Abstract
Mitochondrial genomes of plants are much larger than those of mammals and often contain conserved open reading frames (ORFs) of unknown function. Here, we show that one of these conserved ORFs is actually the gene for ribosomal protein L10 (rpl10) in plant. No rpl10 gene has heretofore been reported in any mitochondrial genome other than the exceptionally gene-rich genome of the protist Reclinomonas americana. Conserved ORFs corresponding to rpl10 are present in a wide diversity of land plant and green algal mitochondrial genomes. The mitochondrial rpl10 genes are transcribed in all nine land plants examined, with five seed plant genes subject to RNA editing. In addition, mitochondrial-rpl10-like cDNAs were identified in EST libraries from numerous land plants. In three lineages of angiosperms, rpl10 is either lost from the mitochondrial genome or a pseudogene. In two of them (Brassicaceae and monocots), no nuclear copy of mitochondrial rpl10 is identifiably present, and instead a second copy of nuclear-encoded chloroplast rpl10 is present. Transient assays using green fluorescent protein indicate that this duplicate gene is dual targeted to mitochondria and chloroplasts. We infer that mitochondrial rpl10 has been functionally replaced by duplicated chloroplast counterparts in Brassicaceae and monocots.
Keywords: chloroplast, GFP, plant mitochondria, ribosomal protein L10, RNA editing
1. Introduction
Plant mitochondrial genomes have several major differences compared with those of vertebrates and other animals: large sizes, the presence of plasmid-like DNA, ongoing and sometimes frequent functional gene transfer to the nucleus, horizontal transfer between more or less distantly related plants, and unusual modes of gene expression such as RNA editing and trans-splicing.1 The relatively large mitochondrial genomes of land plants encode several genes that are not found in the mitochondrial genomes of most other organisms.2 In addition, plant mitochondrial genomes often contain open reading frames (ORFs) of unknown function. Some of these ‘unidentified ORFs’ have been subsequently been identified as functional genes, e.g. atp4, atp8, sdh3, and sdh4.3–5 In addition to these cases, ORFs conserved across plant species have been reported in a number of cases, some of which are transcribed and might be functional.6,7 However, most such conserved ORFs still remain to be assigned to any known genes.
Here, we report a new case of gene identification: the conservation and expression of a gene that encodes ribosomal protein L10 (rpl10) in plant mitochondria. In bacteria, the rpl10 gene is present within a cluster encoding RPL11, RPL1, RPL10, and RPL7/RPL12.8 Cyanobacterial genomes also have an rpl10 gene but no equivalent gene has been found in chloroplast genomes.9 To date, across all of many sequenced mitochondrial genomes of diverse eukaryotes, an rpl10 gene has been identified only in the exceptionally and primitively gene-rich mitochondrial genome of the protist Reclinomonas americana.10,11 The counterpart to this gene is present in the nuclear genome in yeast and mammals.12,13 In contrast, no mitochondrial-type rpl10 gene has yet been reported in either the mitochondrial or nuclear genome of any plant species.
In this study, we present several lines of evidence that together strongly indicate that one of the conserved ORFs present in the mitochondrial genome of diverse plant species corresponds to a functional rpl10 gene. Results from this study also indicate that mitochondrial rpl10 gene has been lost in monocots and some Brassicaceae lineages, and replaced by an extra copy of the nuclear gene that normally encodes chloroplast RPL10 protein.
2. Materials and methods
2.1. Sequence analysis and database search
Sequences homologous to orf168 of Marchantia polymorpha mitochondria DNA14 were searched by Blast algorithm via the National Center for Biotechnology Information (NCBI) website (http://www.ncbi.nlm.nih.gov/). Genes for chloroplast rpl10 were searched in the NCBI database using sequences of Arabidopsis (locus tag: At5g13510) and Oryza (Os03g0284400) as queries. Nucleotide and predicted amino acid sequences were aligned manually with BioEdit ver. 7.0.5.3.15 A sequence alignment of chloroplast RPL10 (Supplementary Fig. S2) was used for construction of a neighbour-joining (NJ) phylogenetic tree after removing gaps and poorly conserved regions. The NJ tree was constructed using ClustalW ver. 1.83 via a DNA Data Bank of Japan website (http://clustalw.ddbj.nig.ac.jp/top-e.html). Bootstrap values were computed from 1000 replicates.
2.2. Plant materials and nucleic acid extraction
Leaves of Chinese cabbage (Brassica rapa, line ANF3-1), cycad (Cycas revoluta), grape (Vitis labrusca cv. Delaware), papaya (Carica papaya), rice (Oryza sativa subsp. japonica cv. Nipponbare), tobacco (Nicotiana tabacum cv. SR1), and tomato [Solanum lycopersicum cv. Saturn (Takii & Seed Co., Kyoto, Japan)] and thalli of liverwort (M. polymorpha) and hornwort (Megaceros flagellaris) were used for plant materials. Total DNA and total RNA were extracted with DNeasy Plant Mini Kit (Qiagen, Valencia, CA, USA) and RNeasy Plant Mini Kit (Qiagen), respectively.
2.3. Reverse transcription–PCR analysis
Total RNA (0.5 µg) was treated with RNase-free DNase I (Roche Diagnostics, Basel, Switzerland). First-strand cDNAs were synthesized using random hexamer primers and the Advantage RT-for-PCR Kit (Takara Bio, Otsu, Japan) as described previously.16 The resulting cDNAs were used for PCR amplifications with a primer pair P1–P2 in Carica, Nicotiana, and Vitis. Primer pairs P1–P3, P1–P4, and P1–P5 were used in Oryza, Brassica, and Cycas, respectively. The PCR products were cloned into a pCR-XL-TOPO vector (Invitrogen, Carlsbad, CA, USA), and 12–14 independent cDNA clones were sequenced for each species to detect any potential RNA editing events.
2.4. Construction and visualization of green fluorescent protein fusion proteins
Sequences encoding the first 71 and 45 residues of chloroplast-like RPL10 in Arabidopsis (At3g12370) and Oryza (Os05g0121500) were amplified by PCR with primer pairs P6–P7 and P8–P9, respectively, and fused in-frame with the 5′-upstream region of a synthetic green fluorescent protein (GFP)17 (kindly provided by Dr Y. Niwa). The resultant constructs were introduced into Arabidopsis epidermal cells using a PDS-1000 particle delivery system (Bio-Rad, Hercules, CA, USA). Mitochondria were labelled with an mt-DsRed construct, in which the GFP ORF in a pWS plasmid (carrying a presequence of Arabidopsis F1-ATPase δ, kindly provided by Prof. W. Sakamoto) was replaced by DsRed (Clontech, Mountain View, CA, USA). Transient expression of the introduced proteins and chloroplast autofluorescence were visualized with a confocal spectral laser scanning microscope Nikon C1Si (Nikon Corporation, Chiyoda-ku, Japan), as reported previously.18
2.5. Oligonucleotide primers
Primers used in this study are as follows. Small letters represent nucleotide deviations to introduce NcoI and SalI restriction sites (underlines).
P1: GGAGTCAGT(C/T)TTCTTT(A/G)AAGATCGAG
P2: CCT(A/C)AGACTCT(A/T)TCTTCCCGACC
P3: GGAGAATCCGCTAGGCTGAGGT
P4: TAGATGAATCGACCTCTGGACAGAT
P5: CTCAGGAAAGGGATAACCTGAGA
P6: GAATCTCGgTCGAcCTCACAGTTTC
P7: TGGCTcCAtgGACTCCCATTTGGTT
P8: GCCGTCGacGATGACGTCCGTG
P9: GAACGGCCcCAtgGCCTCCCACC
3. Results and discussion
3.1. A sequence homologous to orf168 in Marchantia mitochondrial DNA is conserved across diverse plant species
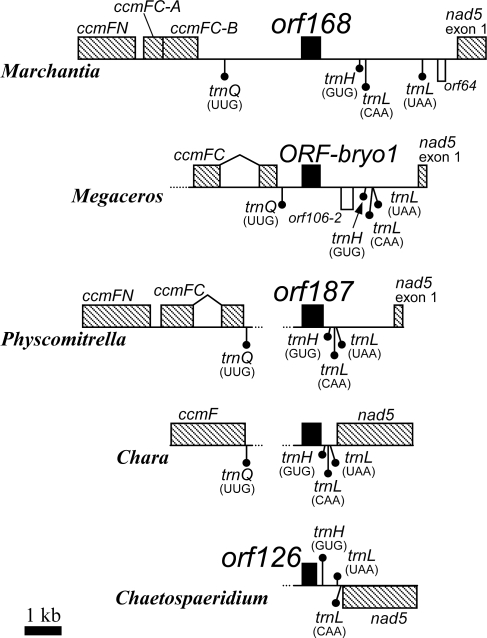
An unidentified orf168 was reported in the Marchantia mitochondrial genome.14 An homologous sequences has since been reported in the mitochondrial genomes of two other bryophytes (Megaceros and Physcomitrella) and two angiosperms (Nicotiana and Vitis), termed ORF-bryo1, orf187, orf159b, and orf159, respectively.7,19–21 These conserved ‘orf168-related sequences’ are currently annotated as unknown ORFs, with no evident correspondence to any characterized proteins. We searched for homologues of the Marchantia orf168 by a Blast search and found 10 additional homologous sequences in the mitochondrial DNAs of diverse plant groups. Altogether, sequences homologous to the orf168 have now been found in 15 plant species: two green algae (Chaetosphaeridium and Chara),22,23 three bryophytes (Marchantia, Megaceros, and Physcomitrella),7,14,20 one gymnosperm (Cycas),24 and nine angiosperms (Bambusa, Brassica, Carica, Helianthus, Nicotiana, Oryza, Solanum, Tripsacum, and Vitis)19,21,25–27 [Allen et al., unpublished (accession no. DQ984517); Carrari et al., unpublished (accession no. FJ374974); Rice et al., unpublished (accession no. EU431224); Lin et al., unpublished (accession no. EU365401)] (Table 1).
Table 1.
List of species, in which rpl10 and its potential counterparts were identified in the mitochondrial DNAs
| Group | Species name | Annotationa | Nucleotide positions (bp)b | Accession number | Reference |
|---|---|---|---|---|---|
| Protist | Reclinomonas americana | rpl10 | 5963–6544 | AF007261 | Lang et al.10 |
| Green alga | Chaetosphaeridium globosum | orf126 | 38 019–38 399 | AF494279 | Turmel et al.22 |
| Chara vulgaris | − | 30 284–30 781 | AY267353 | Turmel et al.23 | |
| Bryophyte | Marchantia polymorpha | orf168 | 184 854–185 360 | M68929 | Oda et al.14 |
| Megaceros aenigmaticus | ORF-bryo1 | 118 155–118 631 | EU660574 | Li et al.7 | |
| Physcomitrella patens | orf187 | 40 074–40 637 | AB251495 | Terasawa et al.20 | |
| Gymnosperm | Cycas taitungensis | − | 48 332–48 838 | AP009381 | Chaw et al.24 |
| Angiosperm | Bambusa oldhamii | −c | 162 094–162 555d | EU365401 | Lin et al. (unpublished) |
| Brassica napus | −c | 219 858–219 707d | AP006444 | Handa26 | |
| Carica papaya | − | 401 684–402 172 | EU431224 | Rice et al. (unpublished) | |
| Helianthus annuus | − | 1095–1583 | AM183222 | Placido et al.27 | |
| Nicotiana tobacum | orf159b | 360 762–360 283 | BA000042 | Sugiyama et al.19 | |
| Oryza sativa | −c | 197 408–196 959d | BA000029 | Notsu et al.25 | |
| Solanum lycopersicum | − | 430 703–430 348e | FJ374974 | Carrari et al. (unpublished) | |
| Tripsacum dactyloides | −c | 384 034–383 929d | DQ984517 | Allen et al. (unpublished) | |
| Vitis vinifera | orf159 | 156 298–155 822 | FM179380 | Goremykin et al.21 |
aMinus (−) means no annotation given in the registered sequence.
bNucleotide positions are indicated with regard to the direction of ORF.
cPseudogene.
dEach nucleotide positions corresponds a region that showed homology to the entire ORF in other angiosperms.
eThe 3′-terminal region of the ORF is missing due to a gap upstream of the position 430 348. The complete ORF has been determined in this study (accession no. AB518477).
The locations of the orf168-homologues in mitochondrial genomes relative to flanking genes are conserved among green algae and bryophytes to a substantial (but variable) degree (Fig. 1), whereas there is no linkage conservation of the orf168-homologue to these genes in seed plants (data not shown). This suggests that an ancestral, green-plant gene cluster including the orf168-homologue was destroyed, and each gene within the cluster was dispersed throughout the genome during the evolution of seed plants, as reported by Li et al.7 and as found for many other mitochondrial genes in angiosperm.
Figure 1.
Mitochondrial genome organization in the vicinity of ORFs that are homologous to orf168 of Marchantia polymorpha mitochondrial DNA. Exons of previously characterized genes, the ORFs potentially encoding the ribosomal protein L10 (RPL10), and other putative ORFs are indicated with hatched, filled, and open boxes, respectively. Group II introns are represented by bent lines. Positions of tRNA gene are marked by filled circles. Genes encoded in sense and antisense strands are placed above and below the horizontal lines, respectively.
cDNA sequences homologous to the orf168 were also found in published EST libraries from a fern (Adiantum), two gymnosperms (Picea and Welwitschia), and a number of angiosperms (e.g. Actinidia, Citrus, Eucalyptus, Gossypium, Malus, Opium, Petunia, Populus, Prunus, Raphanus, Theobroma, and Zinnia) (Supplementary Fig. S1). Although it has not directly shown that these sequences represent transcripts from mitochondrial genes, their high degree of sequence conservation (84–99%; Supplementary Fig. S1) suggests, in light of the generally much lower rate of nucleotide substitutions in plant mitochondrial than nuclear genomes,28,29 that they probably are mitochondrial gene products.
3.2. The orf168-homologues conserved in plant mitochondria are homologous to ribosomal protein L10
An alignment of protein sequences predicted from the orf168-homologues is shown in Fig. 2. These sequences are relatively well conserved despite a few insertions/deletions in their central region. The C-terminal region is more divergent with respect to both sequence and length variation. The sequences in Bambusa, Brassica, and Oryza (and possibly Raphanus) are probably pseudogenes, as they have frame-shift mutations or are severely truncated (Fig. 2, slashes and lower case letters, and also see Supplementary Fig. S1 for Raphanus). Chaetosphaeridium and Tripsacum were omitted from the sequence alignment of Fig. 2 and from further analysis because the homologous sequence in Chaetosphaeridium was greatly diverged and because Tripsacum retains only a short stretch of rpl10 (Table 1).
Figure 2.
Alignment of amino acid sequences between previously characterized RPL10 proteins and the ORFs conserved in plant mitochondria. Upper: RPL10 from Reclinomonas mitochondrion and four eubacteria. Lower: proteins predicted from the Marchantia orf168 and its homologues in plant mitochondria. Identical or similar amino acid residues conserved in >50% sequences are highlighted. Amino acids conserved in two or more non-plant species are marked with asterisks. Gaps are indicated by dashes. Amino acid changes caused by RNA editing events, which were detected by RT–PCR analysis (Cycas, Carica, Nicotiana, Solanum, and Vitis) (this study), or computationally predicted (Megaceros) (accession no. EU660574), are denoted with red letters. Especially, amino acid positions in which the homologies were clearly improved by RNA editing are indicated with filled triangles. Positions of frame-shifts in Bambusa and Oryza are shown with slashes. Note that Brassica lacks two-thirds of the C-terminal conserved region, which is replaced by an unrelated 23 amino acid sequence (shown by lower case letters). The length of each ORF is given in parentheses.
A Blast analysis using the predicted protein sequence of the Physcomitrella ORF187 yielded a hit to the 50S ribosomal protein L10 (RPL10) from the α-proteobacterium Rickettsia (25% identity, 44% similarity) and other bacteria (e.g. Chlamydophila and Methylocella). We believe that this level of similarity is significant, i.e. is indicative of evolutionary homology/common descent, for the following reasons. First, the database search also detected a conserved domain of the ribosomal L10-P0 superfamily (ID: cd00379 at Conserved Domain Database, http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml) within the Physcomitrella sequence. Second, manual sequence alignment between the already-characterized RPL10 (Fig. 2, upper five sequences) and the proteins predicted from the orf168 homologues (Fig. 2, lower) confirmed that several amino acid residues were clearly conserved between the two sequence groups (Fig. 2, asterisks). Third, the average length of the ORF168 homologues in plant mitochondria is (except for pseudogene sequences) 165 amino acids, which is quite similar to the length of bacterial RPL10. Fourth, the level of amino acid identity/similarity between the plant ORFs and bacterial RPL10 proteins is similar to that observed between Reclinomonas and bacterial RPL10 proteins (24% identity, 47% similarity). Finally, this level of conservation is similar to that observed for many mitochondrial ribosomal proteins and their bacterial counterparts (e.g. RPS4 and RPS8;30 also see Table in Hao and Palmer31). Given all this, we conclude that orf168 and its homologues in plant mitochondrial genomes are likely to function as the mitochondrial rpl10 gene in plants. Therefore, these ORFs will be designated as ‘mitochondrial rpl10 genes’ hereafter.
3.3. Plant mitochondrial rpl10 genes are transcribed and RNA edited
The expression of plant mitochondrial rpl10 was examined by reverse transcription (RT)–PCR. Transcripts from rpl10 were detected in all nine species investigated: two bryophytes (Marchantia and Megaceros), one gymnosperm (Cycas), and six angiosperms (Brassica, Carica, Nicotiana, Oryza, Solanum, and Vitis) (data not shown). Sequencing of five of the seed plant cDNAs revealed 3, 9, 5, 5, and 10 sites of C-to-T RNA editing in Cycas, Carica, Nicotiana, Solanum, and Vitis, respectively (Supplementary Fig. S1, Supplementary Table S1). These RNA editing events result in three, seven, four, three, and seven amino acid changes in the predicted proteins, respectively (Fig. 2, red letters), five positions of which clearly improve the similarity in the alignment (Fig. 2, filled triangles). RNA editing events are observed preferentially in protein-coding regions of land plant mitochondria at first and second positions in codons and tend to improve the level of protein sequence conservation.32,33 Seed plant rpl10 shows the same pattern of RNA editing, and therefore these results strongly indicate that the rpl10 gene is probably functional in the mitochondrion of these plants. In contrast, no RNA editing was detectable in the transcripts of Brassica and Oryza, which is consistent with the conclusion drawn in the preceding section that these are probably pseudogenes.
3.4. Loss of functional mitochondrial rpl10 gene and occurrence of an extra chloroplast-type rpl10 gene in angiosperms
As described above, mitochondrial rpl10 gene appears to be a pseudogene in Bambusa, Brassica, and Oryza, whereas no vestige of rpl10 was found in the complete mitochondrial genome sequences of five angiosperms (Arabidopsis, Beta, Sorghum, Triticum, and Zea) and 12 green algae (Chlamydomonas, Chlorogonium, Chlorokybus, Mesostigma, Nephroselmis, Oltmannsiellopsis, Ostreococcus, Pedinomonas, Prototheca, Pseudendoclonium, Scenedesmus, and Volvox) (see Supplementary data for references). Moreover, for three of these plants (Arabidopsis, Oryza, and Sorghum; the latter genome represented by a draft genome sequence), there is no evidence of a mitochondrial-type rpl10 gene in both the nuclear genome and in extensive cDNA datasets. Therefore, it is likely that rpl10 genes of mitochondrial origin have been completely lost in these species. This is somewhat surprising, as the preponderance of genes that have been lost from mitochondrial genomes in angiosperms have been functionally transferred to the nucleus.34
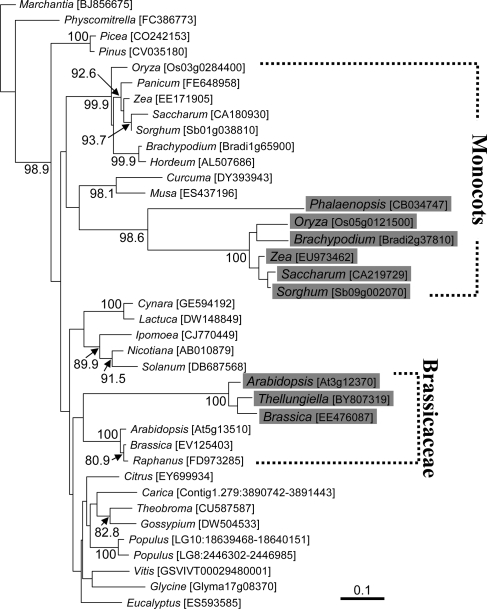
We hypothesized that the RPL10 protein might be supplied by a homologous nuclear gene of chloroplast origin, as found for cases of organellar loss of the rps13 gene.35,36 It has been shown that the rpl10 gene of chloroplast origin has been transferred to the nuclear genome in Arabidopsis and Oryza.37 Our database search showed that sequences homologous to this chloroplast-type rpl10 are present in many other land plants (Supplementary Fig. S2). All of these genes are probably located in the nucleus because the rpl10 gene is absent from all chloroplast genomes sequenced to date in land plants. Interestingly, monocots and Brassicaceae species harbour a second copy of the chloroplast-type rpl10 (Supplementary Fig. S2, lower), in addition to the putatively original copy that is widely present and conserved among land plants (Supplementary Fig. S2, upper). A phylogenetic analysis showed that the ‘extra’ copies of the chloroplast-type rpl10 gene in monocots and Brassicaceae form two independent clusters, each separated from all other examined genes by long branches (Fig. 3, shaded). This result suggests that the extra copy has diverged rapidly compared with the original chloroplast-type gene. Judging from their position in the rpl10 phylogeny, the two extra clades of genes may have originated by independent gene duplication events in monocots and Brassicaceae.
Figure 3.
Independent origins of the mitochondrial rpl10 genes in monocots and Brassicaceae from their respective chloroplast counterparts. An NJ phylogenetic tree is based on chloroplast-type RPL10 protein sequences. Accession numbers of cDNAs and locus tags of genome sequences are shown in brackets following each genus name. When the corresponding sequences are unannotated in the draft genome sequences, their locations are specified by the linkage group, chromosome number, or contig name together with their nucleotide coordinates. The Marchantia sequence was used as an outgroup. The ‘second’ copy of chloroplast RPL10 in monocots and Brassicaceae is shaded. Note that sequences homologous to the second copy of chloroplast RPL10 (At3g12370 and Os05g0121500 in Arabidopsis and Oryza, respectively) were also found in Hordeum, Musa, Panicum, Raphanus, and Triticum, but they are not included in the NJ tree because of incompleteness of their coding regions. Numbers on the nodes are bootstrap values (>80%) from 1000 replicates. Genetic distance is shown with a thick bar.
3.5. The chloroplast-derived rpl10 in Arabidopsis and Oryza undergoes dual-targeting into chloroplasts and mitochondria
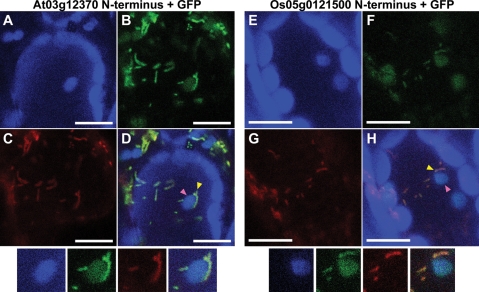
The alignment of the chloroplast-type RPL10 revealed that the N-terminal sequence of the second copy is totally different from that of the original copy (Supplementary Fig. S2) and is also non-homologous even between the monocots and Brassicaceae groups. Because the N-terminal region generally serves as a protein targeting signal, the distinct N-terminal sequences of the second set of RPL10 proteins may imply a difference of their targeting property. Proteome analyses in Arabidopsis and Spinacia have shown that the original, widely present chloroplast-type RPL10 is indeed targeted to chloroplasts, whereas cleavage of the N-terminal transit peptide region has been observed in Spinacia.38,39 In contrast, no such information has been obtained for the second copy of this gene. The second copy may have a short or no cleavable targeting sequence at its N-terminal region, considering the position of the cleavage site of the original copy (Supplementary Fig. S2, bent arrow). Protein localization predictions with Predotar, TargetP, and WoLF Psort40–42 provided contradictory results with respect to this second copy (data not shown). Therefore, we examined its subcellular localization in vivo using GFP. A fusion protein containing GFP and the N-terminal region of the second chloroplast-type RPL10 copy from Arabidopsis was clearly localized to mitochondria (Fig. 4B–D, and yellow arrowhead), suggesting its mitochondrial localization in vivo. In addition, however, GFP signals were also detected in chloroplasts (Fig. 4A, B, D, and pink arrowhead). Similar results were obtained using the Oryza sequence (Fig. 4F–H and E, F, and H, respectively). Therefore, the second chloroplast-derived RPL10 proteins in Arabidopsis and Oryza seem to undergo dual-targeting into both organelles. Dual-targeting has been found for a number of plant organellar proteins,43 including one other ribosomal protein (S16).44
Figure 4.
Subcellular localization in Arabidopsis leaf epidermal cells of GFP fusion proteins having the N-terminal portion of the second chloroplast-like RPL10 copy from Arabidopsis (A–D) and rice (E–H). (A and E) Chloroplast autofluorescence. Note that a number of large particles that did not coincide with the GFP image represent autofluorescence from mesophyll chloroplasts that occur below the epidermal cells. (B and F) GFP fluorescence; (C and G) mt-DsRed fluorescence as a control of mitochondrial targeting; (D and H) merger of the three images (chloroplast autofluorescence, GFP fluorescence, and mt-DsRed fluorescence) (A–C and E–G, respectively). Mitochondria and chloroplasts are indicated by yellow and pink arrowheads, respectively. Scale bar = 10 µm. An enlarged portion of the same small subportion of each image is shown below the set of four full images.
3.6. Evolution of mitochondrial rpl10 in plants
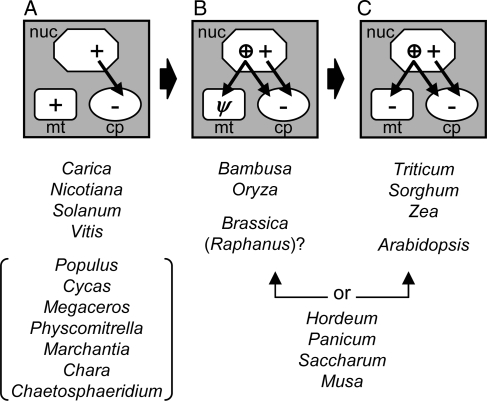
On the basis of the data obtained in this study, we propose a model (Fig. 5) for the evolution of the mitochondrial rpl10 gene. This gene was originally present only in the mitochondrial genome (Fig. 5A). It was lost from the mitochondrial genome early in the evolution of most eukaryotic lineages (e.g. animals, fungi, and most protists), but has been retained in mitochondria of the protist Reclinomonas and certain plants. Indeed, many diverse plants (both land plants and charophytic green algae) still possess an intact and probably functional rpl10 gene in their mitochondrial genomes. In contrast, the chloroplast rpl10 gene was transferred to the nucleus early in eukaryotic evolution, as no green plant chloroplast genomes still contain this gene (GOBASE: The Organelle Genome Database, http://gobase.bcm.umontreal.ca/index.php).
Figure 5.
A model for the evolution of rpl10 genes of mitochondrial and chloroplast origin. Three predicted evolutionary stages with regard to the mitochondrial rpl10 gene (A–C) are shown. The proposed evolutionary direction is indicated with thick arrows. Nucleus, mitochondrion, and chloroplast are represented by an octagon, a rounded rectangle, and an oval, respectively. An intact gene, a pseudogene, and loss of the gene are denoted with a plus (+), ψ, and a minus (−), respectively. The second copy of the nuclear gene encoding chloroplast RPL10 is shown by a circled plus sign. Targeting of proteins produced by the nuclear-encoded genes is indicated with arrows. Genera possibly or ambiguously belong to each of the three stages are shown in parentheses or by a double-headed arrow.
Subsequently, a duplication of the nuclear-located, chloroplast-derived rpl10 gene occurred (actually, probably separate duplications in monocots and in Brassicaceae), whose protein product appears to functionally compensate for mitochondrial RPL10 in certain plants. The mitochondrial rpl10 gene has become a pseudogene in some plants (Fig. 5B) and has been entirely lost from the mitochondrial genome in others (Fig. 5C). Extensive cDNA and nuclear genomic sequence data suggest that monocots and Brassicaceae no longer contain mitochondrial rpl10 in any genome. Our GFP assay demonstrated that the product of an extra copy of the nuclear gene for chloroplast RPL10 is imported into both mitochondria and chloroplasts in Arabidopsis and Oryza. These results strongly suggest that the mitochondrial RPL10 has been functionally replaced, probably twice independently, by the duplicated chloroplast counterpart in monocots and some Brassicaceae lineage. Functionality of the second copy in Raphanus is presently ambiguous, as all four homologous cDNAs of wild radish (R. raphanistrum) that are in GenBank (accession nos. EX746769, EX751093, FD961078, and FD965298) have either a frame-shift mutation or an internal stop codon. In this species, an additional copy of the second copy of the chloroplast-derived rpl10 gene may exist in the nuclear genome. A phylogenetic analysis of all available Brassicaceae cDNA sequences of the second copy does indeed suggest that it has been subject to further duplications in the Brassicaceae, including Raphanus (Supplementary Fig. S3).
During the 17 years since the first complete mitochondrial genome was reported from plants, the liverwort Marchantia polymorpha,14 comparative genomic data have allowed the identification of a number of genes that were previously known only as unidentified ORFs. In the case of ribosomal proteins, 16 ribosomal protein genes were initially identified in the Marchantia mitochondrial genome,30 and this is the first case of the subsequent identification of any new ribosomal protein genes in Marchantia or any other land plant mitochondrial genome. The present study shows that plants are the only group of eukaryote other than Reclinomonas that still retain rpl10 in their mitochondrial genomes, and furthermore, that the evolution of rpl10 within plants has taken some unusual and interesting turns.
Supplementary Data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Funding
This work was partly supported by a Grant-in-Aid for Young Scientists (B) (15780215) from the Ministry of Education, Culture, Sports, Science and Technology of Japan to N.K.
Note added in proof
Another report on rpl10 in plant mitochondria will be published by Jeffrey P. Mower and Linda Bonen in BMC Evol. Biol. These authors also have suggested the functional replacement of mitochondrial rpl10 through duplication of the chloroplast counterparts in crucifers and grasses.
Supplementary Material
Acknowledgements
The authors thank Kyoto Botanical Garden, Prof. J. Hasegawa, Prof. H. Deguchi, Dr S. Morita and Dr H. Motosugi for generous gifts of C. papaya, M. flagellaris, N. tabacum, and V. labrusca samples, respectively. The authors are grateful to Prof. J. D. Palmer for providing many valuable comments, review, and for editorial assistance with the manuscript, Dr Y. Sugiyama for helpful information on RNA editing in N. tabacum, Dr Y. Niwa for providing a GFP construct, Prof. W. Sakamoto for providing a pWS plasmid, and Ms. H. Kasaoka for technical assistance.
Footnotes
Edited by Dr. Katsumi Isono
References
- 1.Knoop V. The mitochondrial DNA of land plants: peculiarities in phylogenetic perspective. Curr. Genet. 2004;46:123–39. doi: 10.1007/s00294-004-0522-8. [DOI] [PubMed] [Google Scholar]
- 2.Kubo T., Newton K.J. Angiosperm mitochondrial genomes and mutations. Mitochondrion. 2008;8:5–14. doi: 10.1016/j.mito.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 3.Burger G., Lang B.F., Reith M., Gray M.W. Genes encoding the same three subunits of respiratory complex II are present in the mitochondrial DNA of two phylogenetically distant eukaryotes. Proc. Natl Acad. Sci. USA. 1996;93:2328–32. doi: 10.1073/pnas.93.6.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gray M.W., Lang B.F., Cedergren R., et al. Genome structure and gene content in protist mitochondrial DNAs. Nucleic Acids Res. 1998;26:865–78. doi: 10.1093/nar/26.4.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heazlewood J.L., Whelan J., Millar A.H. The products of the mitochondrial orf25 and orfB genes are FO components in the plant F1FO ATP synthase. FEBS Lett. 2003;540:201–5. doi: 10.1016/s0014-5793(03)00264-3. [DOI] [PubMed] [Google Scholar]
- 6.Siqueira S.F., Dias S.M.G., Lejeune B., de Souza A.P. Marchantia polymorpha mitochondrial orf identifies transcribed sequence in angiosperm mitochondrial genome. Biochim. Biophys. Acta. 2001;1520:203–11. doi: 10.1016/s0167-4781(01)00273-1. [DOI] [PubMed] [Google Scholar]
- 7.Li L., Wang B., Liu Y., Qiu Y.L. The complete mitochondrial genome sequence of the hornwort Megaceros aenigmaticus shows a mixed mode of conservative yet dynamic evolution in early land plant mitochondrial genomes. J. Mol. Evol. 2009;68:665–78. doi: 10.1007/s00239-009-9240-7. [DOI] [PubMed] [Google Scholar]
- 8.Fujita K., Baba T., Isono K. Genomic Analysis of the genes encoding ribosomal proteins in eight eubacterial species and Saccharomyces cerevisiae. Genome Inform. Ser. Workshop Genome Inform. 1998;9:3–12. [PubMed] [Google Scholar]
- 9.Harris E.H., Boynton J.E., Gillham N.W. Chloroplast ribosomes and protein synthesis. Microbiol. Rev. 1994;58:700–54. doi: 10.1128/mr.58.4.700-754.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lang B.F., Burger G., O'Kelly C.J., et al. An ancestral mitochondrial DNA resembling a eubacterial genome in miniature. Nature. 1997;387:493–7. doi: 10.1038/387493a0. [DOI] [PubMed] [Google Scholar]
- 11.Smits P., Smeitink J.A.M., van den Heuvel L.P., Huynen M.A., Ettema T.J.G. Reconstructing the evolution of the mitochondrial ribosomal proteome. Nucleic Acids Res. 2007;35:4686–703. doi: 10.1093/nar/gkm441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bui D.M., Jarosch E., Schweyen R.J. The yeast ORF YDL202w codes for the mitochondrial ribosomal protein YmL11. Curr. Genet. 1997;31:396–400. doi: 10.1007/s002940050221. [DOI] [PubMed] [Google Scholar]
- 13.Goldschmidt-Reisin S., Kitakawa M., Herfurth E., Wittmann-Liebold B., Grohmann L., Graack H.R. Mammalian mitochondrial ribosomal proteins. N-terminal amino acid sequencing, characterization, and identification of corresponding gene sequences. J. Biol. Chem. 1998;273:34828–36. doi: 10.1074/jbc.273.52.34828. [DOI] [PubMed] [Google Scholar]
- 14.Oda K., Yamato K., Ohta E., et al. Gene organization deduced from the complete sequence of liverwort Marchantia polymorpha mitochondrial DNA. A primitive form of plant mitochondrial genome. J. Mol. Biol. 1992;223:1–7. doi: 10.1016/0022-2836(92)90708-r. [DOI] [PubMed] [Google Scholar]
- 15.Hall T.A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–8. [Google Scholar]
- 16.Kubo N., Harada K., Hirai A., Kadowaki K. A single nuclear transcript encoding mitochondrial RPS14 and SDHB of rice is processed by alternative splicing: common use of the same mitochondrial targeting signal for different proteins. Proc. Natl Acad. Sci. USA. 1999;96:9207–11. doi: 10.1073/pnas.96.16.9207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiu W., Niwa Y., Zeng W., Hirano T., Kobayashi H., Sheen J. Engineered GFP as a vital reporter in plants. Curr. Biol. 1996;6:325–30. doi: 10.1016/s0960-9822(02)00483-9. [DOI] [PubMed] [Google Scholar]
- 18.Arimura S., Fujimoto M., Doniwa Y., et al. Arabidopsis ELONGATED MITOCHONDRIA1 is required for localization of DYNAMIN-RELATED PROTEIN3A to mitochondrial fission sites. Plant Cell. 2008;20:1555–66. doi: 10.1105/tpc.108.058578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sugiyama Y., Watase Y., Nagase M., et al. The complete nucleotide sequence and multipartite organization of the tobacco mitochondrial genome: comparative analysis of mitochondrial genomes in higher plants. Mol. Genet. Genomics. 2005;272:603–15. doi: 10.1007/s00438-004-1075-8. [DOI] [PubMed] [Google Scholar]
- 20.Terasawa K., Odahara M., Kabeya Y., et al. The mitochondrial genome of the moss Physcomitrella patens sheds new light on mitochondrial evolution in land plants. Mol. Biol. Evol. 2007;24:699–709. doi: 10.1093/molbev/msl198. [DOI] [PubMed] [Google Scholar]
- 21.Goremykin V.V., Salamini F., Velasco R., Viola R. Mitochondrial DNA of Vitis vinifera and the issue of rampant horizontal gene transfer. Mol. Biol. Evol. 2008;26:99–110. doi: 10.1093/molbev/msn226. [DOI] [PubMed] [Google Scholar]
- 22.Turmel M., Otis C., Lemieux C. The chloroplast and mitochondrial genome sequences of the charophyte Chaetosphaeridium globosum: insights into the timing of the events that restructured organelle DNAs within the green algal lineage that led to land plants. Proc. Natl Acad. Sci. USA. 2002;99:11275–80. doi: 10.1073/pnas.162203299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turmel M., Otis C., Lemieux C. The mitochondrial genome of Chara vulgaris: insights into the mitochondrial DNA architecture of the last common ancestor of green algae and land plants. Plant Cell. 2003;15:1888–903. doi: 10.1105/tpc.013169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chaw S.M., Shih A.C.C., Wang D., Wu Y.W., Liu S.M., Chou T.Y. The mitochondrial genome of the gymnosperm Cycas taitungensis contains a novel family of short interspersed elements, Bpu sequences, and abundant RNA editing sites. Mol. Biol. Evol. 2008;25:603–15. doi: 10.1093/molbev/msn009. [DOI] [PubMed] [Google Scholar]
- 25.Notsu Y., Masood S., Nishikawa T., et al. The complete sequence of the rice (Oryza sativa L.) mitochondrial genome: frequent DNA sequence acquisition and loss during the evolution of flowering plants. Mol. Genet. Genomics. 2002;268:434–45. doi: 10.1007/s00438-002-0767-1. [DOI] [PubMed] [Google Scholar]
- 26.Handa H. The complete nucleotide sequence and RNA editing content of the mitochondrial genome of rapeseed (Brassica napus L.): comparative analysis of the mitochondrial genomes of rapeseed and Arabidopsis thaliana. Nucleic Acids Res. 2003;31:5907–16. doi: 10.1093/nar/gkg795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Placido A., Damiano F., Sciancalepore M., De Benedetto C., Rainaldi G., Gallerani R. Comparison of promoters controlling on the sunflower mitochondrial genome the transcription of two copies of the same native trnK gene reveals some differences in their structure. Biochim. Biophys. Acta. 2006;1757:1207–16. doi: 10.1016/j.bbabio.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 28.Wolfe K.H., Li W.H., Sharp P.M. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc. Natl Acad. Sci. USA. 1987;84:9054–8. doi: 10.1073/pnas.84.24.9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mower J.P., Touzet P., Gummow J.S., Delph L.F., Palmer J.D. Extensive variation in synonymous substitution rates in mitochondrial genes of seed plants. BMC Evol. Biol. 2007;7:135. doi: 10.1186/1471-2148-7-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takemura M., Oda K., Yamato K., et al. Gene clusters for ribosomal proteins in the mitochondrial genome of a liverwort, Marchantia polymorpha. Nucleic Acids Res. 1992;20:3199–205. doi: 10.1093/nar/20.12.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hao W., Palmer J.D. Fine-scale mergers of chloroplast and mitochondrial genes create functional, transcompartmentally chimeric mitochondrial genes. Proc. Natl Acad. Sci. USA. 2009;106:16728–33. doi: 10.1073/pnas.0908766106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shikanai T. RNA editing in plant organelles: machinery, physiological function and evolution. Cell. Mol. Life Sci. 2006;63:698–708. doi: 10.1007/s00018-005-5449-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jobson R.W., Qiu Y.L. Did RNA editing in plant organellar genomes originate under natural selection or through genetic drift? Biol. Direct. 2008;3:43. doi: 10.1186/1745-6150-3-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adams K.L., Qiu Y.L., Stoutemyer M., Palmer J.D. Punctuated evolution of mitochondrial gene content: high and variable rates of mitochondrial gene loss and transfer to the nucleus during angiosperm evolution. Proc. Natl Acad. Sci. USA. 2002;99:9905–12. doi: 10.1073/pnas.042694899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adams K.L., Daley D.O., Whelan J., Palmer J.D. Genes for two mitochondrial ribosomal proteins in flowering plants are derived from their chloroplast or cytosolic counterparts. Plant Cell. 2002;14:931–43. doi: 10.1105/tpc.010483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mollier P., Hoffmann B., Debast C., Small I. The gene encoding Arabidopsis thaliana mitochondrial ribosomal protein S13 is a recent duplication of the gene encoding plastid S13. Curr. Genet. 2002;40:405–9. doi: 10.1007/s00294-002-0271-5. [DOI] [PubMed] [Google Scholar]
- 37.Bonen L., Calixte S. Comparative Analysis of bacterial-origin genes for plant mitochondrial ribosomal proteins. Mol. Biol. Evol. 2006;23:701–12. doi: 10.1093/molbev/msj080. [DOI] [PubMed] [Google Scholar]
- 38.Yamaguchi K., Subramanian A.R. The plastid ribosomal proteins. Identification of all the proteins in the 50 S subunit of an organelle ribosome (chloroplast) J. Biol. Chem. 2000;275:28466–82. doi: 10.1074/jbc.M005012200. [DOI] [PubMed] [Google Scholar]
- 39.Zybailov B., Rutschow H., Friso G., et al. Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS One. 2008;3:e1994. doi: 10.1371/journal.pone.0001994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Emanuelsson O., Nielsen H., Brunak S., von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000;300:1005–16. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 41.Small I., Peeters N., Legeai F., Lurin C. Predotar: A tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics. 2004;4:1581–90. doi: 10.1002/pmic.200300776. [DOI] [PubMed] [Google Scholar]
- 42.Horton P., Park K.J., Obayashi T., et al. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007;35:W585–7. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carrie C., Giraud E., Whelan J. Protein transport in organelles: dual targeting of proteins to mitochondria and chloroplasts. FEBS J. 2009;276:1187–95. doi: 10.1111/j.1742-4658.2009.06876.x. [DOI] [PubMed] [Google Scholar]
- 44.Ueda M., Nishikawa T., Fujimoto M., et al. Substitution of the gene for chloroplast RPS16 was assisted by generation of a dual targeting signal. Mol. Biol. Evol. 2008;25:1566–75. doi: 10.1093/molbev/msn102. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.