Abstract
Telomeres, the specialized nucleoprotein structures located at linear eukaryotic chromosomal termini, are essential for chromosome stability and are maintained by the special reverse transcriptase named telomerase. In the Saccharomycotina subphylum of budding yeast, telomere repeat sequences and binding factors as well as telomerase components are exceptionally diverse and distinct from those found in other eukaryotes. In this survey, I report a comparative analysis of the domain structures of telomere and telomerase-related factors, which is made possible by the recent sequencing of multiple yeast genomes. This analysis revealed conserved as well as variable aspects of telomere maintenance. Based on these findings, I propose a plausible series of evolutionary events in budding yeast to account for its exceptional telomere structural divergence.
Telomeres and Telomerase: overview and evolutionary diversity
Telomeres are specialized nucleoprotein structures that maintain the integrity of eukaryotic chromosomal termini by protecting them from fusion and recombination, and promoting their replication [1, 2]. Telomeres also play important, though relatively uncharacterized roles in mitosis and meiosis [3]. In most organisms, telomeric DNA consists of short repetitive sequences that are rich in G-residues on the 3’ end-containing strand. These repeats are maintained by a ribonucleoprotein (RNP) known as telomerase, which acts as a reverse transcriptase (RT) [4]. Both telomere binding proteins and telomerase are critical for the maintenance of telomere integrity through multiple cell divisions, which in turn is pivotal in supporting genome stability and promoting cellular life span.
Over the past two decades, the mechanisms of telomere protection and maintenance have been studied using many model systems. The picture that has emerged is one of exceptional evolutionary diversity underpinned by a few conserved factors and interactions. For example, in two of the most well characterized systems, namely Saccharomyces cerevisiae and Homo sapiens, the telomeric repeat sequence, the key telomeric protein components, and the telomerase components, with few exceptions, are all quite distinct (Fig. 1). In fact, the subphylum of budding yeast that includes S. cerevisiae (Saccharomycotina) exhibits arguably the greatest evolutionary diversity with regard to telomeric and telomerase-related factors. In addition to the exceptionally well studied baker’s yeast, this subphylum includes many other closely related Saccharomyces spp, as well as Kluyveromyces, Dabromyces, and Candida spp. The complete genomes of many members of the Saccharomycotina subphylum have recently become available, providing rich opportunities for comparative studies [5, 6]. At least two unusual evolutionary events have occurred in this lineage of budding yeast: emergence of species that translate the CUG codon differently from most other organisms (into Ser rather than Leu); and duplication of the entire genome (WGD) in a specific branch (Fig. 2). Although the relationship of these events to telomeric sequence and protein divergence is unclear, they could have provided new substrates for evolutionary changes.
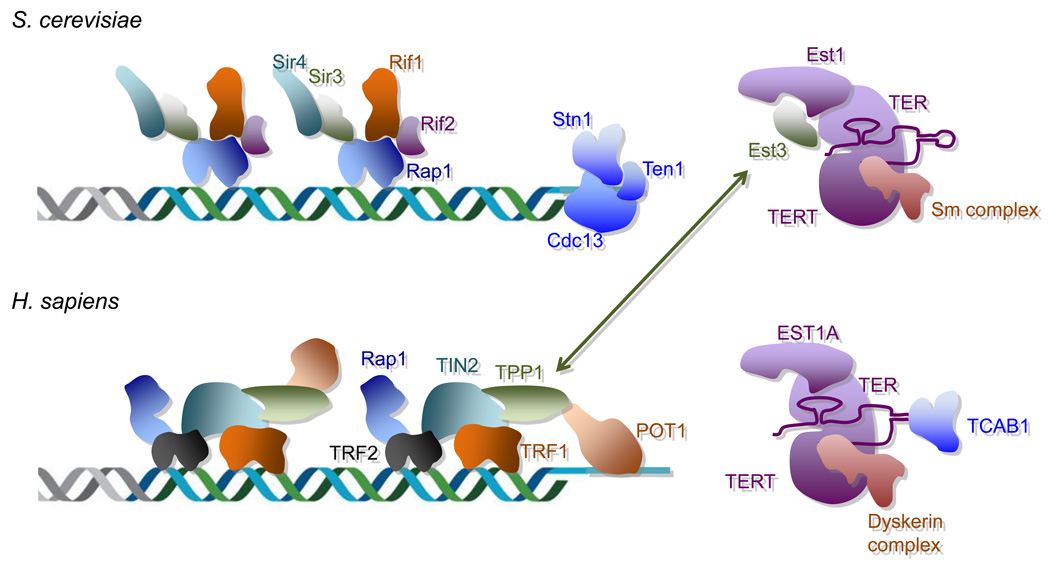
Fig. 1. Comparison of telomeric proteins and telomerase components in budding yeast and humans.
The major components of the telomeric complex and the telomerase complex in budding yeast (S. cerevisiae) and humans (H. sapiens) are illustrated. In yeast, double strand and single strand telomeres are bound by the Rap1 complex and the CST complex, respectively. In humans, telomere DNA is bound by the shelterin complex that contains both double strand and single strand DNA binding proteins (TRF1/2 and POT1, respectively). The telomerase complex in yeast and humans each contains the catalytic reverse transcriptase subunit TERT, the template RNA TER, and several distinct regulatory subunits. The yeast telomerase protein Est3 appears to be homologous to the human telomeric protein TPP1, as indicated by the connecting arrow.
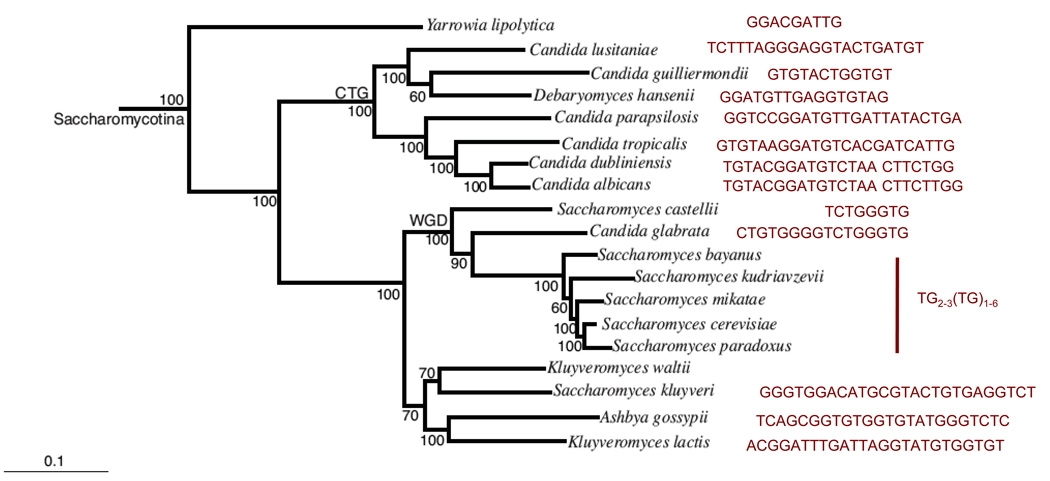
Fig. 2. The phylogenetic relationship of Saccharomycotina species and their telomere repeat unit.
The evolutionary relationships among the Saccharomycotina species and their telomere repeat units are illustrated. The phylogenetic tree was based on comparisons of whole genomes and was reproduced from Fig. 2 of reference 6 [6]. The 0.1 bar designates 10% difference in sequence based on 153 universally distributed fungal genes. The telomere repeat units in these organisms are extremely diverse, ranging in size from 8–25 base pairs. The repeats can also be degenerate (e.g., S. cerevisiae) and non-G-rich (e.g., C. albicans).
In this survey, I first provide a brief overview of telomeric protection and maintenance mechanisms in S. cerevisiae and H. sapiens. Next, I combine a comparative analysis of presumptive orthologues of Saccharomycotina telomeric factors with existing knowledge in order to gain additional insights on the invariant and malleable aspects of telomere maintenance. Special emphases will be placed on structural features of the factors that mediate important telomeric function. In particular, I will attempt to deduce mechanistic conservation (or divergence) of particular pathways/interactions based on structural conservation (or divergence). Finally, I propose a somewhat speculative evolutionary scenario to account for the exceptional variability of telomeric sequences and factors in Saccharomycotina (elements of this scenario have been articulated by others before [7–9]).
Human telomeric proteins and telomerase subunits: a paradigm for organisms with canonical telomere repeats
In humans, the double and single stranded telomeric DNA is bound by a six-protein complex collectively known as Shelterin [10, 11]. As the name implies, a major function of the complex is to shelter the chromosomal ends from machineries that recognize and repair DNA double strand breaks (DSBs). The complex contains two duplex DNA binding proteins (TRF1 and TRF2 (Telomeric Repeat binding Factor 1 and 2)) and a G-tail binding protein (POT1; Protection of Telomere 1) connected through two linker proteins (TIN2 (TRF-Interacting Nuclear protein 2) and TPP1). An additional protein, RAP1 (Repressor Activator Protein 1), is tethered to telomeres through an interaction with TRF2. Mutations in different components of the complex result in distinct telomere dysfunctions, indicating that these proteins are not functionally redundant. By contrast, some subunits do appear to mutually reinforce one another’s association with telomeric DNA [12, 13]. Thus, the functional defects associated with mutations in one component can stem from indirect effects of this component on other subunits of the complex, making it a challenging task to deconvolute the specific mechanisms of individual factors. TRF1 and TRF2 interact with DNA as dimeric molecules through their MYB-like domains. Both proteins are critical for telomere length regulation, whereas TRF2 also has an essential role in preventing telomere fusions. POT1, which is homologous to the Telomere Ending Binding Protein α (TEBPα) from ciliated protozoa, binds single stranded G-tails using two oligosaccharide/oligonucleotides binding (OB) folds and protects the single stranded termini from aberrant reactions [14, 15]. TPP1, which binds POT1 and helps to recruit POT1 to telomere ends, is at least partially homologous to TEBPβ and likewise contains an OB fold [16–19]. In addition to a function in telomere protection, which is anticipated given its role in POT1 recruitment, TPP1 also stimulates processivity in vitro and may recruit telomerase in vivo[18, 20, 21]. Unlike TEBPβ, it has not been shown to contact DNA directly. TIN2 can be considered the central scaffold for shelterin; it interacts with TRF1, TRF2, as well as TPP1, thus helping to stabilize the entire complex [12, 13]. RAP1 is homologous to the major duplex telomere binding protein in yeast, and like TRF2 and its yeast homologue, has been implicated in telomere length control and suppression of non-homologous end joining (NHEJ) [7, 22, 23].
A well-known problem in eukaryotic DNA replication, referred to as the “end replication” problem, is the inability of the chromosomal replication machinery to completely duplicate the ends of linear chromosomes [24, 25]. As a consequence, in many cell populations that undergo repeated cell divisions, telomeres shorten progressively, eventually triggering senescence or apoptosis [26]. The predominant solution to the end replication problem is provided by telomerase, an RNP complex which uses a catalytic Telomerase Reverse Transcriptase (TERT) and a template-containing Telomerase RNA (TER) to extend G-tails. In addition to TERT and TER, human telomerase possesses several other subunits needed for RNP biogenesis and telomere extension. These include proteins involved in Box H/ACA snoRNP maturation (dyskerin, NOP10 (Nucleolar Protein 10), NHP2 (Non-Histone chromosomal Protein 2)) and a Cajal Body-associated factor, TCAB1 [27, 28]. These accessory/regulatory components of telomerase are not well conserved.
Divergence of the Saccharomycotina telomere repeat sequence and telomere binding proteins
The telomere repeat unit appears to be stable in certain fungal lineages (e.g., Ascomycetes and Basidiomycetes) as well as in other phyla (e.g., mammals); these organisms all carry a “canonical” 6-bp repeat 5’-TTAGGG-3’/5’-CCCTAA-3’. Correspondingly, their genomes all appear to encode homologs of the major double and single stranded telomere repeat binding factor found in humans (i.e., TRF1/2 and POT1). By contrast, species in the Saccharomycotina yeast lineage exhibit extraordinary sequence divergence in their telomere repeat unit (Fig. 2). The length of the repeat unit ranges from 8 to 25 bp. In several species, including the well-studied S. cerevisiae, the repeats are degenerate. Furthermore, in contrast to the canonical repeat and many other short repeats found in diverse organisms (e.g., in most ciliated protozoa), the Saccharomycotina telomere repeats are often not rich in deoxyriboguanidine (dG) residues on the 3’-containing strand. These unusual telomere DNA features are accompanied by an unusual complement of telomere repeat binding factors and telomerase subunits. For example, no convincing POT1 homologues can be discerned in the genomes of these organisms. In addition, although a TRF1/2 homologue (TBF1; TTAGGG repeat Binding Factor 1) is often present, it appears to act as a transcription factor and as a subtelomere-binding factor rather than as the telomere-repeat binding factor [29, 30]. Instead, the functions of TRF1/2 and POT1 are assumed by the non-homologous Rap1 and the CST (Cdc13 -Stn1-Ten1) complex [31].
A similar divergence of telomerase subunits is evident. In particular, the telomerase RNAs (TERs) are much larger, typically more than 1,000 nt long, in contrast to the ~200–500 nt TERs usually observed in mammals and ciliates. In addition, the Est3 telomerase subunit (Ever Shorter Telomeres 3) appears to be confined to this branch of budding yeast. Although present in many yeast species, another telomerase protein, Est1, appears to perform a distinct set of functions and interact with distinct factors in budding yeast [32, 33].
In short, the simultaneous presence of unusual telomeric DNA and telomere-related proteins in Saccharomycotina argues strongly for the occurrences of unusual evolutionary events in the common ancestors of these organisms. To gain insight into such events, I undertook a comparative analysis of telomeric and telomerase subunits in these organisms and uncovered unexpected instances of evolutionary plasticity, including loss and acquisition of specific domains and loss of entire genes in certain branches of budding yeast.
The duplex telomere repeat binding factors
Rap1 is the major double strand telomere repeat binding factor in Saccharomycotina. It has been extensively analyzed in S. cerevisiae, where it regulates telomere lengths, telomere position effect, and telomere–telomere fusions [31]. Remarkably, S. cerevisiae Rap1 also binds to other genomic locations, where it mediates additional functions in transcriptional regulation and mating type silencing. Not surprisingly, Rap1 has a complex domain organization befitting its multiplicity of functions (Fig. 3). Multiple sequence alignment of Rap1 homologues ranging from budding yeast and fission yeast to mammals revealed three conserved domains, including an N-terminal BRCT domain, a central MYB domain and an RCT domain (named for Rap1 C-Terminus). The extent of structural conservation is somewhat surprising in light of a major mechanistic distinction that has been described between budding yeast and other Rap1 homologues. Specifically, in fission yeast and mammals, Rap1 lacks direct DNA binding activity, and its telomeric localization depends on specific protein–protein interactions (Fig. 1). This observation immediately suggests that the individual Rap1 domains, although structurally similar in different organisms, must be capable of distinct functions.
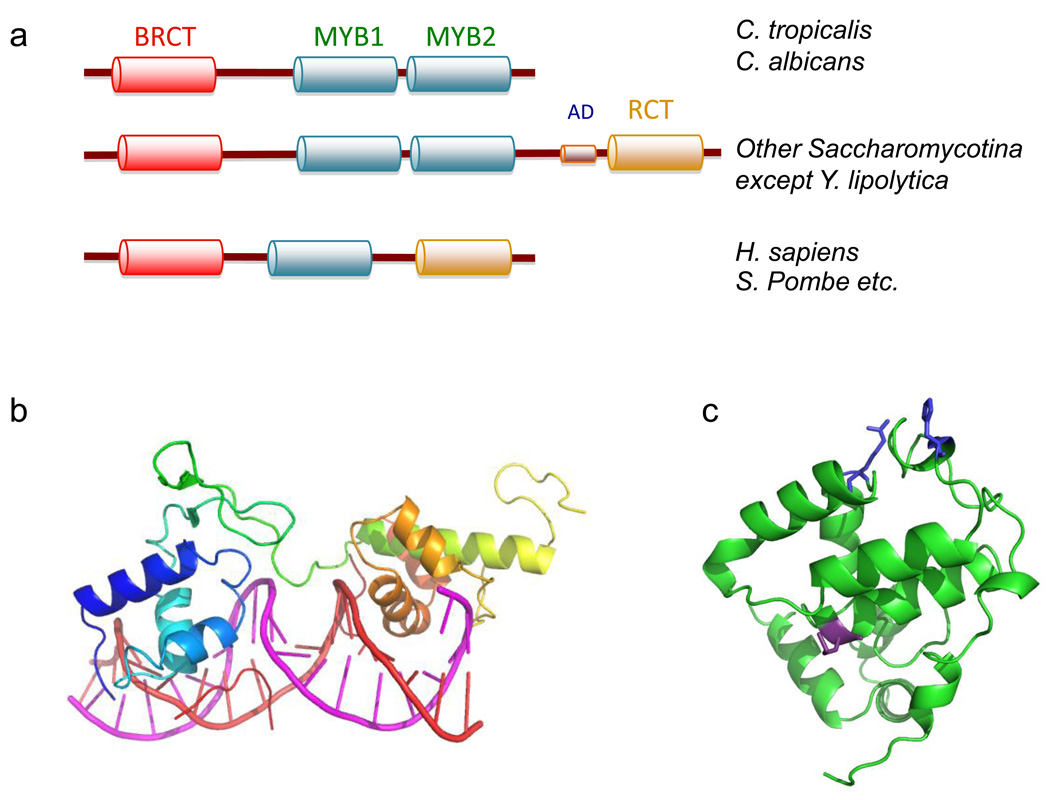
Fig. 3. Domain organization and structure of Rap1.
Sequence alignment and high resolution structural analysis have revealed variations in the domain organization of Rap1 and the molecular bases of its interaction with target DNA and proteins. (a) The domain structures of Rap1 from various Saccharomycotina and other species are illustrated. The BRCT, MYB, and RCT domains are displayed in different colors. Notably, the RCT domain is missing in C. tropicalis and C. albicans, and a single MYB domain is present in non-Saccaromycotina organisms. (b) The crystal structure of the MYB1 and MYB2 domains of S. cerevisiae Rap1 (shown in color spectrum from blue to orange) bound to its target DNA (shown in magenta and red) (PDB ID: 1IGN). Sequence-specific recognition is mediated by the alpha helices of the MYB domains, as well as a connecting linker and a C-terminal tail. (c) The crystal structure of the RCT of S. cerevisiae Rap1 (PDB ID: 3CZ6). The backbone and the residues implicated in Rif2 and Sir3 binding are shown in green, blue and purple, respectively.
One domain that must be functionally flexible is the MYB-like domain. A tandem duplication of this domain can be discerned in Saccharomycotina, whereas only one copy is evident in other yeast and mammals (Fig. 3). In spite of the extraordinary divergence of telomere repeat sequence, the duplicate MYB homology domains appear capable of recognizing the cognate telomere repeat units with high affinity and sequence specificity [34, 35]. The acquisition of two MYB-like domains connected through a flexible linker might have contributed to the versatility and evolutionary malleability of this domain [36].
The RCT (named for Rap1 C-terminal domain) also appears to be evolutionarily malleable. In humans, the Rap1 RCT directs its telomere localization through an interaction with TRF2 [7, 22]. In S. cerevisiae, the RCT has acquired the ability to interact with at least four proteins which have well-defined functions in telomere regulation. Rap1-interacting Factor 1 (Rif1) and Rif2 are required to prevent excessive telomere elongation by telomerase, whereas Silent Information Regulator 3 (Sir3) and Sir4 enable proper telomere silencing. Remarkably, both Rif2 and Sir3 have arisen recently in evolution, following the WGD event that occurred in a Saccharomycotina lineage that includes S. cerevisiae as well as C. glabrata and S. castellii. RIF2 is syntenic with, and corresponds to just the N-terminal region of, Origin Recognition Complex 4 (ORC4), and SIR3 is syntenic with and comparable in length to ORC1 [37, 38]. Because the Rap1 RCT is clearly present in budding yeast that did not undergo WGD, it is tempting to speculate that Orc1 and Orc4 might mediate similar interactions with Rap1 and perform analogous functions in these species. The simultaneous presence of two ORC subunit homologues at telomeres also suggests that the entire ORC complex might, at some point, have been telomere-localized. This notion is supported by the recent observation of ORC proteins at mammalian telomeres [39].
Alignment of Rap1, Rif2, and Sir3 as well as structure-function studies of their interaction determinants provide additional insights into the mechanisms and evolution of these proteins [40–42]. In particular, a recent crystallographic study provided a high-resolution view of the entirely alpha helical RCT of S. cerevisiae Rap1. Consistent with overall structural conservation of the RCT from yeast to mammals, the highly conserved RCT residues are predominantly oriented toward the hydrophobic core and likely are important for the structural integrity of this domain. Site-specific mutagenesis analysis further identified residues within the CTD that are selectively required for interaction with Rif2 (H709 and R747) and Sir3 (M763) [40] (Fig. 3). Consistent with the absence of Rif2 or Sir3 in fission yeast and mammals, the Rap1 residues implicated in Rif2 and Sir3 binding are not conserved in these organisms and are only partially conserved in Saccharomycotina.
In Saccharomycotina, two instances of global structural alteration in Rap1 are evident. Although most members of this clade have retained the three-domain organization, C. albicans and C. tropicalis Rap1 are considerably smaller and lack the RCT (Fig. 3). The interactions mediated by CTD (i.e., with the Rif, Sir or Orc proteins) presumably are absent in these species. The functional consequence of these absences for the Candida spp. is an interesting area for future investigation. Another apparent anomaly is the complete absence of a Rap1 homologue in the Yarrowia lipolytica genome, raising questions about the telomere protection mechanism of this yeast. More intriguingly, a recent study suggests that the transcriptional activation function of S. cerevisiae Rap1 is also a relatively modern invention that emerged in the Saccharomicotina subphylum prior to Kluyveromyces lactis speciation [29]. Thus, aspects of Rap1 function other than telomere regulation were also subject to rapid evolution.
The G-tail binding factors
The Saccharomycotina genomes are distinguished from other genomes by the absence of a convincing Pot1 homologue. Moreover, extensive studies in S. cerevisiae have revealed the existence of an alternative G-tail capping CST complex. Of the three components, Cell Division Cycle 13 (Cdc13) can to recognize telomeric G-tails with high affinity and sequence specificity, whereas Stn1 (Suppressor of temperature-sensitive Cdc13) and Ten1 (Telomeric pathways with Stn1) bind preferentially G-rich DNA with lower affinities [43, 44]. The genes encoding all three proteins are essential for cell viability and hypomorphic alleles of each gene result in extensive telomere degradation, as well as aberrant telomerase and recombination activities at telomeres [31, 45–48]. Aside from telomere protection, S. cerevisiae Cdc13 also promotes the recruitment of telomerase to telomere ends [49]. This function is mediated through an interaction between the Cdc13 recruitment domain (RD) and the telomerase regulatory protein Est1. Furthermore, removal of the Cdc13 C-terminus can induce telomere elongation, pointing to the existence of yet another, negative regulatory function for this protein [50]. Advanced bioinformatic analyses coupled with molecular genetic studies led to the provocative notion that the CST complex resembles structurally the Replication Protein A (RPA) complex, the major non-sequence specific DNA binding activity in eukaryotic cells [43]. In particular, Cdc13, Stn1 and Ten1 were proposed to be homologous to RPA70, RPA32 and RPA14, respectively. Atomic-resolution structures of the RPA subunits demonstrate that these proteins consist of variable numbers of OB fold domains [51]. Domain-swapping experiments provide support for structural and functional similarity between the OB fold domains of RPA32 and Stn1 [43]. By contrast, the relationship between Cdc13 and RPA70 remains unclear [52].
Certain CST homologues in the Saccharomycotina genomes were originally difficult to identify owing to rapid sequence divergence [9]. For example, initial BLAST searches failed to disclose a C. albicans Cdc13 even after the genome was completely sequenced. However, with the availability of more genomic data and the improved ability to construct better profiles and conduct iterative searches, homologues of CST subunits have been identified in all well-characterized Saccharomycotina genomes except that of Y. lipolytica (Fig. 4). The absence of CST as well as Rap1 reinforces the notion that telomere protection in Y. lipolytica might be mediated by rather unconventional factors and mechanisms. Multiple sequence alignments of available CST homologues provide several interesting insights into the structure and function of these proteins (Fig. 4). First, a significant fraction of Cdc13 homologues are noticeably smaller than the S. cerevisiae prototype; they lack certain domains. At least four functional regions have been identified in the 924 amino acid S. cerevisiae Cdc13, including an N-terminal region that interacts with several proteins (N-ter, amino acids 1 to 252), a recruitment domain (RD, amino acids 211 to 331) that mediates the interaction with Est1, a DNA-binding domain (DBD, amino acids 557 to 694) that represents an unusual OB fold with extended loops, and a C-terminal domain that negatively regulates telomere lengths (C-ter) [44, 49, 50, 53]. The subfamlily of smaller Cdc13 proteins (e.g., those from C. albicans, C. tropicalis, etc.) can be aligned well to the DNA-binding and C-terminal domains of S. cerevisiae Cdc13 suggesting that the essential functions of Cdc13 reside in the C-terminal half of the protein. Moreover, the recruitment pathway mediated by the Cdc13–Est1 interaction is likely to be absent in the yeast with small Cdc13, possibly replaced by alternative interactions. In contrast to Cdc13, the domain structures of Stn1 and Ten1 are relatively well conserved in Saccharomycotina. The Stn1 family members have a more similar N-terminal OB fold domain and a more divergent C-terminal domain whereas the Ten1 family members comprise a single OB fold. The Stn1 and Ten1 OB fold domains direct their mutual interaction, possibly accounting for their higher degree of evolutionary conservation [43, 47].
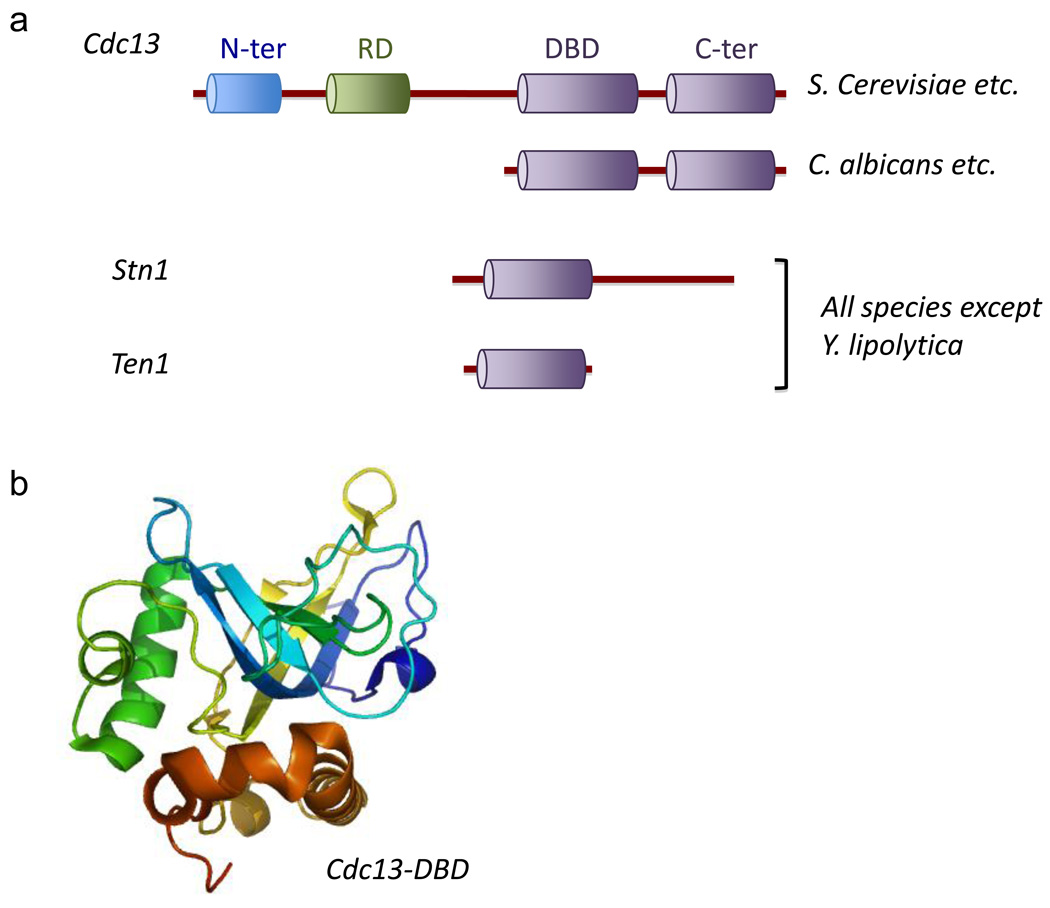
Fig. 4. Domain organizations and structures of the CST complex.
Sequence alignment and high resolution structural analysis have revealed conserved and variable aspects of CST protein domain organizations, as well as the structural basis of DNA recognition by Cdc13. (a) The domain structures of Cdc13, Stn1 and Ten1 from Saccharomycotina yeast are illustrated. Probable OB fold domains are displayed in purple. Notably, the Cdc13 homologues from many Candida species (including C. albicans, C. tropicalis, C. parapsilosis, etc.) are smaller than the S. cerevisiae protein and consist of only the C-terminal domains. (b) The crystal structure of the DNA binding domain of S. cerevisiae Cdc13, which consists of an unusual OB fold, is shown in color spectrum from blue to brown (PDB ID: 1S40).
The telomerase ribonucleoprotein complex
The S. cerevisiae telomerase complex has been extensively analyzed; a complete description of its subunit composition, however, has remained elusive [31, 54]. In addition to TERT and TER, two telomerase-specific regulatory proteins are critical for telomere maintenance. One of the regulatory proteins, Est1, comprises multiple domains and is multifunctional. It interacts with both Cdc13 and telomerase RNA, thereby promoting the recruitment of the telomerase complex to telomere ends [33, 49]. It is also necessary for the incorporation of the other telomerase regulatory protein Est3 [55, 56]. Est3 has been proposed to resemble structurally TPP1 (an OB fold-containing protein), and to activate telomerase function through direct protein-protein interactions [57, 58]. As TPP1 is a component of the mammalian telomeric protein complex, the incorporation of its potential yeast orthologue into the telomerase complex poses fascinating evolutionary questions.
Of the four yeast telomerase components, the TERT polypeptide is most highly conserved, as anticipated from its crucial catalytic function. Comparative sequence analysis and structure function studies have revealed three conserved domains in TERTs ranging from yeast to humans: an N-terminal GQ/TEN domain that exhibits both DNA and RNA binding activity and likely serves as the so-called “anchor site’, a central RNA-binding domain (RBD) that is responsible for stable RNP formation, and a C-terminal reverse transcriptase domain (RT) that catalyzes telomere elongation [4, 59]. This basic domain architecture was retained in all Saccharomycotina TERTs, although the primary amino acid sequences for the GQ/TEN domain exhibit relatively weak similarities (Fig. 5). Given its proposed function in DNA-binding, it is perhaps not surprising that the GQ/TEN domain would evolve species-specific residues to accommodate the diversity of telomere repeat sequence [60, 61]. In contrast to TERT, yeast telomerase RNAs, like those of other phyla, are extremely divergent at the primary sequence level [62–64]. For example, it is not feasible to recognize Candida telomerase RNAs through standard BLAST searches using Saccharomyces TER sequences as queries. However, a recent combined computational and experimental study identified many TERs in the Saccharomycotina genomes, providing a much-needed foundation for detailed analysis [65]. Like TERTs, a number of structural elements in TERs are thought to be universal, including the template that specifies the telomere repeat unit; a TERT-interacting pseudoknot that encloses a triple helix; a 3-way junction/stem-loop necessary for enzymatic function; and a long-range pairing element that might bring the aforementioned structures into close spatial proximity to facilitate telomerase function [63, 66–69]. Preliminary in silico analysis suggests that these structural elements are indeed universally present in Saccharomycotina TERs [65]. One structure that might be unique to budding yeast TERs is the binding target for the telomerase regulatory protein Est1. In S. cerevisiae, a region that is 3’ to the templating domain and that contains several stems is necessary and sufficient for Est1 binding [70]. Comparable stems are thought to exist in all other Saccharomycotina TERs, but their possible role in Est1 binding has not been tested. Indeed, the stem is present even in Candida parapsilosis, a species that lacks an apparent Est1 homolog, thus raising questions concerning its precise function [65]. It is also interesting to note that although two human homologues of Est1 have been reported to associate with telomerase, this association is based not only on RNA–protein interactions, but also protein–protein interactions [71, 72]. This observation highlights the malleability of telomeric functions performed by Est1.
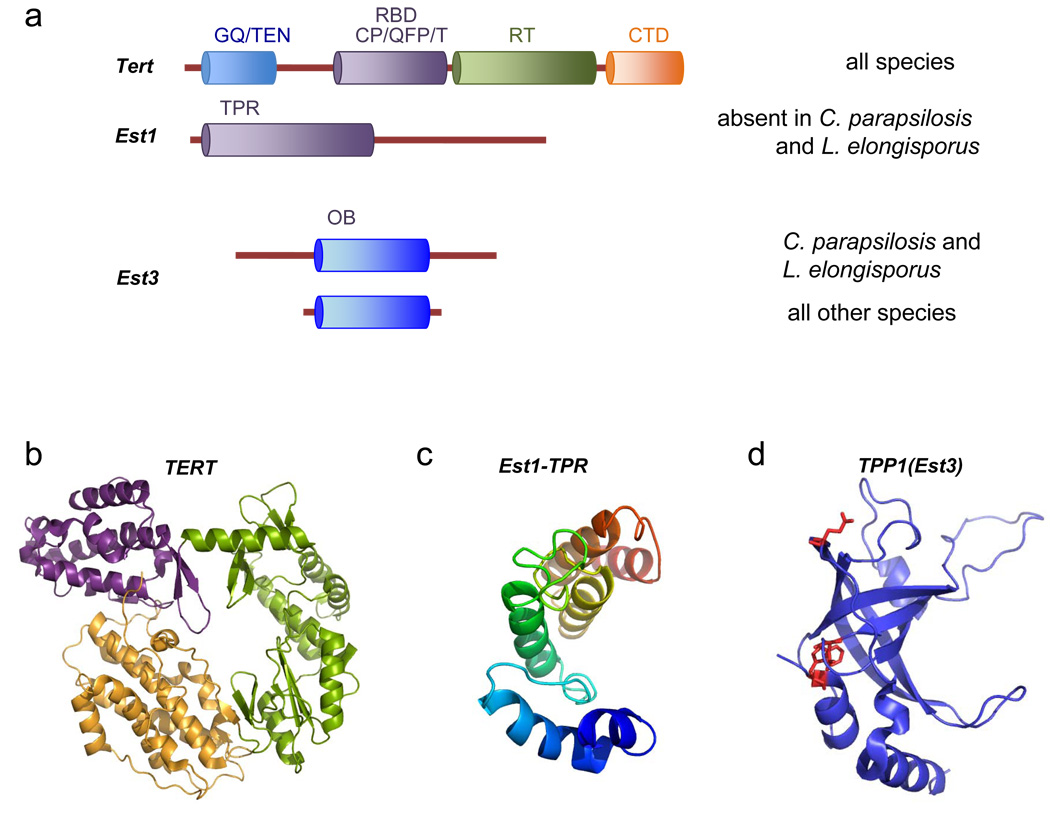
Fig. 5. Domain organizations and structures of the telomerase protein subunits.
Sequence alignment and high resolution structural analysis have revealed conserved and variable aspects of telomerase protein components, and provided insights on their molecular mechanisms. (a) The domain structures of Tert, Est1 and Est3 from Saccharomycotina yeast are illustrated. All yeast TERTs share a conserved structure comprising the GQ/TEN, RNA-binding (RBD), reverse transcriptase (RT) and C-terminal domain (CTD). Est1 homologues possess a TPR-like domain at the N-terminus and a more divergent C-terminal domain, whereas Est3 homologues mostly comprise a single OB fold. Notably, the EST1 gene is missing in the C. parapsilosis and L. elongisporus genomes and the EST3 genes in these organisms possess long N- and C-terminal tails. (b) The crystal structure of the Tert protein from Tribolium castaneum, which lacks the GQ/TEN domain, is displayed (PDB ID: 3DU6). The RBD, RT and CTD domains are shown in purple, green and orange, respectively. (c) A homology model of the TPR-like domain of S. cerevisiae Est1 generated based on the crystal structure of human EST1C (shown in color spectrum from blue to red) (PDB ID: 1YA0). (d) The crystal structure of the OB fold domain of human TPP1 (PDB ID: 2I46), which is proposed to be homologous to Est3. Three residues hypothesized to be important for binding of Est3 to TERT are highlighted in red.
In comparison with the catalytic TERT protein, the sequences of the regulatory proteins Est1 and Est3 in Saccharomycotina are less well conserved, but their basic domain organizations appear invariant with two notable exceptions. The better conserved region of Est1 is near the N-terminus and exhibits sequence and structural similarity to the tetratricopeptide repeat (TPR) motif [73–75]. These motifs consist of multiple copies of anti-parallel alpha helical pairs that together form an interaction surface. Although TPR motifs are generally thought to bind other peptides, the TPR domain of S. cerevisiae Est1 might have an additional function in RNA-binding [76–78] . Interestingly, the C-terminal half of Est1, though less well conserved, is implicated in Cdc13-binding [49, 79]. This observation again reinforces the notion that the Est1–Cdc13 interaction might not be widespread, as suggested by the absence of the Cdc13 RD in most Candida spp. Remarkably, an Est1 homologue cannot be discerned in either the C. parapsilosis or Lodderomyces elogisporus genome [65]. The loss of the EST1 gene in these two genomes is supported by examination of the regions of the genomes that are syntenic to the EST1 loci in other Candida spp. Thus, the function of Est1 in Est3 assembly and telomerase recruitment must have become dispensable or be taken over by other factors in C. parapsilosis and L. elongisporus.
Among the yeast telomerase regulatory proteins, Est3 is the least well conserved. In silico and structure–function analyses have revealed the presence of a TPP1/TEBPβ-like OB fold in this protein, which appears to use a non-nucleic acid binding face for telomerase interaction [57, 58]. Although this basic fold is preserved in all Saccharomycotina Est3, the homologues from C. paraplosis and L. elongisporus are larger, -possessing an ~90 amino acid N-terminal and an ~70 amino acid C-terminal extension. Notably, these large Est3 proteins are encoded by precisely the two genomes that lack recognizable Est1 homologues. It is tempting to speculate that these peculiarities in Est1 and Est3 could be mechanistically related. Perhaps the N- and C-terminal extension of C. parapsilosis and L. elongisporus Est3 strengthened its interaction with telomerase, thus compensating for the loss of Est1. Further studies will be necessary to address this intriguing hypothesis.
The evolution of diverse telomeric repeat sequence and telomere-related proteins in the Saccharomycotina subphylum
The extent of sequence divergence in the telomere repeat unit of budding yeast is quite impressive, all the more so in light of the prevalence of the canonical TTAGGG sequence in most eukaryotic phyla. The yeast telomere sequence diversity is accompanied by a distinct set of telomeric proteins and telomerase regulatory subunits. What plausible events during fungal evolution could have resulted in such sequence and structural divergence? A notion that has gained currency postulates that one or several catastrophic telomerase RNA template mutations in the common ancestor of Saccharomycotina yeast might have arisen and been fixed by subsequent recruitment of Rap1 and Cdc13 to telomeres (Fig. 6)[8, 9, 80]. Although RNA template mutations were clearly the most obvious explanations for telomeric sequence changes, the postulated evolutionary scenario presents several conundrums. First, it is puzzling that spontaneous mutations in telomerase RNA, given their deleterious consequences on telomere capping and hence organismal fitness, would not be immediately selected against and lost during evolution. Second, even if evolutionary pressures favored mutations, it is unclear how the Rap1 and Cdc13 proteins could have evolved such high affinity interactions with such diverse and novel telomere repeat sequences. Third, the reason for the distinctive composition of yeast telomerase RNP is unclear. To address these apparent difficulties, I propose the following modifications to the original scenario. First, instead of spontaneous mutations in telomerase RNA alone, it seems reasonable to postulate certain selection pressures for variant telomerase RNA. For example, it is conceivable that the ancestral RNA, or even the telomeric DNA repeat, was targeted by an ancient pathogen (e.g., a virus) that carried a telomerase RNA or telomere DNA-cleavage activity. Mutations in telomerase RNA that counteract the hypothetical cleavage activity can then provide selective advantages in this unusual setting. Second, the evolution of new binding specificity for Rap1 and Cdc13 might have been relatively gradual. In the case of Rap1, its evolution of DNA binding activity initially might have been facilitated by its telomere localization. It seems reasonable to assume that in the ancestral Saccharomycotina, Rap1 was tethered to telomeres through protein–protein interactions, given the prevalence of this arrangement in nature [7, 81]. In this setting, even low affinity for DNA, which can be provided by a few new hydrogen bonds, could have allowed significant fractions of the molecule to contact DNA directly, thereby providing greater protection, and thus, a selective advantage. Notably, colocalization of proteins, which greatly increases their local concentrations, can amplify the effect on one protein of random mutations in another protein and promote natural selection [82]. The same principle may underlie the evolution of protein–nucleic acid interactions. Perhaps similar to Rap1, the CST complex was also telomere-localized in the ancestral yeast. The loss of POT1 that ensued from telomere repeat alterations might have provided the selection pressure for CST complex to evolve new DNA binding specificities and assume a critical role in telomere protection. Indeed, a STN1 homologue with a proven function in telomere protection has now been identified in Arabidopsis thaliana, an organism that also possesses POT1, arguing for a more widespread role for CST subunits at telomeres [83]. Also worth considering is the potential role of the ancestral yeast repeats in facilitating the transition. If some of these ancestral TTAGGG repeats were retained in the subtelomere region and bound by Tbf1, as in present day S. cerevisiae, the subtelomeric nucleoprotein complex could have performed partial telomeric functions and allowed gradual evolution of a more optimized system [30, 80].
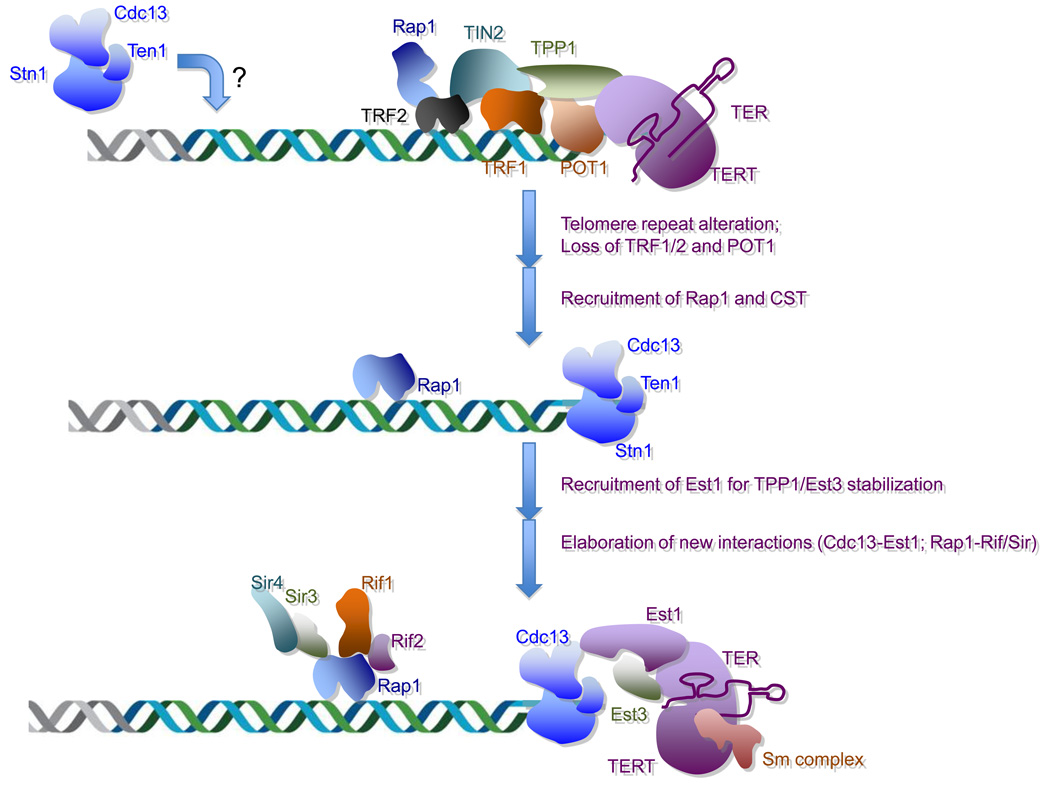
Fig. 6. Evolution of telomeres and telomerases in budding yeast.
A possible series of evolutionary events that lead to the current diversity in telomere and telomerase structures in budding yeast are illustrated. The ancestral yeast is thought to contain a shelterin-like complex as well as a CST complex at telomeres. During evolution, an alteration in the telomere repeat sequence leads to the loss of TRF1, TRF2, POT1, and other associated factors. Subsequently, Rap1 and the CST complex, which were initially telomere-localized, evolved new DNA-binding activities to protect the double and single stranded telomere DNA. In addition, new interactions such as the Cdc13–Est1, Est1–Est3, Rap1–Rif1/2, and Rap1–Sir3/4 interactions, emerged to promote the recruitment and activity of yeast telomerase as well as other aspects of yeast telomere regulation.
Another consequence of telomere sequence alteration is the loss of TPP1. Aside from telomere protection, human TPP1 and its S. pombe homologue Tpz1 are thought to positively regulate telomerase by enhancing its recruitment and activity [18, 21, 84]. Assuming that this was also the case in the ancestral Saccharomycotina, the loss of TPP1 would have impaired telomerase function as well. In light of this consideration, the previously noted structural similarity between TPP1 and Est3 is perhaps not so surprising after all [57, 58]. Specifically, Est3 might have evolved from the ancestral TPP1, and been retained in Saccharomycotina to promote telomerase activity. Furthermore, Est1 might have acquired a new function in Est3 binding in order to stabilize Est3-telomerase association [55, 56]. It is also possible that the Cdc13–Est1 interaction detected in S. cerevisiae evolved to compensate for the loss of the TPP1 recruitment function. Notably, the Cdc13–Est1 and Est1–Est3 interactions appear not to be conserved even in Saccharomycotina, suggesting that alternative compensatory mechanisms are possible. Thus, the challenges presented by the alterations in telomere repeat sequence were met in a variety of ways in members of Saccharomycotina yeast.
A notable aspect of telomere and telomerase remodeling in budding yeast is that distinct changes (e.g., in Rap1, Cdc13, Est1/3) occur in distinct lineages. Thus, each specific solution to the end protection and maintenance problem engendered by the loss of TRF1/2 and POT1 probably conferred too small an advantage for fixation in the entire subphylum. Consequently there appears to have been a continuous search for and emergence of new telomere protection and telomerase recruitment pathways over a long evolutionary time span.
CONCLUDING REMARKS
Although the cause of telomere sequence divergence in Saccharomycotina will probably remain obscure, there seems little question that this sequence divergence was itself responsible for the emergence of atypical telomere protection and maintenance proteins in the budding yeast. The comparative analysis of telomere proteins has provided striking illustrations of evolutionary changes, including the acquisition and loss of entire genes and specific domains, as well as emergence of new and novel interactions. These changes raise many interesting questions concerning the adaptive mechanisms of telomere proteins that allow them to perform the required functions in spite of changes in their binding targets (Box 1). Even more drastic changes than glimpsed here likely occurred in Y. lipolytica, given its lack of discernable TRF, RAP1 and CST homologues. In contrast to the rather stable inheritance of chromosomal replication machinery, the telomere protection and maintenance machineries are clearly more malleable. This difference could in part be due to the existence of alternative telomere maintenance mechanisms that rely on recombination and other cellular processes [85–87]. The tolerance of telomeres to profound evolutionary changes is thus unlikely to be confined to Saccharomycotina. Indeed, many other instances of deviations from the canonical telomere repeat have been described, including that of S. pombe and A. thaliana. Although S. pombe has evidently retained Pot1, it has an unusually large TPP1 homologue that apparently acquired other domains [84]. Could their addition have been in response to challenges posed by the unusual S. pombe telomere repeat? Regardless of the precise answers to such questions, it can be stated with some confidence that continued exploration of telomere evolutionary pathways in different clades will continue to provide important insights on telomere mechanisms.
Box 1: Outstanding Questions
How do the “miniaturized” C. albicans and C. tropicalis Rap1 meet its functional requirement in telomere regulation?
What are the structural bases for recognition of diverse telomere repeat units by yeast Rap1 and Cdc13?
Is the EST1 gene truly lost from the C. parapsilosis and L. elongisporus genomes, and if so, what strategies were developed by these fungi to compensate for its loss?
Have the Candida species with truncated Cdc13 evolved alternative mechanisms of telomerase recruitment?
ACKNOWLEDGEMENT
I thank Wei-Feng Yen for compiling some of the telomere and telomerase protein sequences and members of my laboratory for comments and advice. Works in my laboratory have been supported by NIH (GM062631 and GM069507) and the STARR Cancer Consortium.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Blackburn EH, et al. Telomeres and telomerase: the path from maize, Tetrahymena and yeast to human cancer and aging. Nat Med. 2006;12:1133–1138. doi: 10.1038/nm1006-1133. [DOI] [PubMed] [Google Scholar]
- 2.Cech T. Beginning to understand the end of the chromosome. Cell. 2004:273–279. doi: 10.1016/s0092-8674(04)00038-8. [DOI] [PubMed] [Google Scholar]
- 3.Tomita K, Cooper JP. The meiotic chromosomal bouquet: SUN collects flowers. Cell. 2006;125:19–21. doi: 10.1016/j.cell.2006.03.020. [DOI] [PubMed] [Google Scholar]
- 4.Autexier C, Lue NF. The Structure And Function Of Telomerase Reverse Transcriptase. Annu Rev Biochem. 2006;75:493–517. doi: 10.1146/annurev.biochem.75.103004.142412. [DOI] [PubMed] [Google Scholar]
- 5.Butler G, et al. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature. 2009;459:657–662. doi: 10.1038/nature08064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fitzpatrick DA, et al. A fungal phylogeny based on 42 complete genomes derived from supertree and combined gene analysis. BMC Evol Biol. 2006;6:99. doi: 10.1186/1471-2148-6-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li B, et al. Identification of human Rap1: implications for telomere evolution. Cell. 2000;101:471–483. doi: 10.1016/s0092-8674(00)80858-2. [DOI] [PubMed] [Google Scholar]
- 8.Smogorzewska A, de Lange T. Regulation of telomerase by telomeric proteins. Annu Rev Biochem. 2004;73:177–208. doi: 10.1146/annurev.biochem.73.071403.160049. [DOI] [PubMed] [Google Scholar]
- 9.Teixeira MT, Gilson E. Telomere maintenance, function and evolution: the yeast paradigm. Chromosome Res. 2005;13:535–548. doi: 10.1007/s10577-005-0999-0. [DOI] [PubMed] [Google Scholar]
- 10.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 11.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 12.Kim SH, et al. TIN2 mediates functions of TRF2 at human telomeres. J Biol Chem. 2004;279:43799–43804. doi: 10.1074/jbc.M408650200. [DOI] [PubMed] [Google Scholar]
- 13.Ye JZ, et al. TIN2 binds TRF1 and TRF2 simultaneously and stabilizes the TRF2 complex on telomeres. J Biol Chem. 2004;279:47264–47271. doi: 10.1074/jbc.M409047200. [DOI] [PubMed] [Google Scholar]
- 14.Baumann P, Cech TR. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science. 2001;292:1171–1175. doi: 10.1126/science.1060036. [DOI] [PubMed] [Google Scholar]
- 15.Lei M, et al. Switching human telomerase on and off with hPOT1 protein in vitro. J Biol Chem. 2005;280:20449–20456. doi: 10.1074/jbc.M502212200. [DOI] [PubMed] [Google Scholar]
- 16.Ye JZ, et al. POT1-interacting protein PIP1: a telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes Dev. 2004;18:1649–1654. doi: 10.1101/gad.1215404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu D, et al. PTOP interacts with POT1 and regulates its localization to telomeres. Nat Cell Biol. 2004;6:673–680. doi: 10.1038/ncb1142. [DOI] [PubMed] [Google Scholar]
- 18.Wang F, et al. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445:506–510. doi: 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
- 19.Houghtaling BR, et al. A dynamic molecular link between the telomere length regulator TRF1 and the chromosome end protector TRF2. Curr Biol. 2004;14:1621–1631. doi: 10.1016/j.cub.2004.08.052. [DOI] [PubMed] [Google Scholar]
- 20.Hockemeyer D, et al. Telomere protection by mammalian Pot1 requires interaction with Tpp1. Nat Struct Mol Biol. 2007;14:754–761. doi: 10.1038/nsmb1270. [DOI] [PubMed] [Google Scholar]
- 21.Xin H, et al. TPP1 is a homologue of ciliate TEBP-beta and interacts with POT1 to recruit telomerase. Nature. 2007;445:559–562. doi: 10.1038/nature05469. [DOI] [PubMed] [Google Scholar]
- 22.Li B, de Lange T. Rap1 affects the length and heterogeneity of human telomeres. Mol Biol Cell. 2003;14:5060–5068. doi: 10.1091/mbc.E03-06-0403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bae NS, Baumann P. A RAP1/TRF2 complex inhibits nonhomologous end-joining at human telomeric DNA ends. Mol Cell. 2007;26:323–334. doi: 10.1016/j.molcel.2007.03.023. [DOI] [PubMed] [Google Scholar]
- 24.Olovnikov A. Principles of marginotomy in template synthesis of polynucleotides. Doklady Akad. Nauk SSSR. 1971;201:1496–1499. [PubMed] [Google Scholar]
- 25.Watson J. Origin of concatemeric T7 DNA. Nat New Biol. 1972;239:197–201. doi: 10.1038/newbio239197a0. [DOI] [PubMed] [Google Scholar]
- 26.Allsopp R, et al. Telomere length predicts replicative capacity of human fibroblasts. Proc Natl Acad Sci U S A. 1992;89:10114–10118. doi: 10.1073/pnas.89.21.10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Venteicher AS, et al. A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science. 2009;323:644–648. doi: 10.1126/science.1165357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fu D, Collins K. Purification of human telomerase complexes identifies factors involved in telomerase biogenesis and telomere length regulation. Mol Cell. 2007;28:773–785. doi: 10.1016/j.molcel.2007.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hogues H, et al. Transcription factor substitution during the evolution of fungal ribosome regulation. Mol Cell. 2008;29:552–562. doi: 10.1016/j.molcel.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berthiau AS, et al. Subtelomeric proteins negatively regulate telomere elongation in budding yeast. Embo J. 2006;25:846–856. doi: 10.1038/sj.emboj.7600975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lundblad V. Budding yeast telomeres. In: de Lange T, et al., editors. Telomeres and Telomerase. 2nd edn. Cold Spring Harbor Laboratory Press; 2006. [Google Scholar]
- 32.Lundblad V. Telomere replication: an est fest. Curr Biol. 2003;13:pR439–pR441. doi: 10.1016/s0960-9822(03)00365-8. [DOI] [PubMed] [Google Scholar]
- 33.Taggart AK, Zakian VA. Telomerase: what are the Est proteins doing? Curr Opin Cell Biol. 2003;15:275–280. doi: 10.1016/s0955-0674(03)00040-1. [DOI] [PubMed] [Google Scholar]
- 34.Wahlin J, Cohn M. Saccharomyces cerevisiae RAP1 binds to telomeric sequences with spatial flexibility. Nucleic Acids Res. 2000;28:2292–2301. doi: 10.1093/nar/28.12.2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wahlin J, et al. DNA binding and telomere length regulation of yeast RAP1 homologues. J Mol Biol. 2003;332:821–833. doi: 10.1016/s0022-2836(03)00850-7. [DOI] [PubMed] [Google Scholar]
- 36.Konig P, et al. The crystal structure of the DNA-binding domain of yeast RAP1 in complex with telomeric DNA. Cell. 1996;85:125–136. doi: 10.1016/s0092-8674(00)81088-0. [DOI] [PubMed] [Google Scholar]
- 37.van Hoof A. Conserved functions of yeast genes support the duplication, degeneration and complementation model for gene duplication. Genetics. 2005;171:1455–1461. doi: 10.1534/genetics.105.044057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marcand S, et al. Multiple pathways inhibit NHEJ at telomeres. Genes Dev. 2008;22:1153–1158. doi: 10.1101/gad.455108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deng Z, et al. The origin recognition complex localizes to telomere repeats and prevents telomere-circle formation. Curr Biol. 2007;17:1989–1995. doi: 10.1016/j.cub.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 40.Feeser EA, Wolberger C. Structural and functional studies of the Rap1 C-terminus reveal novel separation-of-function mutants. J Mol Biol. 2008;380:520–531. doi: 10.1016/j.jmb.2008.04.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moretti P, et al. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev. 1994;8:2257–2269. doi: 10.1101/gad.8.19.2257. [DOI] [PubMed] [Google Scholar]
- 42.Moretti P, Shore D. Multiple interactions in Sir protein recruitment by Rap1p at silencers and telomeres in yeast. Mol Cell Biol. 2001;21:8082–8094. doi: 10.1128/MCB.21.23.8082-8094.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao H, et al. RPA-like proteins mediate yeast telomere function. Nat Struct Mol Biol. 2007;14:208–214. doi: 10.1038/nsmb1205. [DOI] [PubMed] [Google Scholar]
- 44.Hughes TR, et al. Identification of the single-strand telomeric DNA binding domain of the Saccharomyces cerevisiae Cdc13 protein. Proc Natl Acad Sci U S A. 2000;97:6457–6462. doi: 10.1073/pnas.97.12.6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bertuch AA, Lundblad V. The maintenance and masking of chromosome termini. Curr Opin Cell Biol. 2006;18:247–253. doi: 10.1016/j.ceb.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 46.Grandin N, et al. Ten1 functions in telomere end protection and length regulation in association with Stn1 and Cdc13. Embo J. 2001;20:1173–1183. doi: 10.1093/emboj/20.5.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Petreaca RC, et al. Chromosome end protection plasticity revealed by Stn1p and Ten1p bypass of Cdc13p. Nat Cell Biol. 2006;8:748–755. doi: 10.1038/ncb1430. [DOI] [PubMed] [Google Scholar]
- 48.Puglisi A, et al. Distinct roles for yeast Stn1 in telomere capping and telomerase inhibition. EMBO J. 2008;27:2328–2339. doi: 10.1038/emboj.2008.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pennock E, et al. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell. 2001;104:387–396. doi: 10.1016/s0092-8674(01)00226-4. [DOI] [PubMed] [Google Scholar]
- 50.Chandra A, et al. Cdc13 both positively and negatively regulates telomere replication. Genes Dev. 2001;15:404–414. doi: 10.1101/gad.861001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bochkarev A, Bochkareva E. From RPA to BRCA2: lessons from single-stranded DNA binding by the OB-fold. Curr Opin Struct Biol. 2004;14:36–42. doi: 10.1016/j.sbi.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 52.Mitton-Fry RM, et al. Structural basis for telomeric single-stranded DNA recognition by yeast Cdc13. J Mol Biol. 2004;338:241–255. doi: 10.1016/j.jmb.2004.01.063. [DOI] [PubMed] [Google Scholar]
- 53.Hsu CL, et al. Interaction of Saccharomyces Cdc13p with Pol1p, Imp4p, Sir4p and Zds2p is involved in telomere replication, telomere maintenance and cell growth control. Nucleic Acids Res. 2004;32:511–521. doi: 10.1093/nar/gkh203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nugent CI, Lundblad V. The telomerase reverse transcriptase: components and regulation. Genes & Dev. 1998;12:1073–1085. doi: 10.1101/gad.12.8.1073. [DOI] [PubMed] [Google Scholar]
- 55.Osterhage JL, et al. Proteasome-dependent degradation of Est1p regulates the cell cycle-restricted assembly of telomerase in Saccharomyces cerevisiae. Nat Struct Mol Biol. 2006;13:720–728. doi: 10.1038/nsmb1125. [DOI] [PubMed] [Google Scholar]
- 56.Hsu M, et al. Mutual dependence of Candida albicans Est1p and Est3p in telomerase assembly and activation. Eukaryot Cell. 2007;6:1330–1338. doi: 10.1128/EC.00069-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu EY, et al. A proposed OB-fold with a protein-interaction surface in Candida albicans telomerase protein Est3. Nat Struct Mol Biol. 2008;15:985–989. doi: 10.1038/nsmb.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee J, et al. The Est3 protein associates with yeast telomerase through an OB-fold domain. Nat Struct Mol Biol. 2008;15:990–997. doi: 10.1038/nsmb.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Collins K. The biogenesis and regulation of telomerase holoenzymes. Nat Rev Mol Cell Biol. 2006;7:484–494. doi: 10.1038/nrm1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lue NF, Li Z. Modeling and structure function analysis of the putative anchor site of yeast telomerase. Nucleic Acids Res. 2007;35:5213–5222. doi: 10.1093/nar/gkm531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jacobs SA, et al. Crystal structure of the essential N-terminal domain of telomerase reverse transcriptase. Nat Struct Mol Biol. 2006;13:218–225. doi: 10.1038/nsmb1054. [DOI] [PubMed] [Google Scholar]
- 62.Dandjinou A, et al. A phylogenetically based secondary structure for the yeast telomerase RNA. Curr Biol. 2004;14:1148–1158. doi: 10.1016/j.cub.2004.05.054. [DOI] [PubMed] [Google Scholar]
- 63.Chen J, Greider C. An emerging concensus for telomerase RNA structure. Proc Natl Acad Sci U S A. 2004;101:14683–14684. doi: 10.1073/pnas.0406204101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zappulla D, Cech T. Yeast telomerase RNA: a flexible scaffold for protein subunits. Proc Natl Acad Sci U S A. 2004;101:10024–10029. doi: 10.1073/pnas.0403641101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gunisova S, et al. Identification and comparative analysis of telomerase RNAs from Candida species reveal conservation of functional elements. RNA. 2009;15:546–559. doi: 10.1261/rna.1194009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lin J, et al. A universal telomerase RNA core structure includes structured moifs required for binding the telomerase reverse transcriptase protein. Proc Natl Acad Sci U S A. 2004;101:14713–14718. doi: 10.1073/pnas.0405879101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brown Y, et al. A critical three-way junction is conserved in budding yeast and vertebrate telomerase RNAs. Nucleic Acids Res. 2007;35:6280–6289. doi: 10.1093/nar/gkm713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shefer K, et al. A triple helix within a pseudoknot is a conserved and essential element of telomerase RNA. Mol Cell Biol. 2007;27:2130–2143. doi: 10.1128/MCB.01826-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen J, et al. A critical stem-loop structure in the CR4-CR5 domain of mammalian telomerase RNA. Nucleic Acids Res. 2002;30:592–597. doi: 10.1093/nar/30.2.592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Seto AG, et al. A bulged stem tethers Est1p to telomerase RNA in budding yeast. Genes Dev. 2002;16:2800–2812. doi: 10.1101/gad.1029302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Snow B, et al. Functional conservation of the telomerase protein est1p in humans. Curr Biol. 2003;13:698–704. doi: 10.1016/s0960-9822(03)00210-0. [DOI] [PubMed] [Google Scholar]
- 72.Redon S, et al. Protein RNA and protein protein interactions mediate association of human EST1A/SMG6 with telomerase. Nucleic Acids Res. 2007;35:7011–7022. doi: 10.1093/nar/gkm724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Reichenbach P, et al. A human homolog of yeast est1 associates with telomerase and uncaps chromosome ends when overexpressed. Curr Biol. 2003;13:568–574. doi: 10.1016/s0960-9822(03)00173-8. [DOI] [PubMed] [Google Scholar]
- 74.Singh S, et al. Analysis of telomerase in Candida albicans: potential role in telomere end protection. Eukaryot Cell. 2002;1:967–977. doi: 10.1128/EC.1.6.967-977.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fukuhara N, et al. SMG7 is a 14-3-3-like adaptor in the nonsense-mediated mRNA decay pathway. Mol Cell. 2005;17:537–547. doi: 10.1016/j.molcel.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 76.D'Andrea L, Regan L. TPR proteins: the versatile helix. Trends Biochem Sci. 2003;28:655–662. doi: 10.1016/j.tibs.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 77.Evans S, Lundblad V. The Est1 subunit of Saccharomyces cerevisiae telomerase makes multiple contributions to telomere length maintenance. Genetics. 2002;162:1101–1115. doi: 10.1093/genetics/162.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yu EY, et al. Regulation of telomere structure and functions by subunits of the INO80 chromatin remodeling complex. Mol Cell Biol. 2007;27:5639–5649. doi: 10.1128/MCB.00418-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Qi H, Zakian VA. The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase alpha and the telomerase-associated est1 protein. Genes Dev. 2000;14:1777–1788. [PMC free article] [PubMed] [Google Scholar]
- 80.Brun C, et al. Proteins that bind to double-stranded regions of telomeric DNA. Trends Cell Biol. 1997;7:317–324. doi: 10.1016/S0962-8924(97)01092-1. [DOI] [PubMed] [Google Scholar]
- 81.Kanoh J, Ishikawa F. spRap1 and spRif1, recruited to telomeres by Taz1, are essential for telomere function in fission yeast. Curr Biol. 2001;11:1624–1630. doi: 10.1016/s0960-9822(01)00503-6. [DOI] [PubMed] [Google Scholar]
- 82.Kuriyan J, Eisenberg D. The origin of protein interactions and allostery in colocalization. Nature. 2007;450:983–990. doi: 10.1038/nature06524. [DOI] [PubMed] [Google Scholar]
- 83.Song X, et al. STN1 protects chromosome ends in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2008;105:19815–19820. doi: 10.1073/pnas.0807867105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Miyoshi T, et al. Fission yeast Pot1-Tpp1 protects telomeres and regulates telomere length. Science. 2008;320:1341–1344. doi: 10.1126/science.1154819. [DOI] [PubMed] [Google Scholar]
- 85.McEachern MJ, Haber JE. Break-induced replication and recombinational telomere elongation in yeast. Annu Rev Biochem. 2006;75:111–135. doi: 10.1146/annurev.biochem.74.082803.133234. [DOI] [PubMed] [Google Scholar]
- 86.McEachern M, Haber J. Telomerase-independent telomere maintenance in yeast. In: de Lange T, et al., editors. Telomeres and Telomerase. 2nd edn. Cold Spring Harbor Laboratory Press; 2006. pp. 199–224. [Google Scholar]
- 87.Zubko MK, Lydall D. Linear chromosome maintenance in the absence of essential telomere-capping proteins. Nat Cell Biol. 2006;8:734–740. doi: 10.1038/ncb1428. [DOI] [PubMed] [Google Scholar]