Abstract
Hepatitis C virus (HCV) modulates cellular lipid metabolism to enhance its replication. HCV circulates in the blood in association with lipoproteins. HCV infection is associated with enhanced lipogenesis, reduced secretion and β-oxidation of lipids. HCV-induced imbalance in lipid homeostasis leads to steatosis. Many lipids are crucial for viral life cycle, and inhibitors of cholesterol/fatty acid biosynthetic pathways inhibit viral replication, maturation and secretion. HCV negatively modulates the synthesis and secretion of very low-density lipoproteins (VLDL). The components involved in VLDL assembly are also required for HCV morphogenesis/secretion, suggesting that HCV coopts the VLDL secretory pathway for its own secretion. This review highlights HCV-altered lipid metabolic events that aid in the viral life cycle and ultimately promote liver disease pathogenesis.
Human hepatitis C virus (HCV) infects about 2–3% of the world’s population. HCV infection leads to chronic hepatitis in up to 60–80% of infected individuals [1]. HCV infection is associated with liver steatosis, fibrosis, cirrhosis, and hepatocellular carcinoma (HCC) [2]. Like other positive-strand RNA viruses, HCV requires alteration of intracellular membrane architecture to facilitate its genomic replication [3]. The formation of replication competent ribonucleoprotein (RNP) complexes, with subsequent assembly and release of infectious virions involves membrane reorganization, intracellular trafficking and recruitment of crucial viral and host cofactors. Consistent with this, RNA interference and proteomic analyses identified host proteins involved in membrane biogenesis, vesicular organization and intracellular trafficking to be crucial for HCV replication and morphogenesis [4–7]. Among the host cofactors most notably, a lipid kinase, phosphotidylinositol 4-kinase (PI4K), is shown to be required for efficient HCV replication [5–7]. PI4K-specific siRNAs reduced the accumulation of altered membranous structures conducive for HCV RNA replication in infected cells [5]. Genomic analysis of HCV genotype 1a infected chimpanzees showed a positive correlation between upregulation of genes involved in lipid metabolism and onset of viremia [8]; furthermore, 30% of total proteins associated with HCV RNP complexes are functionally involved in lipid metabolism [9]. From these observations, it is evident that upregulation of host lipid metabolism to enhance the availability of important lipid constituents and membrane fluidity is crucial for establishing efficient HCV RNA replication machinery. Saturated and mono-unsaturated fatty acids required to maintain membrane structure and fluidity stimulate HCV replication, whereas polyunstaturated fatty acids (PUFAs), that perturb membrane fluidity inhibit HCV replication [10, 11].
Inhibitors of cholesterol and fatty acid biosynthetic pathways have been effectively used to inhibit HCV replication [11–14]. Inhibition of VLDL assembly and secretion also affected virion morphogenesis and secretion, leading to the notion that HCV may co-opt/hijack the VLDL secretion pathway for virion maturation/secretion [9, 15, 16]. The reliance of HCV for its replication, morphogenesis and secretion on host lipid metabolic pathways necessitates their modulation by HCV to create a lipid-rich intracellular environment favorable for its multiplication. HCV influences host lipid metabolism at three levels: enhanced lipogenesis, impaired degradation and impaired export [2]. These detrimental alterations in lipid metabolism incurred during HCV infection manifest as the pathological basis for some of the HCV-associated maladies, most notably steatosis and metabolic syndromes such as insulin resistance, obesity, and hepatocellular carcinoma [2]. Steatosis, or accumulation of hepatocellular lipid droplets, and altered serum lipid profiles are common consequences of HCV infection induced altered lipid homeostasis [17, 18]. The current therapy against HCV, a combination of pegylated-interferon α and ribavirin, is only partially effective, being both toxic and genotype-specific. Anti-HCV therapies targeting HCV proteins have been developed; however, rapidly mutating HCV genome results in evolution of drug-resistant viral mutants. Due to a considerable cross talk between HCV and host lipid metabolism, targeting components of host lipid metabolic pathways holds promise as an effective anti-HCV therapeutic strategy. This review highlights the role of HCV in regulating host lipid metabolism, with emphasis on lipoprotein assembly and how these alterations affect viral infectious process and liver disease pathogenesis.
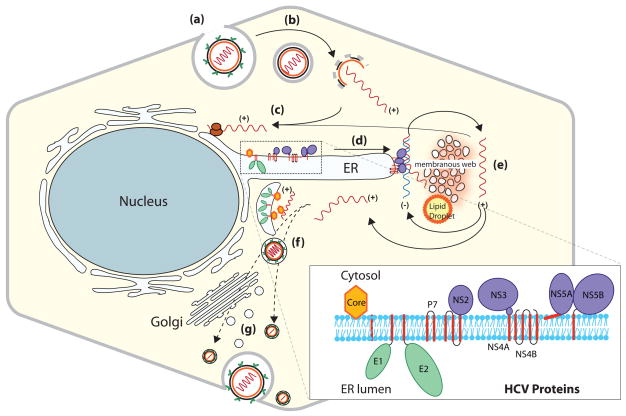
The HCV genome is a 9.6-kb of single-stranded positive sense RNA that unlike eukaryotic mRNA lacks the 5′ cap and 3′ polyA tail. The 5′ UTR contains an internal ribosome entry site (IRES), which directs cap-independent translation of a polyprotein precursor of ~3000 amino acids [19]. The polyprotein is processed by host signal peptidases and viral proteases into mature structural (core, E1, and E2) and nonstructural (NS) proteins (p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B) (Figure 1, inset) [20, 21]. All the NS proteins are associated or tethered to the endoplasmic reticulum (ER) membrane following their synthesis [3] (Figure 1, inset). HCV RNA genome replicates within the RNP complexes assembled on the ER derived membranous structures [3, 21] (Figure 1). HCV exhibits molecular heterogeneity and is grouped into six genotypes, which display different geographical distribution and response to treatment [22]. Research on the molecular mechanisms of HCV replication and pathogenesis were hampered by the lack of an efficient cell culture system or a suitable animal model. However in 2005, an in vitro cell culture based HCV (JFH1, genotype 2a) infection system was reported [23–25]. This strain of HCV productively infects human hepatoma cell line Huh-7 and more robustly infects the HuH-7 derived sub-clones, Huh-7.5 and Huh-7.5.1. This in vitro model of HCV infection is currently used to investigate various aspects of HCV virology and subsequent intracellular events relevant to disease pathogenesis.
Figure 1. HCV life Cycle.
The viral life cycle is illustrated in steps i–vii. (i) HCV enters hepatocytes via putative receptors. (ii) pH-dependent fusion of the viral envelope and uncoating of genomic RNA occurs in endosomes, followed by (iii) IRES-mediated translation on the rough endoplasmic reticulum (ER). HCV proteins and their association in the ER are shown in the inset; next, (iv) assembly of ribonucleoprotein complexes (RNP) occurs, and (v) these RNP complexes engage in RNA synthesis to produce (+) polarity viral RNAs. RNA synthesis is believed to occur in the HCV-induced membranous structures termed ‘membranous web’. (vi) + polarity RNAs are encapsidated, and (vii) HCV maturation and release ensues. HCV virions traffic through the Golgi or bypass the Golgi network. The mechanistic details of steps ‘vi and vii’ are not fully characterized.
Hepatocytes are the major sites of HCV infection. HCV entry into hepatocytes is mediated by direct interaction with CD81, scavenger receptor class B member 1 (SR-B1) and other accessory factors, which include low-density lipoprotein receptor (LDLr) and glucosaminoyglycans [26, 27, 28]. After binding to the cell surface via its putative receptors, HCV is transferred to tight junctions, where the virus interacts with claudin-1 [29] and occludin [30, 31] prior to gaining entry into the hepatocytes [26–28]. The HCV viral particles found in patients’ sera as lipoprotein complexes infect hepatocytes by the lipoprotein uptake route independent of viral envelope proteins. They initially bind to cell surface glucosaminoglycans followed by subsequent interaction with LDLr and/or SRB1 [32, 33]. The SRB1 and LDLr-mediated uptake of HCV pseudoparticles or serum derived HCV virions is competitively inhibited by serum LDL and VLDL [32, 33]. Hence, lipoprotein levels in patients’ sera determine the efficacy of treatment. Indeed, it has been reported that patients with high serum LDL and cholesterol levels respond readily to anti-HCV treatment compared to those with low levels [34].
HCV-associated steatosis and lipid metabolism
Liver steatosis (fatty liver) or accumulation of hepatocellular lipid droplets, storage sites of cytosolic neutral lipids, is the prominent histological phenotype of HCV infection occurring in 73% of patients infected with genotype 3 and in 50% of patients infected with other genotypes [2, 17]. Steatosis appears to be a direct consequence of viral protein expression in genotype 3 infection, whereas in other genotype infections, it is associated with metabolic syndromes like insulin resistance and obesity [2]. However, HCV-associated regulation of host lipid metabolism does not entirely appear to be genotype-specific.
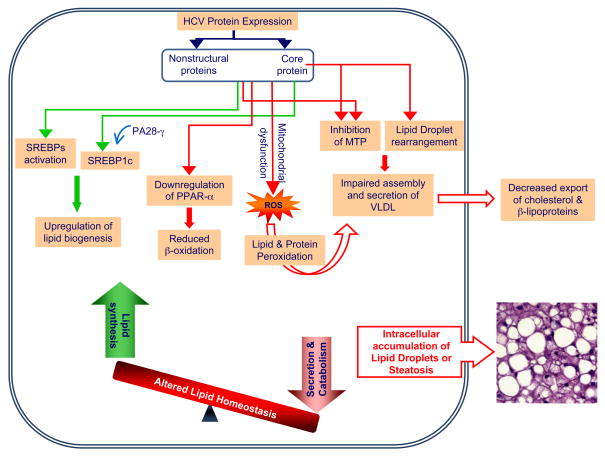
HCV modulates lipid homeostasis by increasing lipogenesis via SREBP activation [35] and reducing oxidation and lipid export [2] (Figure 2). SREBPs are ER membrane bound transcription factors that activate genes encoding enzymes of cholesterol and fatty acid biosynthesis [36]. There are three isoforms designated SREBP-1a, SREBP-1c and SREBP-2 [37]. SREBP-1a activates all SREBP target genes, whereas SREBP-1c and SREBP-2 activate genes involved in fatty acid and cholesterol metabolism, respectively [37]. Inhibiting SREBP activation by treatment with 25-hydroxycholesterol blocks HCV replication [8]. Similarly, fatty acid synthase (FAS), an enzyme primarily involved in de novo synthesis of fatty acids, is upregulated during HCV infection, and inhibition of FAS activity also inhibits HCV replication and release [8, 38]. These observations highlight the importance of upregulating de novo synthesis of fatty acids and cholesterol to enhance the availability of important lipid constituents for establishment of efficient HCV replication.
Figure 2. HCV induced alterations in lipid metabolism and steatosis.
HCV alters cellular lipid metabolism to create a lipid rich intracellular environment to facilitate its own multiplication. HCV activates sterol regulatory element binding proteins (SREBPs), the master regulators of cholesterol/fatty acid biosynthesis. HCV core protein in the presence of proteasome activator PA28γ activates SREBP1c. HCV induces the downregulation of peroxisome proliferator-activated receptor α (PPAR-α), a transcription factor required for genes involved in β-oxidation and transport of fatty acids. HCV downregulates very-low-density lipoprotein (VLDL) particle secretion by inhibiting the activity of microsomal triglyceride transfer protein (MTP) activity. HCV core protein-induced rearrangement and aggregation of lipid droplets also interferes with VLDL assembly. Mitochondrial dysfunction and generation of reactive oxygen species (ROS) during HCV infection perturbs important cellular functions like VLDL assembly by promoting peroxidation of important enzymes and lipids. These events disturb lipid homeostasis leading to the intracellular accumulation of lipid droplets, which manifests as steatosis, the prominent pathological phenotype associated with HCV infection.
HCV promotes ER stress induced unfolded protein response (UPR) in infected hepatocytes [39, 40]. Recent studies reveal a critical role of the UPR in hepatic lipid metabolism [41–43]. X-box-binding protein 1 (XBP1), a key regulator of the ER stress induced unfolded protein response (UPR), was recently characterized as an important factor in hepatic lipogenesis independent of its putative role in UPR signaling [43]. The ER stress-induced C/EBP-homologous protein (CHOP) negatively modulates CCAAT/enhancer binding protein α (C/EBPα), a transcription factor crucial for lipid homeostasis and hepatic steatosis [41]. Interestingly, CHOP expression is elevated in HCV infection and sensitizes cells to oxidant injury [44]. Similarly, ER stress-induced eukaryotic initiation factor 2α (eIF2α) phosphorylation also triggers hepatic lipogenesis and promotes steatosis [42]. These findings suggest differential regulation of genes involved in lipid homeostasis by mediators of ER stress response; however, further studies are warranted to understand how HCV governs the ER stress response to promote its lipogenic and steatogenic potential to facilitate its life cycle.
HCV infection is also associated with reduced serum cholesterol and β-lipoprotein levels [2]. Upon successful antiviral treatment, cholesterol and β-lipoprotein levels are restored to normalcy, suggesting that HCV gene expression is responsible for their altered levels [2]. HCV can also lead to hepatic steatosis by down-regulating fatty acid breakdown and cholesterol export. Oxidative stress induced during HCV infection via mitochondrial dysfunction generates ROS, which can alter functions of proteins and lipids via peroxidation and hence negatively influence important cellular processes like fatty acid oxidation and export [39]. The significance of impaired lipid degradation in HCV-associated steatosis is substantiated by reduced mRNA levels of peroxisome proliferator-activated receptor α (PPARα) in HCV infected patients with steatosis when compared to those without steatosis [45, 46]. PPARα is highly expressed in hepatocytes and crucial for lipid homeostasis via its ability to upregulate genes involved in β-oxidation and transport of fatty acids [47]. Interestingly, a recent study using PPARα homozygous, heterozygous and null mice with liver specific expression of HCV core protein indicates that persistent PPARα activation is essential for development of severe hepatic steatosis and its progression into hepatocellular carcinoma. This study argues for a more complex role of PPARα in HCV-associated pathogenesis [48]. Among the viral proteins, HCV core protein has been shown individually to affect a wide variety of cellular functions including lipogenesis, steatosis and β-oxidation (Box 1).
Association of HCV virion with lipoproteins
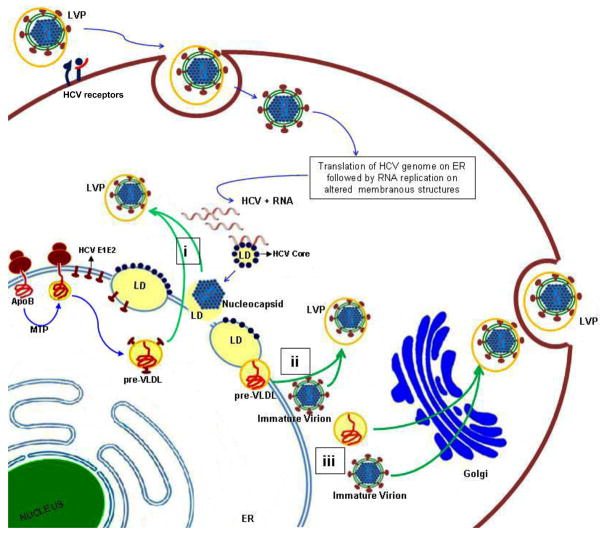
HCV particles circulate in the blood of hepatitis C patients as a heterogenous population of varying buoyant densities ranging between 1.03 to 1.2 g/ml [49–51]. The low-density particles are efficient in transmitting infection to chimpanzees and cultured hepatocytes [52, 53]. These infectious particles are rich in triglycerides, apolipoprotein B-100 (apoB), and apolipoprotein E (apoE), and physiochemically resemble VLDL particles, hence termed lipo-viral particles (LVPs) [50, 53]. Antibodies against human apoB and apoE efficiently precipitated the infectious low-density HCV particles (LVPs) from patients’ sera suggesting a close interaction between the HCV virion and VLDL [50]. Similarly, infectious virions derived from in vitro cultures also display physiochemical properties resembling the LVPs found in patients’ sera [16, 54]. In the in vitro model of HCV infection, immature intracellular virions are less infectious and denser than the highly infectious secreted virions [54]. These studies indicate that the infectious virions assembled in the cell presumably acquire the low-density configuration and their enhanced infectivity stature by associating with host lipoproteins prior to their secretion into the extracellular milieu (Figure 3). Several criteria, including detergent resistance of apoB and HCV virion interaction [50], necessity of LVP delipidation for recognition by HCV anti-core antibodies [53], and localization of HCV-nucleocapsid structures in the LVPs [51, 53], collectively suggest that viral particles and VLDL are integral components of LVPs (Figure 3).
Figure 3. HCV co-opts the VLDL secretion pathway.
HCV entry is mediated by putative receptors, but lipo-viral particle (LVP) entry into hepatocytes may be also mediated by low-density lipoprotein receptors (LDLr) and glycosoaminoglycans (GAGs). Virus binding and internalization is followed by translation of the HCV RNA genome. Details of HCV morphogenesis are not fully characterized. The HCV core protein binds to lipid droplets (LDs), and this interaction is essential for HCV genome replication and virion assembly (depicted in figure as blue dots bound to LDs). It is also presumed that HCV co-opts the very-low-density lipoprotein (VLDL) secretory pathway to facilitate its exit. The first step of VLDL assembly involves the co-translational lipidation of apoB by microsomal triglyceride transfer protein (MTP) generating a pre-VLDL particle. The pre-VLDL then matures into VLDL by fusing with large triglyceride rich droplets, presumed to occur in post-ER compartments or the Golgi. The newly assembled immature HCV virions probably fuse with the preVLDL particle prior to or during the second maturation step of VLDL generating LVPs that are secreted into the extra-cellular milieu via the VLDL route. We propose three different possibilities of how LVPs are assembled. (i) Pre-VLDL particles during their maturation process may incorporate the HCV envelope protein present on the ER membrane. The lipid droplet tethered to HCV nucleocapsid then fuses with these pre-VLDL particles carrying HCV envelope proteins, forming LVPs. (ii) Pre-VLDL and immature HCV virions fuse with each other in post-ER compartments. Lastly, it is possible that (iii) the nascent VLDL particles and immature HCV virions fuse with each other during their transit through the Golgi network.
Virion assembly and secretion: Role of VLDL
Little is known about HCV morphogenesis. However, circumstantial evidence suggests that HCV virion and VLDL particles share a common route of secretion, and reagents that abrogate VLDL secretion also block HCV secretion. VLDL is assembled and secreted by hepatocytes. VLDL contains a central core rich in neutral lipids, triglycerides and cholesterol, surrounded by a phospholipid monolayer rich in apolipoproteins [55]. VLDL assembly occurs in two steps, the first step involves the co-translational lipidation of apoB-100 by MTP in the lumen of ER [56] (Figure 3). Unlipidated apoB-100 is targeted for degradation by the ubiquitin proteasome machinery [55, 56]. This partially lipidated pre-VLDL particle acquires a majority of triglycerides in the second maturation step, involving the subsequent fusion of pre-VLDL particle with a large triacylglycerol rich droplet [55]. Although the exact mechanism and intracellular location (ER, post-ER or Golgi) of this second lipidation event are still unclear, it appears to be dependent on the GTP binding protein ARF-1 (ADP ribosylation factor-1) and phospholipase D activities [57]. The ER to Golgi transport of VLDL prior to its eventual secretion occurs in specialized ER-derived, COP-II dependent transport vesicles known as the VLDL transport vesicle (VTVs) [58].
RNA interference-mediated suppression of apoB-100 [9, 15] or apoE [16] inhibits HCV virion secretion. Similar inhibition of virion secretion was observed by inhibiting MTP activity [9, 15, 59]. Targeted inhibition of acyl-CoA synthetase-3 (ACSL3), an enzyme crucial for VLDL assembly and secretion also blocked HCV virion secretion [60]. The reagents that impair VLDL assembly also abrogated HCV secretion, although HCV RNA replication was unaffected [9, 15, 59, 60], suggesting that functional components of VLDL assembly and secretion are required by HCV for its secretion. Apart from inhibiting virion secretion, disruption in apoB biosynthesis also resulted in reduced accumulation of intracellular infectious virions, suggesting that intracellular apoB levels function as a major determinant of HCV virion morphogenesis [15]. Proteomic analysis of membrane vesicles containing HCV RNP complexes obtained from HCV-expressing cells revealed an enrichment of VLDL assembly components such as MTP, apoB, and apoE in these vesicles, although HCV RNA replication is independent of VLDL assembly [9].
It is apparent from these observations that apart from aiding virion secretion, VLDL assembly is also relevant to the HCV maturation process. Colocalization of HCV replication and VLDL assembly components indicate close proximity between the intracellular regions involved in viral genome replication and virion assembly [9, 61]. The overlap between the VLDL and HCV-virion assembly and secretion routes is further substantiated by non-proteasomal degradation of intracellular high-density HCV particles upon failure to associate with VLDL [15]. These observations indicate that the newly assembled nascent virions probably undergo an additional maturation step where they acquire the low-density configuration by associating with VLDL and concomitantly enhance their chances of egress from cell by piggy-backing on the VLDL secretion system [15]. Recently it was shown that HCV envelope glycoproteins expressed in HepG2 and apoB secreting Caco-2 cells are efficiently secreted into the extracellular milieu along with VLDL [62]. Thus, it appears that envelope proteins have an intrinsic capacity to co-opt the VLDL secretion pathway even in the absence of other HCV proteins. Although not completely characterized, this inherent feature of envelope proteins to hitch a ride with VLDL strengthens the notion that HCV virion assembly and secretion processes utilize the VLDL pathway.
The exact mechanism(s) involved in the association of HCV particles with VLDL is not clear and needs to be fully characterized. These two different pathways could merge either in the ER, in the post-ER compartments, or during their transit via the Golgi (Figure 3). Oxysterol-binding protein (OSBP) that is functionally involved in ER-to-Golgi ceramide transport has been recently implicated in HCV maturation/secretion when recruited to the Golgi compartment [63]. The endo H-sensitivity of HCV replication complex associated apoB [9], and high density of replication complex associated membranes compared to density of Golgi-clusters suggest that VLDL components associated with HCV replication complex have not yet traversed the Golgi secretion pathway [51]. Both isoforms of apolipoprotein B, apoB-100 and apoB-48, have been shown to be associated with LVPs [64]. ApoB-48 is the truncated form of ApoB100 and is a predominant protein of lipoprotein particles called chylomicrons, which are assembled and secreted by intestinal enterocytes [65]. These reports suggest that enterocytes can secrete LVPs, similar to hepatocytes. The significance of these observations, however, needs to be further investigated. A recent study demonstrates that the average diameter of LVPs in a patient’s serum is about 55 nm [51], which is much smaller than a chylomicron and contradicts the notion supporting the intestinal origin of LVPs.
The reliance of HCV assembly and secretion on the VLDL pathway requires changes in cellular lipid metabolism to ensure a lipid-rich microenvironment with a reduced rate of VLDL secretion to enhance the feasibility of co-assembling with VLDL. Reduced MTP activity and an apparent hypobetalipoproteinemia is observed in chronic hepatitis C patients, and negatively correlates with hepatic steatosis and viral load especially in genotype 3 infections [66, 67]. Cells harboring sub-genomic replicon RNAs that encode only the HCV NS proteins also display reduced MTP activity and VLDL secretion [68]. It was recently revealed that HCV core protein induces redistribution of lipid droplets around the microtubule-organizing center, preceded by accumulation of core protein on the LD surface [69] (Figure 3). It has been demonstrated that association between HCV core protein and LDs is necessary for the release of infectious virions 61. HCV core protein interacts with the lipid droplets through its hydrophobic domain D2, specifically via a valine at position 147, which is a major determinant of enhanced assembly and release of virions [70, 71]. The core protein-mediated redistribution of LDs around the perinuclear region probably enhances the proximity between sites of HCV RNA replication and virion assembly [69]. Since the majority of triglycerides incorporated into VLDL particles are derived from lipid droplets, it is possible that this redistribution of LDs in propinquity to the sites of HCV replication enhance the chances of fusion or incorporation of nascent HCV virion or nucleocapsid into the VLDL. In addition, such an aggregation and redistribution might negatively influence VLDL assembly by perturbing the availability of LDs, a primary source of lipids required for VLDL synthesis [69]. Interestingly, a recent report showed that lipoprotein particles isolated from HCV patient sera differentially modulated lipid synthesis in human monocyte-derived macrophages in comparison to lipoproteins obtained from normal subjects [72]. In light of these observations, it is tempting to speculate that HCV infection influences the biochemical composition of lipoproteins and eventually affects lipid metabolism [72]. Serum lipid profiles of chronic HCV patients exhibit low levels of total cholesterol (TC), high-density lipoprotein cholesterol (HDL-C) and low-density lipoprotein cholesterol (LDL-C) compared to uninfected controls [18]. The levels were significantly lower in HCV patients displaying steatosis compared to those without steatosis [18]. Furthermore, patients infected with HCV genotype 3a show higher incidence of steatosis and more pronounced reduction in serum lipids (hypocholesteremia) [18]. These HCV related alterations in lipid profiles are important for the HCV life cycle and correlate positively with response to anti-HCV treatments, specifically in genotype 3a infection [2].
Future Directions
Host lipid metabolic pathways and the VLDL pathway facilitate replication, assembly, secretion and HCV entry. The association of HCV with VLDL may help the virus evade host immune defense by masking its putative antigenic moieties from immune recognition. Growing insight into the details of HCV mediated modulations of host lipid metabolism may eventually unravel uncharacterized facets of lipid/fatty acid biosynthetic pathways, including VLDL assembly. Elucidating mechanistic details underlying the similitude between viral and host lipid metabolic pathways will help identify potential host cell factors that are required for HCV and the infectious processes. In addition, this would also enable the design of potential multifaceted therapeutic modules to curb HCV infection, reduce serum levels of atherosclerotic lipoproteins and reduce susceptibility to metabolic syndrome. Thus, therapeutic interventions targeting host factors involved in lipid metabolism to control HCV infection holds great promise for the future.
Box 1: HCV Core induced alterations in lipid metabolism
The HCV core protein is a 21kDa basic protein, whose primary function is packaging viral RNA to produce nucleocapsids. Of the HCV proteins, the core protein has been implicated in a wide variety of cellular functions. The most pronounced effect of expressing HCV core protein in transgenic mice is steatosis [73]. Induction of steatosis was more evident by accumulation of large lipid droplets upon expression of HCV genotype 3 core protein, which differs from the core protein of other genotypes by having a phenylalanine at position 164 instead of a tyrosine [74, 75]. This phenylalanine residue lies in the α-helix that sits between the ER phospholipid monolayers where lipid droplet biogenesis is presumed to occur. Since phenylalanine is more hydrophobic than tyrosine, perhaps it can enhance the affinity of core protein to lipids resulting in increased generation of lipid droplets [70]. The core protein enhances lipogenesis by triggering activation of transcription factor RxRα (retinoid X receptor agr;)and fatty acid synthase (FAS), both crucial for the de novo synthesis of fatty acids. Indeed, expression of the highly steatogenic HCV genotype 3a core protein maximally triggered FAS promoter activity compared to other genotypes [76]. HCV core protein from several genotypes induced proteolytic cleavage of SREBPs [35]. Core protein also activated SREBP1c via LXR agr;/RXR agr; pathway, with assistance from the nuclear proteasome activator PA28-γ. In PA28-γ(−/−) mice, core expression did not lead to hepatic steatosis, even though it did localize to the hepatocyte nucleus. Core expression in PA28-γ(+/+) mice, however, led to hepatic steatosis [77]. HCV core expressing HepG2 cells or transgenic mice displayed reduced levels of PPAR agr; mRNA [45, 78], suggesting that HCV core also alters lipid metabolism by down-regulating PPARagr;, essential for genes involved in β-oxidation and secretion of fatty acids [47]. HCV core expressing transgenic mice displayed reduced MTP activity, resulting in reduced assembly and secretion of VLDL particles [73], and this inhibition was reversed by hepatic overexpression of ApoAII [73]. In contrast, high levels of ApoAII in chronic HCV patients were not protective but promoted the development of liver steatosis [79].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shepard CW, et al. Global epidemiology of hepatitis C virus infection. Lancet Infect Dis. 2005;5:558–567. doi: 10.1016/S1473-3099(05)70216-4. [DOI] [PubMed] [Google Scholar]
- 2.Negro F, Sanyal AJ. Hepatitis C virus, steatosis and lipid abnormalities: clinical and pathogenic data. Liver Int. 2009;29(Suppl 2):26–37. doi: 10.1111/j.1478-3231.2008.01950.x. [DOI] [PubMed] [Google Scholar]
- 3.Dubuisson J, et al. Interaction of hepatitis C virus proteins with host cell membranes and lipids. Trends Cell Biol. 2002;12:517–523. doi: 10.1016/s0962-8924(02)02383-8. [DOI] [PubMed] [Google Scholar]
- 4.Mannova P, et al. Modification of host lipid raft proteome upon hepatitis C virus replication. Mol Cell Proteomics. 2006;5:2319–2325. doi: 10.1074/mcp.M600121-MCP200. [DOI] [PubMed] [Google Scholar]
- 5.Berger KL, et al. Roles for endocytic trafficking and phosphatidylinositol 4-kinase III alpha in hepatitis C virus replication. Proc Natl Acad Sci U S A. 2009;106:7577–7582. doi: 10.1073/pnas.0902693106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tai AW, et al. A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host Microbe. 2009;5:298–307. doi: 10.1016/j.chom.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vaillancourt FH, et al. Identification of a lipid kinase as a host factor involved in hepatitis C virus RNA replication. Virology. 2009;387:5–10. doi: 10.1016/j.virol.2009.02.039. [DOI] [PubMed] [Google Scholar]
- 8.Su AI, et al. Genomic analysis of the host response to hepatitis C virus infection. Proc Natl Acad Sci U S A. 2002;99:15669–15674. doi: 10.1073/pnas.202608199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang H, et al. Hepatitis C virus production by human hepatocytes dependent on assembly and secretion of very low-density lipoproteins. Proc Natl Acad Sci U S A. 2007;104:5848–5853. doi: 10.1073/pnas.0700760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leu GZ, et al. Anti-HCV activities of selective polyunsaturated fatty acids. Biochem Biophys Res Commun. 2004;318:275–280. doi: 10.1016/j.bbrc.2004.04.019. [DOI] [PubMed] [Google Scholar]
- 11.Kapadia SB, Chisari FV. Hepatitis C virus RNA replication is regulated by host geranylgeranylation and fatty acids. Proc Natl Acad Sci U S A. 2005;102:2561–2566. doi: 10.1073/pnas.0409834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ikeda M, et al. Different anti-HCV profiles of statins and their potential for combination therapy with interferon. Hepatology. 2006;44:117–125. doi: 10.1002/hep.21232. [DOI] [PubMed] [Google Scholar]
- 13.Aizaki H, et al. Critical role of virion-associated cholesterol and sphingolipid in hepatitis C virus infection. J Virol. 2008;82:5715–5724. doi: 10.1128/JVI.02530-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amemiya F, et al. Targeting lipid metabolism in the treatment of hepatitis C virus infection. J Infect Dis. 2008;197:361–370. doi: 10.1086/525287. [DOI] [PubMed] [Google Scholar]
- 15.Gastaminza P, et al. Cellular determinants of hepatitis C virus assembly, maturation, degradation, and secretion. J Virol. 2008;82:2120–2129. doi: 10.1128/JVI.02053-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang KS, et al. Human apolipoprotein e is required for infectivity and production of hepatitis C virus in cell culture. J Virol. 2007;81:13783–13793. doi: 10.1128/JVI.01091-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Negro F. Hepatitis C virus and liver steatosis: when fat is not beautiful. J Hepatol. 2004;40:533–535. doi: 10.1016/j.jhep.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 18.Siagris D, et al. Serum lipid pattern in chronic hepatitis C: histological and virological correlations. J Viral Hepat. 2006;13:56–61. doi: 10.1111/j.1365-2893.2005.00655.x. [DOI] [PubMed] [Google Scholar]
- 19.Wang C, Siddiqui A. Structure and function of the hepatitis C virus internal ribosome entry site. Curr Top Microbiol Immunol. 1995;203:99–115. doi: 10.1007/978-3-642-79663-0_5. [DOI] [PubMed] [Google Scholar]
- 20.Appel N, et al. From structure to function: new insights into hepatitis C virus RNA replication. J Biol Chem. 2006;281:9833–9836. doi: 10.1074/jbc.R500026200. [DOI] [PubMed] [Google Scholar]
- 21.Tellinghuisen TL, et al. Studying hepatitis C virus: making the best of a bad virus. J Virol. 2007;81:8853–8867. doi: 10.1128/JVI.00753-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simmonds P. Genetic diversity and evolution of hepatitis C virus--15 years on. J Gen Virol. 2004;85:3173–3188. doi: 10.1099/vir.0.80401-0. [DOI] [PubMed] [Google Scholar]
- 23.Wakita T, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhong J, et al. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci U S A. 2005;102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindenbach BD, et al. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 26.von Hahn T, Rice CM. Hepatitis C virus entry. J Biol Chem. 2008;283:3689–3693. doi: 10.1074/jbc.R700024200. [DOI] [PubMed] [Google Scholar]
- 27.Dubuisson J, et al. Early steps of the hepatitis C virus life cycle. Cell Microbiol. 2008;10:821–827. doi: 10.1111/j.1462-5822.2007.01107.x. [DOI] [PubMed] [Google Scholar]
- 28.Burlone ME, Budkowska A. Hepatitis C virus cell entry: role of lipoproteins and cellular receptors. J Gen Virol. 2009;90:1055–1070. doi: 10.1099/vir.0.008300-0. [DOI] [PubMed] [Google Scholar]
- 29.Evans MJ, et al. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature. 2007;446:801–805. doi: 10.1038/nature05654. [DOI] [PubMed] [Google Scholar]
- 30.Ploss A, et al. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature. 2009;457:882–886. doi: 10.1038/nature07684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu S, et al. Tight junction proteins claudin-1 and occludin control hepatitis C virus entry and are downregulated during infection to prevent superinfection. J Virol. 2009;83:2011–2014. doi: 10.1128/JVI.01888-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maillard P, et al. The interaction of natural hepatitis C virus with human scavenger receptor SR-BI/Cla1 is mediated by ApoB-containing lipoproteins. FASEB J. 2006;20:735–737. doi: 10.1096/fj.05-4728fje. [DOI] [PubMed] [Google Scholar]
- 33.Molina S, et al. The low-density lipoprotein receptor plays a role in the infection of primary human hepatocytes by hepatitis C virus. J Hepatol. 2007;46:411–419. doi: 10.1016/j.jhep.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 34.Gopal K, et al. Correlation between beta-lipoprotein levels and outcome of hepatitis C treatment. Hepatology. 2006;44:335–340. doi: 10.1002/hep.21261. [DOI] [PubMed] [Google Scholar]
- 35.Waris G, et al. Hepatitis C virus induces proteolytic cleavage of sterol regulatory element binding proteins and stimulates their phosphorylation via oxidative stress. J Virol. 2007;81:8122–8130. doi: 10.1128/JVI.00125-07. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Horton JD, et al. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Horton JD, et al. SREBPs: transcriptional mediators of lipid homeostasis. Cold Spring Harb Symp Quant Biol. 2002;67:491–498. doi: 10.1101/sqb.2002.67.491. [DOI] [PubMed] [Google Scholar]
- 38.Yang W, et al. Fatty acid synthase is up-regulated during hepatitis C virus infection and regulates hepatitis C virus entry and production. Hepatology. 2008;48:1396–1403. doi: 10.1002/hep.22508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tardif KD, et al. Hepatitis C virus, ER stress, and oxidative stress. Trends Microbiol. 2005;13:159–163. doi: 10.1016/j.tim.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 40.Sir D, et al. Induction of incomplete autophagic response by hepatitis C virus via the unfolded protein response. Hepatology. 2008;48:1054–1061. doi: 10.1002/hep.22464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rutkowski DT, et al. UPR pathways combine to prevent hepatic steatosis caused by ER stress-mediated suppression of transcriptional master regulators. Dev Cell. 2008;15:829–840. doi: 10.1016/j.devcel.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oyadomari S, et al. Dephosphorylation of translation initiation factor 2alpha enhances glucose tolerance and attenuates hepatosteatosis in mice. Cell Metab. 2008;7:520–532. doi: 10.1016/j.cmet.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee AH, et al. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science. 2008;320:1492–1496. doi: 10.1126/science.1158042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ciccaglione AR, et al. Activation of the ER stress gene gadd153 by hepatitis C virus sensitizes cells to oxidant injury. Virus Res. 2007;126:128–138. doi: 10.1016/j.virusres.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 45.Dharancy S, et al. Impaired expression of the peroxisome proliferator-activated receptor alpha during hepatitis C virus infection. Gastroenterology. 2005;128:334–342. doi: 10.1053/j.gastro.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 46.de Gottardi A, et al. Peroxisome proliferator-activated receptor-alpha and -gamma mRNA levels are reduced in chronic hepatitis C with steatosis and genotype 3 infection. Aliment Pharmacol Ther. 2006;23:107–114. doi: 10.1111/j.1365-2036.2006.02729.x. [DOI] [PubMed] [Google Scholar]
- 47.Staels B, et al. Mechanism of action of fibrates on lipid and lipoprotein metabolism. Circulation. 1998;98:2088–2093. doi: 10.1161/01.cir.98.19.2088. [DOI] [PubMed] [Google Scholar]
- 48.Tanaka N, et al. PPARalpha activation is essential for HCV core protein-induced hepatic steatosis and hepatocellular carcinoma in mice. J Clin Invest. 2008;118:683–694. doi: 10.1172/JCI33594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Andre P, et al. Hepatitis C virus particles and lipoprotein metabolism. Semin Liver Dis. 2005;25:93–104. doi: 10.1055/s-2005-864785. [DOI] [PubMed] [Google Scholar]
- 50.Nielsen SU, et al. Association between hepatitis C virus and very-low-density lipoprotein (VLDL)/LDL analyzed in iodixanol density gradients. J Virol. 2006;80:2418–2428. doi: 10.1128/JVI.80.5.2418-2428.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nielsen SU, et al. Characterization of hepatitis C RNA-containing particles from human liver by density and size. J Gen Virol. 2008;89:2507–2517. doi: 10.1099/vir.0.2008/000083-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Agnello V, et al. Hepatitis C virus and other flaviviridae viruses enter cells via low density lipoprotein receptor. Proc Natl Acad Sci U S A. 1999;96:12766–12771. doi: 10.1073/pnas.96.22.12766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Andre P, et al. Characterization of low- and very-low-density hepatitis C virus RNA-containing particles. J Virol. 2002;76:6919–6928. doi: 10.1128/JVI.76.14.6919-6928.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gastaminza P, et al. Differential biophysical properties of infectious intracellular and secreted hepatitis C virus particles. J Virol. 2006;80:11074–11081. doi: 10.1128/JVI.01150-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shelness GS, Ledford AS. Evolution and mechanism of apolipoprotein B-containing lipoprotein assembly. Curr Opin Lipidol. 2005;16:325–332. doi: 10.1097/01.mol.0000169353.12772.eb. [DOI] [PubMed] [Google Scholar]
- 56.Hussain MM, et al. Microsomal triglyceride transfer protein and its role in apoB-lipoprotein assembly. J Lipid Res. 2003;44:22–32. doi: 10.1194/jlr.r200014-jlr200. [DOI] [PubMed] [Google Scholar]
- 57.Gibbons GF, et al. Synthesis and function of hepatic very-low-density lipoprotein. Biochem Soc Trans. 2004;32:59–64. doi: 10.1042/bst0320059. [DOI] [PubMed] [Google Scholar]
- 58.Siddiqi SA. VLDL exits from the endoplasmic reticulum in a specialized vesicle, the VLDL transport vesicle, in rat primary hepatocytes. Biochem J. 2008;413:333–342. doi: 10.1042/BJ20071469. [DOI] [PubMed] [Google Scholar]
- 59.Nahmias Y, et al. Apolipoprotein B-dependent hepatitis C virus secretion is inhibited by the grapefruit flavonoid naringenin. Hepatology. 2008;47:1437–1445. doi: 10.1002/hep.22197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yao H, Ye J. Long chain acyl-CoA synthetase 3-mediated phosphatidylcholine synthesis is required for assembly of very low density lipoproteins in human hepatoma Huh7 cells. J Biol Chem. 2008;283:849–854. doi: 10.1074/jbc.M706160200. [DOI] [PubMed] [Google Scholar]
- 61.Miyanari Y, et al. The lipid droplet is an important organelle for hepatitis C virus production. Nat Cell Biol. 2007;9:1089–1097. doi: 10.1038/ncb1631. [DOI] [PubMed] [Google Scholar]
- 62.Icard V, et al. Secretion of hepatitis C virus envelope glycoproteins depends on assembly of apolipoprotein B positive lipoproteins. PLoS ONE. 2009;4:e4233. doi: 10.1371/journal.pone.0004233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Amako Y, et al. Role of Oxysterol Binding Protein in Hepatitis C Virus infection. J Virol. 2009;83 doi: 10.1128/JVI.00958-09. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Diaz O, et al. Preferential association of Hepatitis C virus with apolipoprotein B48-containing lipoproteins. J Gen Virol. 2006;87:2983–2991. doi: 10.1099/vir.0.82033-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gibbons GF, et al. Mobilisation of triacylglycerol stores. Biochim Biophys Acta. 2000;1483:37–57. doi: 10.1016/s1388-1981(99)00182-1. [DOI] [PubMed] [Google Scholar]
- 66.Mirandola S, et al. Liver microsomal triglyceride transfer protein is involved in hepatitis C liver steatosis. Gastroenterology. 2006;130:1661–1669. doi: 10.1053/j.gastro.2006.02.035. [DOI] [PubMed] [Google Scholar]
- 67.Petit JM, et al. Hepatitis C virus-associated hypobetalipoproteinemia is correlated with plasma viral load, steatosis, and liver fibrosis. Am J Gastroenterol. 2003;98:1150–1154. doi: 10.1111/j.1572-0241.2003.07402.x. [DOI] [PubMed] [Google Scholar]
- 68.Domitrovich AM, et al. Hepatitis C virus nonstructural proteins inhibit apolipoprotein B100 secretion. J Biol Chem. 2005;280:39802–39808. doi: 10.1074/jbc.M510391200. [DOI] [PubMed] [Google Scholar]
- 69.Boulant S, et al. Hepatitis C virus core protein induces lipid droplet redistribution in a microtubule- and dynein-dependent manner. Traffic. 2008;9:1268–1282. doi: 10.1111/j.1600-0854.2008.00767.x. [DOI] [PubMed] [Google Scholar]
- 70.Boulant S, et al. Structural determinants that target the hepatitis C virus core protein to lipid droplets. J Biol Chem. 2006;281:22236–22247. doi: 10.1074/jbc.M601031200. [DOI] [PubMed] [Google Scholar]
- 71.Shavinskaya A, et al. The lipid droplet binding domain of hepatitis C virus core protein is a major determinant for efficient virus assembly. J Biol Chem. 2007;282:37158–37169. doi: 10.1074/jbc.M707329200. [DOI] [PubMed] [Google Scholar]
- 72.Napolitano M, et al. Very low density lipoprotein and low density lipoprotein isolated from patients with hepatitis C infection induce altered cellular lipid metabolism. J Med Virol. 2007;79:254–258. doi: 10.1002/jmv.20793. [DOI] [PubMed] [Google Scholar]
- 73.Perlemuter G, et al. Hepatitis C virus core protein inhibits microsomal triglyceride transfer protein activity and very low density lipoprotein secretion: a model of viral-related steatosis. Faseb J. 2002;16:185–194. doi: 10.1096/fj.01-0396com. [DOI] [PubMed] [Google Scholar]
- 74.Abid K, et al. An in vitro model of hepatitis C virus genotype 3a-associated triglycerides accumulation. J Hepatol. 2005;42:744–751. doi: 10.1016/j.jhep.2004.12.034. [DOI] [PubMed] [Google Scholar]
- 75.Piodi A, et al. Morphological changes in intracellular lipid droplets induced by different hepatitis C virus genotype core sequences and relationship with steatosis. Hepatology. 2008;48:16–27. doi: 10.1002/hep.22288. [DOI] [PubMed] [Google Scholar]
- 76.Jackel-Cram C, et al. Up-regulation of fatty acid synthase promoter by hepatitis C virus core protein: genotype-3a core has a stronger effect than genotype-1b core. J Hepatol. 2007;46:999–1008. doi: 10.1016/j.jhep.2006.10.019. [DOI] [PubMed] [Google Scholar]
- 77.Moriishi K, et al. Critical role of PA28gamma in hepatitis C virus-associated steatogenesis and hepatocarcinogenesis. Proc Natl Acad Sci U S A. 2007;104:1661–1666. doi: 10.1073/pnas.0607312104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yamaguchi A, et al. Hepatitis C virus core protein modulates fatty acid metabolism and thereby causes lipid accumulation in the liver. Dig Dis Sci. 2005;50:1361–1371. doi: 10.1007/s10620-005-2788-1. [DOI] [PubMed] [Google Scholar]
- 79.Petit JM, et al. Apolipoprotein-AII concentrations are associated with liver steatosis in patients with chronic hepatitis C. Dig Dis Sci. 2007;52:3431–3434. doi: 10.1007/s10620-006-9719-7. [DOI] [PubMed] [Google Scholar]