modular proteins with an N-terminal Trx-like module as well as one (ScGrx3 and ScGrx4), two (HsGrx3 (also called PICOT, for PKC (protein kinase C) Interacting Cousin Of Thioredoxin) or three (plant GrxS17) Grx domains (Figure 2) [12,14]. Some other fusion proteins with domains of known (e.g. HesB, rhodanese and ferredoxin-thioredoxin reductase) or unknown functions (e.g. the N-terminal domain of plant GrxS16 isoforms) are found in specific organisms. The distribution of other Grx classes is restricted to particular kingdoms. Class III, specific to higher plants, includes Grxs with CCxx active sites. Class IV contains hybrid proteins comprising a Grx domain (Cxx[C/S] active site) fused to two domains of unknown function. They are found in most eukaryotic photosynthetic organisms and some non-mammalian animals. Classes V and VI are specific to cyanobacteria and some other bacteria and include elongated proteins which contain a Grx domain (CPWG and CPW[C/S] active sites respectively) and a domain of unknown function.
Figure 2. Domain organisation of class I and II Grxs.
The classification of Grxs from photosynthetic organisms (terrestrial plants, algae, cyanobacteria) into 6 classes is likely to be the most complex classification as there are about 30 to 40 Grx genes in higher plants. This figure illustrates the correspondence for other model organisms (Escherichia coli (Ec), Homo sapiens (Hs) and Saccharomyces cerevisiae (Sc)) for classes I and II. These two classes contain Grxs that have been identified either as containing an Fe–S cluster, as participating in iron–sulphur cluster assembly or as participating in iron sensing. Grx domains for members of classes I and II are represented in violet and red, respectively. Additional domains include a Trx-like module (green), a domain of unknown function (grey), a HesB domain (blue), a rhodanese domain (yellow) and a ferredoxin-thioredoxin reductase domain corresponding to the catalytic subunit (pink). The full-length mature proteins, devoid of their targeting sequences, have been depicted here.
Involvement of CGFS Grxs in Fe–S cluster assembly
A role for the mitochondrial yeast Grx5p in Fe–S cluster biogenesis was initially suggested from studies of a grx5 deletion mutant. These yeast cells displayed deficient cluster assembly for at least two Fe–S proteins (aconitase and succinate dehydrogenase) leading to impaired respiratory growth and increased sensitivity to oxidative stress as a result of the accumulation of free iron in the cell [17,18]. Later, it was proposed, based on radiolabeled 55Fe immunoprecipitation experiments, that Grx5p most likely facilitates the transfer of preassembled clusters from a U-type ISC scaffold proteins (Isu1p) to acceptor proteins or, through bioinformatic predictions, that it is involved either in regulating the Nfs1 cysteine desulphurase or in the assembly of Fe–S clusters on scaffold proteins [19,20]. The specific interaction of Grx5p with the A-type ISC scaffold protein Isa1p, but not Isa2p, demonstrated by yeast two-hybrid experiments, further supports the view that Grxs participate in the initial steps of cluster assembly or in the transfer of preassembled clusters [21]. Furthermore, complementation experiments of the yeast grx5 deletion strain demonstrated that most CGFS Grxs from prokaryotic or eukaryotic sources other than yeast, when targeted to the yeast mitochondrial matrix, can functionally substitute for Grx5p, suggesting that this role might be conserved throughout evolution [22,23].
CGFS Grxs contain a labile Fe–S cluster
A series of recent papers indicate that the recombinant versions of bacterial, human, yeast and plant Grx5 orthologs produced in E. coli are able to incorporate labile Fe–S clusters [22,24]. It is possible that, owing to their labile nature as well as technical limitations, the presence of Fe–S clusters complexed to these CGFS Grxs has yet to be confirmed in vivo. Analytical and spectroscopic (UV-visible absorption/circular dichroism (CD), resonance Raman and Mössbauer) analyses of anaerobically purified proteins or proteins re-purified after in vitro cysteine desulphurase-mediated cluster reconstitution of apoproteins indicated that two plant CGFS Grxs (the plastidial GrxS14 and GrxS16) could incorporate one [2Fe–2S]2+ cluster per homodimer with complete cysteinyl ligation [22]. Cysteine mutagenesis studies on these Grxs, the requirement for glutathione in GrxS14 reconstitution experiments, as well as the recent determination of the 3D structure of the E. coli Grx4 holodimer, indicates that the [2Fe–2S] cluster is ligated by the active site cysteines of two Grx monomers and two glutathiones (Figure 3) [22,25,26]. The [2Fe–2S]2+ clusters of these CGFS Grxs are oxidatively and reductively labile as evidenced by cluster degradation upon exposure to air and following anaerobic reduction with dithionite [22].
Figure 3. Comparison of the 3D structures of poplar GrxC1, human Grx2 and E. coli Grx4.
The dimeric structures of poplar GrxC1 (PDB: 2E7P) (a) human Grx2 (PDB: 2HT9) (b) and E. coli Grx4 (PDB: 2E7P) (c) are presented with the α1 and α2 helices of protomer A in the same orientation. Except for a few differences concerning the length of α-helices and their orientation, the structures of poplar GrxC1 and human Grx2 are nearly superimposable. By contrast, although each protein contains a subunit-bridging [2Fe–2S] cluster which is ligated by the N-terminal active site cysteine of the two Grx monomers and two glutathione molecules, the orientation of protomer B in E. coli Grx4 is rotated by 90° compared to the situation in poplar GrxC1 and human Grx2. This different orientation, which leads to new interactions between protomers, has been proposed to contribute to the cluster lability in EcGrx4 [25]. The Fe–S cluster, the ligating cysteines and glutathione molecules are shown in stick representation. Sulfur atoms are colored in yellow, iron atoms in brown. Figures were made using PyMol (http://www.pymol.org).
Possible physiological roles for chloroplastic CGFS Grxs
The ability of CGFS Grxs to assemble [2Fe–2S] clusters points to a potential role as a scaffold protein for de novo Fe–S cluster biosynthesis. However, a scaffold protein must also be competent for intact cluster transfer to a physiologically relevant acceptor protein. Hence, the recent demonstration using CD spectroscopy that chloroplastic GrxS14 can rapidly and stoichiometrically transfer its [2Fe–2S] cluster intact to the apoform of a plant-type [2Fe–2S] ferredoxin (Fdx) provides additional support for this proposal [22]. Alternatively, the ability to efficiently transfer [2Fe–2S] clusters to acceptor proteins is also consistent with a role for CGFS Grxs in cluster trafficking, i.e. mediating the transfer of preformed Fe–S clusters to scaffold proteins or from scaffold to acceptor proteins. Such a role is supported by yeast genetics studies which indicated a role for Grx5p in facilitating cluster transfer from Isu1p to mitochondrial acceptor proteins [19]. Hence, to address these questions, there is a pressing need for in vivo experimentation, especially for the characterization of plant knock-out mutants for grxS14 and grxS16. Alternatively, the use of different protein combinations could allow in vitro Fe–S transfer studies to provide information about potential roles for direct Fe–S transfer or Fe–S trafficking. Such combinations include (i) cluster-loaded forms of chloroplastic scaffold proteins (e.g. SufA, NifU-like proteins (NFU1-3), and the ABC transporter SufB) together with apoforms of CGFS Grxs to address the possibility that CGFS Grxs receive Fe–S clusters from various types of scaffold proteins; (ii) holoforms of CGFS Grxs and apoforms of a wide range of physiologically relevant acceptor Fe–S proteins to demonstrate the scaffold function of CGFS Grxs; and (iii) apoforms of CGFS Grxs with cluster-loaded scaffold proteins together with acceptor apoproteins to address the effect of CGFS Grxs on the rates of cluster transfer (Figure 1) [6]. Spectroscopic methods that are sensitive to the nature, properties and ligation of biological Fe–S centres (UV-visible absorption/CD, resonance Raman, Mössbauer and electron paramagnetic resonance (EPR)) can be used to monitor the kinetics and mechanism of interprotein Fe–S cluster transfer (Box 1).
Box 1. Assessing the kinetics and mechanism of interprotein Fe–S cluster transfer
In addition to the characterization of the nature and properties of Fe–S clusters assembled in Grxs, a major experimental challenge is to measure the specificity and kinetics of cluster transfer from donor proteins to acceptor proteins. If the acceptor protein has an Fe–S cluster-dependent activity, the time course of cluster incorporation can be conveniently monitored by activity assays of the reaction mixture as a function of time. However, controls involving the addition of iron chelators and/or parallel Mössbauer studies to monitor the fate of the Fe–S cluster on the donor protein in the presence or absence of the acceptor protein are required to demonstrate intact cluster transfer as opposed to degradation of the cluster on the donor protein and reassembly on the acceptor protein. UV-visible absorption is generally not effective for monitoring cluster transfer due to the similarity of the spectroscopic signatures of [4Fe–4S] and [2Fe–2S] clusters as a function of protein environment. By contrast, UV-visible CD is much more sensitive to the protein environment of both [4Fe–4S] and [2Fe–2S] clusters; indeed it has already proven to be a useful method for monitoring the kinetics of cluster transfer experiments involving CGFS Grxs [22]. For donor and acceptor proteins that have distinctive resonance Raman or Mössbauer properties for their [4Fe–4S] or [2Fe–2S] centres, freeze-quench kinetic studies can be effective for monitoring cluster transfer. This approach involves rapid mixing of the cluster transfer reactants, flash freezing after specific reaction times in an isopentane-dry ice bath, and assessing the cluster composition of the donor and acceptor proteins using Mössbauer or resonance Raman spectroscopy. The extent of cluster transfer can also be monitored by analytical or UV-visible absorption after protein separation using affinity or gel filtration chromatography, but this method is time-consuming and subject to increased error owing to the protein separation steps.
Regulation of the SUF machinery
The interaction of the yeast Trx-Grx hybrid proteins, Grx3p and Grx4p, with BolA, a member of a protein family of unknown function, suggests that a similar interaction between chloroplastic CGFS Grxs and SufE1 might exist [27]. SufE1, an essential component of the SUF machinery, contains an N-terminal traditional SufE domain fused to a BolA domain [28]. In E. coli, SufE is a sulphurtransferase which mediates sulphur transfer from the SufS cysteine desulphurase to SufB, a potential Fe–S cluster scaffold protein [9]. Hence, by interacting with SufE1, CGFS Grxs could also either receive the sulphur required for the assembly of their own Fe–S clusters or participate in Fe–S cluster transfer from SufE1 to SufB. Alternatively, the interaction of CGFS Grxs with SufE1 might be a general way to regulate the chloroplastic machinery depending on the Fe or Fe–S cluster status of this compartment. For example, under replete iron conditions, the presence of an Fe–S cluster on GrxS14 or GrxS16 might promote an interaction with SufE1 in order to inhibit the SUF machinery. As some other plastidial SufE proteins lack a BolA domain, this mode of regulation would only be operative for a specific part of the SUF system. Hence, in addition to a potential role in Fe–S cluster biosynthesis or trafficking, CGFS Grxs might also serve as sensors of the chloroplastic Fe or Fe–S cluster status.
Additional regulatory roles for Fe–S cluster-bound glutaredoxins
Independent of their roles in Fe–S cluster biogenesis, Grxs contribute to the regulation of iron-responsive genes in yeast, termed the iron regulon. Those containing stable Fe–S clusters could serve as a redox or iron sensor. Finally, through their involvement in the mitochondrial Fe–S cluster machinery, Grxs also influence another iron-dependent process, haem synthesis.
Iron sensing and regulation of iron homeostasis
In S. cerevisiae, two CGFS Grxs, Grx3p and Grx4p, form a complex with the iron-responsive transcription factor Aft1p (activator of ferrous transport 1) either in the cytosol or in the nucleus, thereby inhibiting the expression of iron-responsive genes [29,30]. The role of these Grxs was later refined following their identification in a cytosolic complex comprising two other proteins called Fra1p and Fra2p (Fe repressor of activation-1 and 2), which correspond to an aminopeptidase P-like protein and a BolA-like protein, respectively [27]. Mutation of any one of these four genes or a defect in the mitochondrial Fe–S cluster biogenesis machinery has a similar phenotype involving constitutive expression of iron regulon genes. It will be important to determine the roles of Fra1p and Fra2p in this complex and whether the incorporation of an Fe–S cluster in these Grxs is required for their function. From these biochemical and genetic experiments, the present view concerning iron regulation in yeast is that the Grx3–Grx4–BolA–aminopeptidase complex acts as a sensor of the status of mitochondrial Fe–S cluster biogenesis by forming a cytosolic Fe- or Fe–S cluster-dependent complex with Aft1p, thereby inhibiting Atf1p nuclear translocation and subsequent transcriptional activation of the iron regulon [27].
The absence of Grx3/4p orthologs in bacteria coupled with the absence of Aft1p orthologs in non-yeast organisms, suggests that the Aft1p-dependent iron sensing pathway is likely restricted to a few yeast species [14,31,32]. For example, in bacteria and in animals, the iron sensors which regulate genes involved in iron metabolism are the ferric uptake regulator (Fur) and iron regulatory proteins (IRP1 or IRP2), respectively, and there is no evidence that either of these systems are regulated through an interaction with Grx3/4 orthologs [2]. In plants, the cellular iron-responsive factor has yet to be identified, although key transcription factors that regulate the expression of genes involved in iron uptake, reduction and transport have been described [33,34]. Further work is needed, both in prokaryotes and eukaryotes, to determine whether the CGFS Grxs–BolA interaction is widespread and whether it is linked to the regulation of iron-related mechanisms.
Redox sensing under oxidative stress conditions
Some class I Grxs contain stable, albeit reductively-labile [2Fe–2S] clusters. They are located in various sub-cellular compartments: cytosol (poplar GrxC1, CGYC active site), mitochondria (human Grx2, CSYC active site; and Trypanosoma brucei Grx1, CAYS active site) or endoplasmic reticulum (S. cerevisiae Grx6p, CSYS active site) [26,35–37]. In addition, alternative splicing leads to the expression of human Grx2 in several compartments (Grx2a in mitochondria, Grx2b and Grx2c in both the nucleus and cytosol) [38]. The [2Fe–2S] cluster in poplar GrxC1 and human Grx2 bridges the homodimeric Grx and is coordinated by the N-terminal active site cysteine of two monomers and two external glutathione molecules (Figure 3) [26,39,40]. Poplar GrxC1 is neither able to complement the yeast grx5 deletion strain nor to transfer its [2Fe–2S] cluster to acceptor proteins [22,26]. Moreover, the [2Fe–2S] clusters on both poplar GrxC1 and human Grx2 are stabilized in vitro by GSH and destabilized by glutathione disulphide (GSSG). Therefore, the [2Fe–2S] clusters on these proteins could serve as sensors of the cellular redox status, especially under oxidative stress conditions where the GSH:GSSG ratio is reduced [36].
Regulation of haem biosynthesis
A grx5 zebrafish mutant and a human patient presenting a mutation in an intronic region of the GRX5 gene resulting in a decrease, by a factor 12, in Grx5 expression, both exhibit hypochromic anemia, a defect linked to a deficiency in haem or globin synthesis in erythrocytes [41,42]. In these organisms, it appears that the defects in Fe–S cluster assembly resulting from the absence of this mitochondrial Grx trigger the activation of cytosolic IRP1 which in turn blocks the synthesis of aminolaevulinate synthase 2, the first enzyme of haem biosynthesis. Alternative explanations could be that the deficiency in mitochondrial Fe–S cluster biogenesis completely disturbs cellular iron distribution (which is frequently observed when a gene from this pathway is disrupted) or prevents the functioning of ferrochelatase, a mitochondrial [2Fe–2S] cluster-containing enzyme which catalyzes the last step of haem biosynthesis in vertebrates [43]. By contrast, in plants, the entire sirohaem biosynthetic pathway is localized in chloroplasts; plants also express a [2Fe–2S] cluster-containing sirohydrochlorin ferrochelatase which is susceptible to regulation by Fe–S cluster-bound chloroplastic Grxs [44].
New CGFS Grx partners and functions related to metal homeostasis
The identification of the physiological partners of Fe–S cluster-containing Grxs would provide greater insight into understanding the metabolic pathways and processes in which they participate. In silico genomic analyses can help identify such putative functional or physical interacting proteins. STRING 8 (search tool for the retrieval of interacting genes/proteins) is a useful tool for analyzing the presence of fusion proteins, gene clustering, i.e. the conservation of gene order in presumptive bacterial operons (the genes can be either co-directional or in a counter orientation), the co-occurrence of genes in a large panel of organisms (currently 630 can be analyzed) across all kingdoms and the co-expression of genes, i.e. variation of the transcript levels under the same conditions [45]. Based on the above-mentioned criteria, STRING attributes a score between 0 and 1 to indicate the strength of the prediction. These genomic interaction-detection methods have proven to be complementary to high-throughput experimental approaches such as yeast two-hybrid experiments, whose results are also compiled in the STRING database [46–48].
This type of analysis has been helpful in predicting Grx partners. As an example, it was unexpectedly reported that ferredoxin-thioredoxin reductase, a chloroplastic [4Fe-4S]-containing enzyme devoted to the reduction of Trxs, can also reduce the apoform of a Chlamydomonas reinhardtii CGFS Grx [49]. Indeed, in various proteobacteria and archaea, CGFS Grx-encoding genes are adjacent to ferredoxin-thioredoxin reductase-encoding genes and they undergo a gene fusion in Desulfotalea psychrophila LSv54 (Figure 2) [50]. As the apoforms of CGFS Grxs display deglutathionylation activity, it is noteworthy that not all of the predicted protein partners are Fe–S containing proteins; indeed, some might simply be regulated via glutathionylation [49].
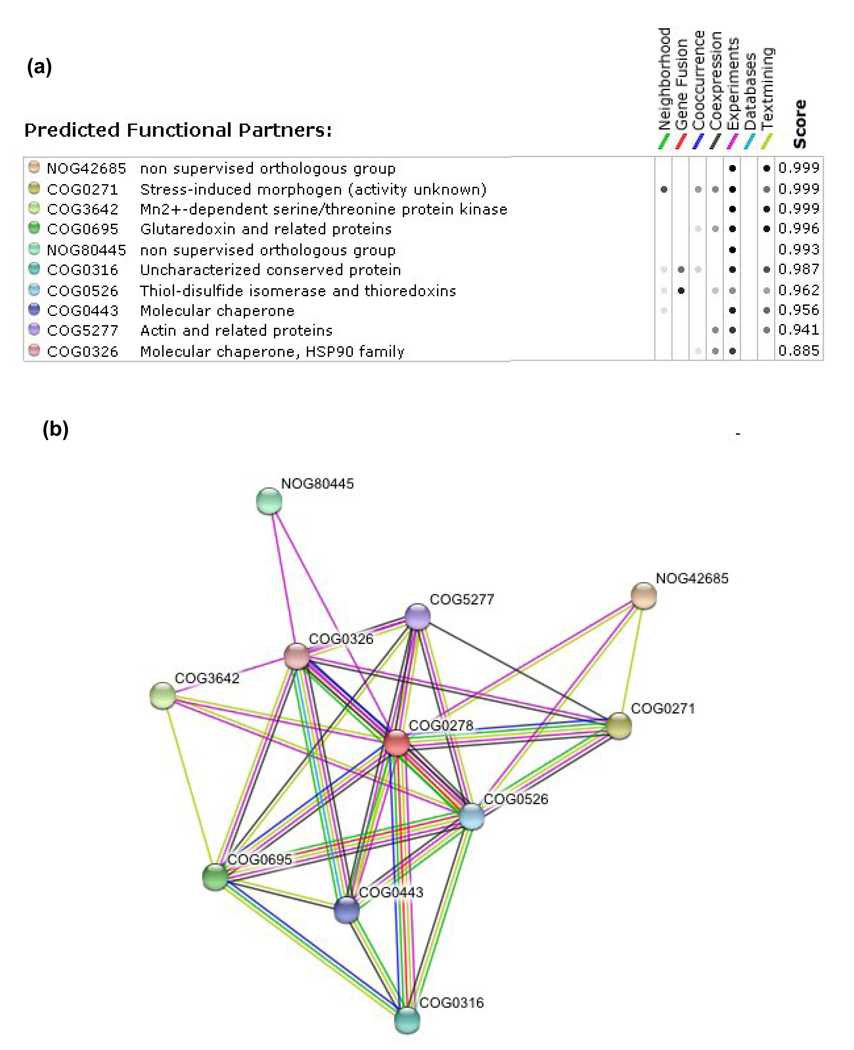
Out of the ten best hits obtained via the STRING analysis, five confirm that CGFS Grxs are linked to Fe–S cluster biogenesis or iron sensing (Figure 4). The two strongest scores correspond to proteins (Aft1p and BolA) experimentally recognized as CGFS Grx interactors [27,29,30,51–53]. In addition, the genes encoding CGFS Grxs and BolA-type proteins are frequently found in adjacent positions in many prokaryote organisms; moreover, the gene co-occurrence is strong [14,54]. Indeed, with very few exceptions, the two genes are present together in the same subset of microorganisms and are both absent elsewhere [14]. These observations suggest that the Grx–BolA interaction might constitute a general requirement for iron status control in organisms in which the two genes are present. However, as CGFS Grxs and BolA are encoded by multigenic families (there are three or more bolA genes and two to five CGFS Grx genes in most eukaryotes) and targeted to various sub-cellular compartments, the molecular determinants of these interactions require further exploration [12,14,22,32,55]. For example, it is possible that the N-terminal Trx-like extension present in yeast Grx3p and Grx4p is required for their interaction with BolA as, in bacteria, the bolA gene is adjacent to genes encoding CGFS Grxs which lack this extension.
Figure 4. Putative CGFS Grx target proteins retrieved using the STRING webtool.
An in silico genomic analysis based notably on gene fusion, gene clustering, gene co-occurrence and gene co-expression, and combined to experimental data, was used to predict putative functional or physical interacting proteins. The 10 most confident putative CGFS Grx-interacting proteins (COG0278) (a) and their representation as a Grx network (b) are listed along with the 7 criteria which were used in the analysis. In this network, the number of connections, which represents the different criteria used, is used as a correlation for the strength of the interaction, unless experimental data exist for such interaction. NOG42685 corresponds to Aft1p transcription factor, COG0271 to BolA proteins, COG0316 to IscA/HesB/YfhF scaffold proteins, COG0443 to HscA/Hsp70/Ssq1p chaperones, COG0326 to HscB/Hsp90/Jac1p chaperones, COG3642 to a family of serine/threonine protein kinases with Bud32p as representative, COG0695 to class I Grxs, NOG80445 to phosphatidylinositol 4,5-bisphosphate protein, COG0526 to Trxs with Grx3/4p and plant GrxS17 as representatives and COG5277 to actin.
Several other hits confirm the involvement of CGFS Grxs in Fe–S cluster biogenesis. First, in several proteobacteria of the Xanthomonadale and Myxococcale order, a Grx domain is included in a tripartite fusion protein (COG0316) of unknown function (Figure 2, Figure 4) [31]. An N-terminal Grx module is fused to a module with homology to A-type scaffold proteins, but lacking the conserved cysteines normally required for the binding of an Fe–S centre, and a sulphurtransferase of the rhodanese superfamily. Second, two molecular chaperone families, Hsp70 (also referred to as HscA or Ssq1, COG0443) and Hsp90 (HscB or Jac1, COG0326), which facilitate the transfer of Fe–S clusters preassembled on Isu/IscU proteins to acceptor proteins, are predicted as potential Grx partners [7,19,56]. This prediction essentially comes from large scale yeast two-hybrid experiments and from the observation that Ssq1p overexpression in a yeast grx5 deletion strain suppressed its phenotypic defects [18,57]. Co-occurrence has also been identified between genes encoding CGFS Grxs and major proteins involved in Fe–S cluster biogenesis, i.e. the Yah1p ferredoxin, the Isa1p and Isa2p scaffold proteins, the Yfh1p frataxin and Nfu1p [21]. In addition, in some microbial genomes, CGFS Grx-encoding genes are also clustered with ferredoxin-encoding genes (Figure 5).
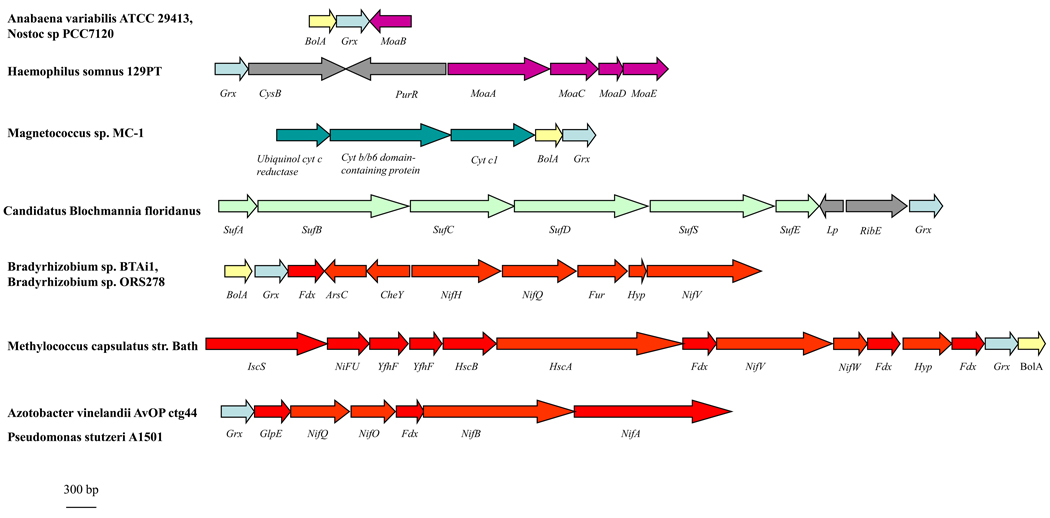
Figure 5. Gene clustering between CGFS Grx-encoding genes and iron- or molybdenum-related genes.
Based on the genome analysis of approximately 2500 sequenced microorganisms available using the “protein clusters” tool available at the NCBI webpage and the Microbial Genome Database (MGDB, http://mbgd.genome.ad.jp/), we list genes involved in metal homeostasis which are either directly adjacent to CGFS Grx-encoding genes or belong to a putative operon containing a CGFS Grx gene. The size of the genes is drawn to scale. Hyp stands for genes which encode hypothetical proteins, CysB codes for a LysR-type transcriptional regulator, PurR for the master regulatory protein of purine nucleotide biosynthesis, Lp for lipoprotein, RibE for the subunit alpha of a flavin synthase, ArsC for an arsenate reductase of the C-type, CheY for a transcriptional regulatory protein, Fdx for a ferredoxin, Fur for the ferric uptake regulator and GlpE for a protein of the rhodanese/sulphurtransferase family. MoaA, B, C, D, E encode proteins belonging to the molybdenum biosynthetic pathway, SufA, B, C, D, E, S and NifA, B, H, Q, O, U, V, W encode proteins belonging to the SUF and NIF assembly machinery, respectively. IscS encode a cysteine desulphurase, Yfhf a scaffold proteins and HscA and B encode two chaperone proteins.
The analysis of gene clustering in a larger set of microbial genomes enabled the identification of other new putative Grx partners or functions related to iron or molybdenum, which were not retrieved from the STRING analysis. For example, the CGFS grx gene, in the presence or absence of bolA gene, is present in putative suf and nif operons (Figure 5). Only one example, found in the endosymbiont Candidatus Blochmania floridanus, supports a possible relation between Grx and the SUF system and the grx gene is separated from the sufABCDSE genes by two genes with no clear relationship with Fe–S cluster assembly. Therefore, the putative interaction of Grxs with some components of the SUF machinery, as suggested in plants, should be viewed with caution. By contrast, there is stronger support for an association between CGFS grx genes and genes involved in the NIF Fe–S cluster assembly system in several nitrogen-fixing bacteria, e.g. Bradyrhizobium sp BTAi1, Methylococcus capsulatus, Azotobacter vinelandii AvOPctg44 and Pseudomonas stutzeri A1501 (Figure 5). The nif gene cluster usually comprises somewhere between 20 to 30 genes which can be distributed among several chromosomal positions and in different transcriptional directions [58]. These NIF proteins (many of which contain Fe–S clusters) are essential for the maturation of the complex heterometallic nitrogenase enzyme, which comprises several Fe–S cofactors, a [4Fe–4S] cluster in the Fe protein and a double cubane P-cluster along with the FeMoco cluster in the FeMo protein [58]. The presence of CGFS grx genes in the nif operon suggests that CGFS Grxs might be required for the correct assembly of nitrogenase cofactors, either directly via assembly of the [4Fe–4S] cluster, the P-cluster or the FeMoco cluster, or indirectly through the assembly of Fe–S clusters in some NIF Fe–S assembly proteins.
The presence of CGFS grx genes in close proximity to some genes encoding proteins involved in molybdenum cofactor (Moco) synthesis suggests that Grxs might be directly or indirectly involved in Moco biosynthesis (Figure 5) [59]. For instance, the gene encoding the radical S-adenosylmethionine enzyme MoaA, which catalyzes the first step of bacterial Moco biosynthesis and contains two [4Fe-4S] clusters, is found in close proximity to a grx gene [60].
A final example is the clustering of grx and bolA genes with three genes encoding subunits of the ubiquinol cytochrome c reductase (bc1) complex in Magnetococcus sp. In this case, the Grx might be important for maturation of the Rieske-type [2Fe-2S] centre (one iron ligated by two cysteines and one ligated by two histidine residues) or for glutathionylation of critical cysteine residues, as observed for mitochondrial complex II [61].
Concluding remarks and future perspectives
The involvement of CGFS Grxs in iron homeostasis has only recently emerged from studies showing that they are involved in mitochondrial, and most likely also chloroplastic, Fe–S cluster biogenesis as well as overall iron regulation in yeast. They also appear to be involved in channelling iron for haem or Fe–S cluster biosynthesis in vertebrate animals. The more stable [2Fe–2S] clusters present in class I Grxs appear to play a role in the oxidative stress response. However, such a role might be counterproductive if the sensing mechanism involves cluster degradation and liberation of iron susceptible to producing hydroxyl radicals through a Fenton reaction. Clearly there is pressing need for further characterizing the ligation, stability and type of Fe–S clusters that can be assembled on Grxs, identifying the physiological partners of Grxs which contain Fe–S clusters, and investigating the functions of cluster-bound Grxs in specific organisms and organelles using both in vivo and in vitro approaches (Box 2).
Box 2. Remaining questions
Can CGFS Grxs incorporate Fe–S centres other than [2Fe–2S] clusters, and can these clusters be transferred to acceptor proteins? The restoration of aconitase functionality, which requires the presence of a [4Fe–4S], by expressing CGFS Grxs into the yeast grx5 deletion strain, suggests that these Grxs might also incorporate [4Fe–4S] clusters in a manner similar to many other scaffold proteins [7].
What is the role of single domain yeast Grx5p orthologs in mitochondrial Fe–S cluster biogenesis? Do they function as general or specific scaffold proteins for the assembly and delivery of Fe–S clusters, or do they facilitate the transfer of preassembled clusters from scaffold proteins to acceptor proteins?
What are the roles of Grxs with stable Fe–S clusters? Plant GrxC1 orthologs are cytosolic proteins that apparently cannot transfer their [2Fe–2S] clusters to apo Fe–S proteins. Mammalian mitochondria contain two Fe–S cluster-containing Grxs, Grx2 and the yeast Grx3/4p ortholog, PICOT. It was recently proposed that glutathione depletion could influence important pathological events associated with Parkinson’s disease via its effects on Grx2 activity and mitochondrial Fe–S biogenesis [62].
The presence of an N-terminal extension conserved in plant GrxS16 orthologs, but not in GrxS14 orthologs, raises the possibility that these two plant chloroplastic Fe–S cluster-containing Grxs perform different functions.
There is a critical need for direct in vivo evidence for the presence of Fe–S clusters in both class I and II Grxs. As the apoproteins exhibit thiol-disulphide reductase and/or deglutathionylation activities, it will be important to separate both functions [49]. Such analyses are complicated by the fact that oxidative stress response (a known Grx function) and iron metabolism are intimately linked [13,63].
Acknowledgements
Our work is supported by grants from the National Institutes of Health (GM62524 to M.K.J) and from the ANR programs (GNP05010G and JC07_204825) to N.R and J.C. We also acknowledge all colleagues who have contributed to the work described here.
Glossary
- Electron paramagnetic resonance (EPR)
Spectroscopic technique for identifying, characterizing and quantifying chemical species that have one or more unpaired electrons, such as organic free radicals and transition metals centres.
- Glutaredoxins
Proteins which catalyze GSH-disulphide oxidoreductions. However, some specific isoforms can ligate Fe–S clusters and are involved in Fe–S cluster biogenesis, most likely independently of their thiol-disulphide reductase activity.
- Glutathionylation
The covalent attachment of glutathione on a cysteine residue via a disulphide bridge. It is considered as a post-translational regulatory modification and/or as a transient protective mechanism for critical cysteine residues.
- Iron–sulphur cluster
Cluster constituted by iron and inorganic sulphide. The most common types of Fe–S clusters, attached via cysteinate ligation into the polypeptide, are the [2Fe–2S] and the cubane [4Fe–4S] forms. Alternatively, some enzymes contain cubane-type [3Fe–4S] clusters and Rieske type [2Fe–2S] clusters which have two histidine ligands at one Fe site.
- ISC
The primary system for general Fe–S cluster biosynthesis in bacteria. It generally comprises a transcriptional regulator IscR, a cysteine desulphurase IscS, scaffold proteins IscU and IscA, chaperone proteins hscA and hscB and an electron donor Fdx. This system also constitutes the eukaryotic mitochondrial Fe–S cluster biosynthesis pathway.
- Mössbauer spectroscopy
Spectroscopic technique based on the resonant emission and absorption of gamma rays, which can detect subtle changes in the electronic environment of 57Fe nuclei. The ability of Mössbauer to quantify and identify all types of Fe in a sample makes this technique essential for meaningful quantitative investigations of Fe–S proteins.
- NIF
This system is specifically required for the maturation of Fe–S-containing proteins, especially nitrogenase, in nitrogen-fixing organisms. The operon generally contains the cysteine desulphurase NifS and two potential scaffold proteins, nifIscA and NifU.
- Resonance Raman spectroscopy
Spectroscopic technique for selectively investigating the vibrational properties of chromophoric metal centres in biological samples.
- SUF
This Fe–S cluster biosynthesis system is found in many bacteria which possess the ISC system, and it operates under iron limitation or oxidative stress conditions. It has also been conserved in plant chloroplasts. For many archaea and bacteria, including cyanobacteria, it represents the sole Fe–S cluster assembly system. In most organisms, the proteins involved are a cysteine desulphurase SUFS, a scaffold protein SUFA and a few other components: SUFB, SUFC, SUFD form a complex which most likely provides energy for this process, and SUFE is a sulphurtransferase associated with SUFS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lill R, Muhlenhoff U. Maturation of iron-sulfur proteins in eukaryotes: mechanisms, connected processes, and diseases. Annu Rev Biochem. 2008;77:669–700. doi: 10.1146/annurev.biochem.76.052705.162653. [DOI] [PubMed] [Google Scholar]
- 2.Rouault TA, Tong WH. Iron-sulfur cluster biogenesis and human disease. Trends Genet. 2008;24:398–407. doi: 10.1016/j.tig.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beinert H, et al. Iron-sulfur clusters: nature's modular, multipurpose structures. Science. 1997;277:653–659. doi: 10.1126/science.277.5326.653. [DOI] [PubMed] [Google Scholar]
- 4.Johnson MK, Smith AD. Iron-sulfur proteins. Encyclopedia of Inorganic chemistry. 2005:2589–2619. [Google Scholar]
- 5.Johnson DC, et al. Structure, function, and formation of biological iron-sulfur clusters. Annu Rev Biochem. 2005;74:247–281. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- 6.Balk J, Lobreaux S. Biogenesis of iron-sulfur proteins in plants. Trends Plant Sci. 2005;10:324–331. doi: 10.1016/j.tplants.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 7.Bandyopadhyay S, et al. Iron-sulfur cluster biosynthesis. Biochem Soc Trans. 2008;36:1112–1119. doi: 10.1042/BST0361112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fontecave M, et al. Mechanisms of iron-sulfur cluster assembly: the SUF machinery. J Biol Inorg Chem. 2005;10:713–721. doi: 10.1007/s00775-005-0025-1. [DOI] [PubMed] [Google Scholar]
- 9.Layer G, et al. SufE transfers sulfur from SufS to SufB for iron-sulfur cluster assembly. J Biol Chem. 2007;282:13342–13350. doi: 10.1074/jbc.M608555200. [DOI] [PubMed] [Google Scholar]
- 10.Holmgren A. Hydrogen donor system for Escherichia coli ribonucleoside-diphosphate reductase dependent upon glutathione. Proc Natl Acad Sci U S A. 1976;73:2275–2279. doi: 10.1073/pnas.73.7.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fernandes AP, Holmgren A. Glutaredoxins: glutathione-dependent redox enzymes with functions far beyond a simple thioredoxin backup system. Antioxid Redox Signal. 2004;6:63–74. doi: 10.1089/152308604771978354. [DOI] [PubMed] [Google Scholar]
- 12.Herrero E, de la Torre-Ruiz MA. Monothiol glutaredoxins: a common domain for multiple functions. Cell Mol Life Sci. 2007;64:1518–1530. doi: 10.1007/s00018-007-6554-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rouhier N, et al. The role of glutathione in photosynthetic organisms: emerging functions for glutaredoxins and glutathionylation. Annu Rev Plant Biol. 2008;59:143–166. doi: 10.1146/annurev.arplant.59.032607.092811. [DOI] [PubMed] [Google Scholar]
- 14.Couturier J, et al. Evolution and diversity of glutaredoxins in photosynthetic organisms. Cell Mol Life Sci. 2009;66:2539–2557. doi: 10.1007/s00018-009-0054-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fahey RC. Novel thiols of prokaryotes. Annu Rev Microbiol. 2001;55:333–356. doi: 10.1146/annurev.micro.55.1.333. [DOI] [PubMed] [Google Scholar]
- 16.Rouhier N, et al. Genome-wide analysis of plant glutaredoxin systems. J Exp Bot. 2006;57:1685–1696. doi: 10.1093/jxb/erl001. [DOI] [PubMed] [Google Scholar]
- 17.Rodriguez-Manzaneque MT, et al. Grx5 glutaredoxin plays a central role in protection against protein oxidative damage in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:8180–8190. doi: 10.1128/mcb.19.12.8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodriguez-Manzaneque MT, et al. Grx5 is a mitochondrial glutaredoxin required for the activity of iron/sulfur enzymes. Mol Biol Cell. 2002;13:1109–1121. doi: 10.1091/mbc.01-10-0517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muhlenhoff U, et al. Components involved in assembly and dislocation of iron-sulfur clusters on the scaffold protein Isu1p. EMBO J. 2003;22:4815–4825. doi: 10.1093/emboj/cdg446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alves R, et al. Predictive reconstruction of the mitochondrial iron-sulfur cluster assembly metabolism. II. Role of glutaredoxin Grx5. Proteins. 2004;57:481–492. doi: 10.1002/prot.20228. [DOI] [PubMed] [Google Scholar]
- 21.Vilella F, et al. Evolution and cellular function of monothiol glutaredoxins: involvement in iron-sulphur cluster assembly. Comp Funct Genomics. 2004;5:328–341. doi: 10.1002/cfg.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bandyopadhyay S, et al. Chloroplast monothiol glutaredoxins as scaffold proteins for the assembly and delivery of [2Fe-2S] clusters. EMBO J. 2008;27:1122–1133. doi: 10.1038/emboj.2008.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Molina-Navarro MM, et al. Prokaryotic and eukaryotic monothiol glutaredoxins are able to perform the functions of Grx5 in the biogenesis of Fe/S clusters in yeast mitochondria. FEBS Lett. 2006;580:2273–2280. doi: 10.1016/j.febslet.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 24.Picciocchi A, et al. CGFS-type monothiol glutaredoxins from the cyanobacterium Synechocystis PCC6803 and other evolutionary distant model organisms possess a glutathione-ligated [2Fe-2S] cluster. Biochemistry. 2007;46:15018–15026. doi: 10.1021/bi7013272. [DOI] [PubMed] [Google Scholar]
- 25.Iwema T, et al. Structural basis for delivery of the intact [Fe2S2] cluster by monothiol glutaredoxin. Biochemistry. 2009;48:6041–6043. doi: 10.1021/bi900440m. [DOI] [PubMed] [Google Scholar]
- 26.Rouhier N, et al. Functional, structural, and spectroscopic characterization of a glutathione-ligated [2Fe-2S] cluster in poplar glutaredoxin C1. Proc Natl Acad Sci U S A. 2007;104:7379–7384. doi: 10.1073/pnas.0702268104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumanovics A, et al. Identification of FRA1 and FRA2 as genes involved in regulating the yeast iron regulon in response to decreased mitochondrial iron-sulfur cluster synthesis. J Biol Chem. 2008;283:10276–10286. doi: 10.1074/jbc.M801160200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu XM, Moller SG. AtSufE is an essential activator of plastidic and mitochondrial desulfurases in Arabidopsis. EMBO J. 2006;25:900–909. doi: 10.1038/sj.emboj.7600968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ojeda L, et al. Role of glutaredoxin-3 and glutaredoxin-4 in the iron regulation of the Aft1 transcriptional activator in Saccharomyces cerevisiae. J Biol Chem. 2006;281:17661–17669. doi: 10.1074/jbc.M602165200. [DOI] [PubMed] [Google Scholar]
- 30.Pujol-Carrion N, et al. Glutaredoxins Grx3 and Grx4 regulate nuclear localisation of Aft1 and the oxidative stress response in Saccharomyces cerevisiae. J Cell Sci. 2006;119:4554–4564. doi: 10.1242/jcs.03229. [DOI] [PubMed] [Google Scholar]
- 31.Alves R, V E, Sorribas A, Herrero E. Evolution based on domain combinations: the case of glutaredoxins. BMC Evol Biol. 2009;9:66. doi: 10.1186/1471-2148-9-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morel M, et al. Comparison of the thiol-dependent antioxidant systems in the ectomycorrhizal Laccaria bicolor and the saprotrophic Phanerochaete chrysosporium. New Phytol. 2008;180:391–407. doi: 10.1111/j.1469-8137.2008.02498.x. [DOI] [PubMed] [Google Scholar]
- 33.Jeong J, Guerinot ML. Homing in on iron homeostasis in plants. Trends Plant Sci. 2009;14:280–285. doi: 10.1016/j.tplants.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 34.Pilon M, et al. Essential transition metal homeostasis in plants. Curr Opin Plant Biol. 2009;12:347–357. doi: 10.1016/j.pbi.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 35.Comini MA, et al. Monothiol glutaredoxin-1 is an essential iron-sulfur protein in the mitochondrion of African trypanosomes. J Biol Chem. 2008;283:27785–27798. doi: 10.1074/jbc.M802010200. [DOI] [PubMed] [Google Scholar]
- 36.Lillig CH, et al. Characterization of human glutaredoxin 2 as iron-sulfur protein: a possible role as redox sensor. Proc Natl Acad Sci U S A. 2005;102:8168–8173. doi: 10.1073/pnas.0500735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mesecke N, et al. Two novel monothiol glutaredoxins from Saccharomyces cerevisiae provide further insight into iron-sulfur cluster binding, oligomerization, and enzymatic activity of glutaredoxins. Biochemistry. 2008;47:1452–1463. doi: 10.1021/bi7017865. [DOI] [PubMed] [Google Scholar]
- 38.Lonn ME, et al. Expression pattern of human glutaredoxin 2 isoforms: identification and characterization of two testis/cancer cell-specific isoforms. Antioxid Redox Signal. 2008;10:547–557. doi: 10.1089/ars.2007.1821. [DOI] [PubMed] [Google Scholar]
- 39.Feng Y, et al. Structural insight into poplar glutaredoxin C1 with a bridging iron-sulfur cluster at the active site. Biochemistry. 2006;45:7998–8008. doi: 10.1021/bi060444t. [DOI] [PubMed] [Google Scholar]
- 40.Johansson C, et al. Reversible sequestration of active site cysteines in a 2Fe-2S-bridged dimer provides a mechanism for glutaredoxin 2 regulation in human mitochondria. J Biol Chem. 2007;282:3077–3082. doi: 10.1074/jbc.M608179200. [DOI] [PubMed] [Google Scholar]
- 41.Camaschella C, et al. The human counterpart of zebrafish shiraz shows sideroblastic-like microcytic anemia and iron overload. Blood. 2007;110:1353–1358. doi: 10.1182/blood-2007-02-072520. [DOI] [PubMed] [Google Scholar]
- 42.Wingert RA, et al. Deficiency of glutaredoxin 5 reveals Fe-S clusters are required for vertebrate haem synthesis. Nature. 2005;436:1035–1039. doi: 10.1038/nature03887. [DOI] [PubMed] [Google Scholar]
- 43.Ajioka RS, et al. Biosynthesis of heme in mammals. Biochim Biophys Acta. 2006;1763:723–736. doi: 10.1016/j.bbamcr.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 44.Raux-Deery E, et al. Identification and characterization of the terminal enzyme of siroheme biosynthesis from Arabidopsis thaliana: a plastid-located sirohydrochlorin ferrochelatase containing a 2FE-2S center. J Biol Chem. 2005;280:4713–4721. doi: 10.1074/jbc.M411360200. [DOI] [PubMed] [Google Scholar]
- 45.Jensen LJ, et al. STRING 8--a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dandekar T, et al. Conservation of gene order: a fingerprint of proteins that physically interact. Trends Biochem Sci. 1998;23:324–328. doi: 10.1016/s0968-0004(98)01274-2. [DOI] [PubMed] [Google Scholar]
- 47.Enright AJ, et al. Protein interaction maps for complete genomes based on gene fusion events. Nature. 1999;402:86–90. doi: 10.1038/47056. [DOI] [PubMed] [Google Scholar]
- 48.Marcotte EM, et al. Detecting protein function and protein-protein interactions from genome sequences. Science. 1999;285:751–753. doi: 10.1126/science.285.5428.751. [DOI] [PubMed] [Google Scholar]
- 49.Zaffagnini M, et al. Biochemical characterization of glutaredoxins from Chlamydomonas reinhardtii reveals the unique properties of a chloroplastic CGFS-type glutaredoxin. J Biol Chem. 2008;283:8868–8876. doi: 10.1074/jbc.M709567200. [DOI] [PubMed] [Google Scholar]
- 50.Jacquot JP, et al. Structural and evolutionary aspects of thioredoxin reductases in photosynthetic organisms. Trends Plant Sci. 2009;14:336–343. doi: 10.1016/j.tplants.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 51.Ho Y, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 52.Ito T, et al. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci U S A. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Uetz P, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 54.Huynen MA, et al. Combining data from genomes, Y2H and 3D structure indicates that BolA is a reductase interacting with a glutaredoxin. FEBS Lett. 2005;579:591–596. doi: 10.1016/j.febslet.2004.11.111. [DOI] [PubMed] [Google Scholar]
- 55.Zhou YB, et al. hBolA, novel non-classical secreted proteins, belonging to different BolA family with functional divergence. Mol Cell Biochem. 2008;317:61–68. doi: 10.1007/s11010-008-9809-2. [DOI] [PubMed] [Google Scholar]
- 56.Chandramouli K, Johnson MK. HscA and HscB stimulate [2Fe-2S] cluster transfer from IscU to apoferredoxin in an ATP-dependent reaction. Biochemistry. 2006;45:11087–11095. doi: 10.1021/bi061237w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McClellan AJ, et al. Diverse cellular functions of the Hsp90 molecular chaperone uncovered using systems approaches. Cell. 2007;131:121–135. doi: 10.1016/j.cell.2007.07.036. [DOI] [PubMed] [Google Scholar]
- 58.Rubio LM, Ludden PW. Biosynthesis of the iron-molybdenum cofactor of nitrogenase. Annu Rev Microbiol. 2008;62:93–111. doi: 10.1146/annurev.micro.62.081307.162737. [DOI] [PubMed] [Google Scholar]
- 59.Schwarz G, Mendel RR. Molybdenum cofactor biosynthesis and molybdenum enzymes. Annu Rev Plant Biol. 2006;57:623–647. doi: 10.1146/annurev.arplant.57.032905.105437. [DOI] [PubMed] [Google Scholar]
- 60.Hanzelmann P, Schindelin H. Crystal structure of the S-adenosylmethionine-dependent enzyme MoaA and its implications for molybdenum cofactor deficiency in humans. Proc Natl Acad Sci U S A. 2004;101:12870–12875. doi: 10.1073/pnas.0404624101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen YR, et al. Mitochondrial complex II in the post-ischemic heart: oxidative injury and the role of protein S-glutathionylation. J Biol Chem. 2007;282:32640–32654. doi: 10.1074/jbc.M702294200. [DOI] [PubMed] [Google Scholar]
- 62.Lee DW, et al. A Disruption in Iron-Sulfur Center Biogenesis via Inhibition of Mitochondrial Dithiol Glutaredoxin 2 May Contribute to Mitochondrial and Cellular Iron Dysregulation in Mammalian Glutathione-Depleted Dopaminergic Cells: Implications for Parkinsons Disease. Antioxid Redox Signal. 2009 doi: 10.1089/ars.2009.2489. doi:10.1089/ars.2009.2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Toledano MB, et al. The system biology of thiol redox system in Escherichia coli and yeast: differential functions in oxidative stress, iron metabolism and DNA synthesis. FEBS Lett. 2007;581:3598–3607. doi: 10.1016/j.febslet.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 64.Ye H, et al. CpNifS-dependent iron-sulfur cluster biogenesis in chloroplasts. New Phytol. 2006;171:285–292. doi: 10.1111/j.1469-8137.2006.01751.x. [DOI] [PubMed] [Google Scholar]