Abstract
Nuclear DNA of metazoans is organized in supercoiled loops anchored to a proteinaceous substructure known as the nuclear matrix (NM). DNA is anchored to the NM by non-coding sequences known as matrix attachment regions (MARs). There are no consensus sequences for identification of MARs and not all potential MARs are actually bound to the NM constituting loop attachment regions (LARs). Fundamental processes of nuclear physiology occur at macromolecular complexes organized on the NM; thus, the topological organization of DNA loops must be important. Here, we describe a general method for determining the structural DNA loop organization in any large genomic region with a known sequence. The method exploits the topological properties of loop DNA attached to the NM and elementary topological principles such as that points in a deformable string (DNA) can be positionally mapped relative to a position-reference invariant (NM), and from such mapping, the configuration of the string in third dimension can be deduced. Therefore, it is possible to determine the specific DNA loop configuration without previous characterization of the LARs involved. We determined in hepatocytes and B-lymphocytes of the rat the DNA loop organization of a genomic region that contains four members of the albumin gene family.
Keywords: DNA topology, loop attachment regions, matrix attachment regions, nuclear matrix, nucleotype
1. Introduction
In the interphase nucleus of metazoan cells, the DNA is organized in supercoiled loops anchored to a nuclear substructure commonly known as the nuclear matrix (NM) which is a non-soluble complex of ribonucleoproteins obtained after extracting the nucleus with non-ionic detergents, high salt and treatment with DNase.1,2 The exact composition of the NM is a matter of debate as some 400 proteins have been associated with this structure.3 The DNA is anchored to the NM by means of non-coding sequences of variable length known as matrix attachment regions (MARs). Yet, there is no consensus sequence for a priori identification of MARs, although they are generally rich in AT and repetitive sequences, and map to regions where the DNA is intrinsically curved or kinked and has a propensity for base unpairing.4 MARs are operationally classified into structural-constitutive, resistant to high-salt extraction and functional-facultative, non-resistant to high-salt extraction.5,6 Therefore, not all potential MARs are actually bound to the NM constituting true loop attachment regions (LARs).5 There is evidence that when multiple copies of a specific MAR are present, these are used in a selective fashion, indicating the adaptability of the MAR sequence to serve as anchor only under certain conditions.7 The higher-order structure of interphase and metaphase chromosomes is likely to be maintained by constitutive MARs.4,8 It has been suggested that the dynamic selectivity in the use of MARs as DNA anchors would modulate both the DNA loop average length and the stability of the topological relationships between DNA and the NM during development and cell differentiation.9,10 Only a very limited number of proteins have been identified that participate in sequence-specific binding of DNA to the NM.2,4 However, saturation experiments indicate the existence of some 150 000 salt-resistant DNA-binding sites per NM in rat hepatocytes,11 whereas the average DNA loop size in adult rat hepatocytes is 80–90 kb,12 and this figure is compatible with roughly 66 000 DNA loops per rat diploid genome.13 Therefore, given that there are no MAR consensus sequences, these facts imply that such DNA–NM interactions are the result of indirect readout effects between DNA and NM proteins and thus not equivalent to the direct readout interactions between transcription factors and specific DNA sequences.14 Such sequence-independent DNA–protein interactions depend on DNA shape and overall DNA mechanical properties such as curvature, helical twist and bending and torsional flexibilities,15 therefore the local topology (spatial configuration) of the DNA involved must be a determining factor.16 Fundamental processes of nuclear physiology such as DNA replication, transcription and processing of primary transcripts occur at macromolecular complexes or factories organized on the NM.17–19 Thus, the topological relationship between the DNA loops and the NM appears to be very important for appropriate nuclear physiology. Indeed, correct repair of DNA damage must include the recovery of both the double helix integrity and the complex three-dimensional DNA topology; otherwise, the cell will not survive.20,21
Varied approaches have been used for establishing the DNA loop organization in a given region of the genome, based on the experimental22–26 or the in silico identification of MARs27 which has been shown to be quite unreliable in the absence of experimental confirmation,28 and more recently on a combination of both approaches coupled to high-throughput analytical techniques.29,30 All these methods have their specific advantages and pitfalls, but all of them have as priority the identification of the actual MARs (LARs) in a given DNA region so as to infer from such data the possible DNA loop organization. However, considering that DNA is a continuous helical string that becomes looped by binding to the NM, thus establishing a topological higher-order in third-dimensional space, it follows from elementary topological considerations that by mapping the relative positions to the NM of several points (i.e. DNA sequences) along a given string of DNA whose whole sequence is known, it will be possible to establish the spatial configuration of the loops formed by such a DNA string without the need of characterizing the LARs involved. We have previously developed a method for mapping the position of any DNA sequence relative to the NM based on the direct use of PCR on NM-bound templates.31 An extension of this method, described hereunder, was used for determining the structural DNA loop organization in a region of 162 kb in chromosome 14 of the rat where four closely related genes that are members of the albumin gene family are located. Such a characterization was done in nucleoids which consist of the NM plus the naked DNA loops attached to it, obtained from freshly isolated hepatocytes and naïve B-lymphocytes from the rat and so the resulting DNA loop organization very likely corresponds to the actual in vivo organization.
2. Materials and methods
2.1. Animals
Male Wistar rats weighing 200–250 g were used in accordance with the official Mexican norm for production, care and use of laboratory animals (NOM-062-ZOO-1999).
2.2. Primary cells
Primary rat hepatocytes were obtained from livers of male Wistar rats, using the protocol described previously.6 Briefly, the livers were washed in situ by perfusion with PBS without Ca2+ and Mg2+ (PBS-A) at 37°C for 5 min at 15 ml/min. Next, the tissue was perfused with a solution of collagenase IV, Sigma (0.025% collagenase with 0.075% CaCl2 in HEPES buffer, pH 7.6) for 8 min. Viable hepatocytes were counted in a haemocytometer and used immediately for preparing nucleoids (see below). Primary naïve B-lymphocytes were obtained from the spleen of male Wistar rats. Briefly, the spleen was fragmented and filtered in PBS-A. Total lymphocytes were isolated in preformed gradients of isotonic Percoll (25%) by centrifugation at 2100 rpm 15 min at room temperature. Subsequently, naïve B-lymphocytes were purified by magnetic separation (MACS Separation columns, Miltenyi Biotec) with anti-CD43-specific beads (rat CD43 microbeads, Miltenyi Biotec) according to the manufacturer's instructions. Viable B-lymphocytes were counted in a haemocytometer and used immediately for preparing nucleoids.
2.3. Preparation of nucleoids
The DNA loops plus the nuclear substructure constitute a ‘nucleoid’, a very large nucleoprotein aggregate generated by gentle lysis of a cell at pH 8 in non-ionic detergent and the presence of high-salt concentration. Nucleoids were prepared as described previously.21,31 Briefly, freshly isolated and washed hepatocytes and naïve B-lymphocytes are suspended in ice-cold PBS-A, and aliquots of 50 µl containing 3 × 105 hepatocytes or 5 × 105 lymphocytes are gently mixed with 150 µl of a lysis solution containing 2.6 M NaCl, 1.3 mM EDTA, 2.6 mM Tris, 0.6% Triton-X 100 (pH 8.0). After 20 min for hepatocytes and 15 min for native B-lymphocytes (at 4°C), the mixture is washed in 14 ml of PBS-A at 4°C for 4 min at 3000 rpm (1500 g). The pellet is recovered in a volume ranging from 200 to 300 µl.
2.4. DNase I digestion of nucleoid samples
The washed nucleoids are pooled for setting up the DNase I digestion curves (1.8 × 106 nucleoids of hepatocytes or 2.5 × 106 nucleoids of naïve B-lymphocytes in 1.2 ml of PBS-A) and mixed with 5 ml of DNase I digestion buffer (10 mM MgCl2, 0.1 mM dithiothreitol, 50 mM Tris at pH 7.2). Digestions were carried out at 37°C with 0.5 U/ml DNase I (Sigma). Each digestion time-point aliquot contains 3 × 105 nucleoids of hepatocytes or 5 × 105 nucleoids of B-lymphocytes. Digestion reactions were stopped by adding 200 µl of stop buffer (final EDTA concentration of 30 mM). The stop buffer contains 0.2 M EDTA and 10 mM Tris at pH 7.5. After digestion with DNase I, the NM-bound DNA was determined by spectrometry on aliquots of partially digested nucleoid samples that were washed and further handled as described previously.31 The final nucleoid pellet was re-suspended in 200 µl of double-distilled H2O to be used directly as a template for PCR.
2.5. Genomic DNA primers
Distinct sets of primers were designed for establishing the topological positions relative to the NM of 15 small DNA sequences located along 162 kb of the genomic region containing four members of the rat albumin gene family. Primer pairs were designed approximately 10 kb each in order to establish rather regular intervals along the region studied. We took into account important considerations in the oligonucleotide design so as to get primers with high specificity but able to perform efficient DNA amplification under the same PCR conditions (reactants concentrations, temperatures, times of each step and number of cycles). It is important to remark that MARs are not suitable targets for high-quality primer design, because their characteristic composition (richness of A-T tracks and repetitive DNA) prevents an acceptable design. Therefore, both potential and actual MARs within the chosen region are likely to be excluded by our primer-design algorithms. All primer sets were designed with a length of 20–25 bp, G-C content between 50% and 55% (with a difference <3%), Tm of 55–60°C (with a difference <2°C), and PCR products of 250–550 bp (Table 1). Secondary structures with ΔG < −1 kcal/mol and dimmers/duplexes with ΔG < −2 kcal/mol were avoided. Additionally, the specificity of each primer set was confirmed by the NCBI BLAST algorithm.
Table 1.
Sets of primers and corresponding amplicon sizes defining 15 target sequences along the 162-kb genomic region containing members of the rat albumin gene family
| Amplicon | Forward primer | Reverse primer | Amplicon length (bp) |
|---|---|---|---|
| a | TGGCAAACATACGCAAGGGA | GCGAAACACACCCCTGGAAA | 275 |
| b | GAGGACAGTTAGTGCTGTAGGGTTG | CCTCCAACGAAGTTCCCAGAAT | 547 |
| c | TCCTTTGTAACCAGGCAAGTGG | CCCATTTCCCAGATCCTTCACTCT | 374 |
| d | CCCAGGGTCAGAGTATATCAGTGC | CGCTGAACGTATGTCTGAGTCA | 305 |
| e | TGGTAGGCAGAGATGTGAGGAAAG | CCTGTTGTCCTAATGCTGGTCCTA | 382 |
| f | CTGATCTTCAGGCAATATGGCAGG | TTGGCTGATGTCGTCTGGACA | 393 |
| g | AAGGATAGGTGCTTGGCTGACA | GCCCTAACCCTGTGTGTGTATCTTG | 504 |
| h | GATCACGTAACAACCCTGTCAGCT | CCTTCACAGCACCCGTCATACA | 263 |
| i | GGTGCTGGGAATTTGACTAAGGC | TAAACTCAGGTGACAGGCTACGGC | 465 |
| j | AGGAACCAGGGAATCGAGTGCT | AACTTGCGGGTGTTCTCTCCTT | 392 |
| k | TGTCAGCATGATGGTGGCTCA | CTCGATTTGCCATGTCCTGTCT | 252 |
| l | GGGCTGGGTCCATATTGCTTGA | ATGCTTTGGGCTTGCCTGAAG | 373 |
| m | ACGACTTCCCTTCCTATCCACAG | GTAGAAAGTCGTTCTGGTTGCCAC | 234 |
| n | CCCTAATCTTGCTGTGGTTTGG | TGAGAGCTGGGCAGACATCAA | 355 |
| o | GGTGACAGTTGACAGAGAGCCTTC | GCTCCATGCTGACCTTGAAGTCTA | 272 |
2.6. PCR amplification
Fifty nanograms of NM-bound DNA was used as a template for PCR. PCR was carried out using 0.7 U GoTaq DNA polymerase (Promega), 2.5 mM MgCl2, 0.2 mM of each dNTP and 0.1 µM of each primer. Amplification was performed in an Applied Biosystems 2720 thermocycler and the same amplification program was used for all pairs of primers: initial denaturising step at 94°C for 5 min, denaturising step at 94°C for 45 s, annealing at 56°C for 30 s and extension at 72°C for 1 min for 35 cycles, with a final extension at 72°C for 10 min. The identity of all the amplicons was confirmed by restriction analysis with the appropriate restriction enzymes. Amplicons were electrophoresed on 2% agarose gels and visualized using ethidium bromide staining (0.5 µl/ml), recorded and analyzed using a Kodak 1D Image Analysis Software 3.5 system. Amplicons were scored as positive or negative on partially digested nucleoid samples, depending on whether they are detectable by the software using the default settings.
2.7. Computer-aided prediction of MAR sequences
The potential MARs in the region studied were identified by in silico prediction using four different freely available software: MarFinder,32 MARScan,33 ChrClass34 and SMARtest.27
3. Results and discussion
3.1. Rationale of the method
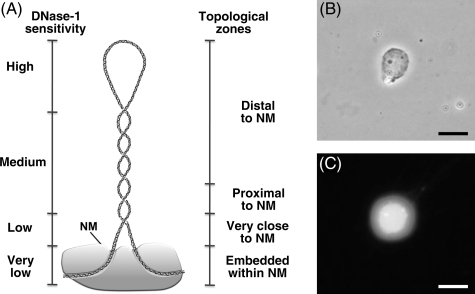
Our method exploits the topological properties of the average DNA loop that result from the fact that such loops are topologically constrained by being anchored to the NM, thus being equivalent to closed DNA circles. Under such a condition, the DNA molecule undergoes structural stress resulting from two factors: the covalently linked backbones of the DNA strands are helicoidal but rigid, and the low-energy hydrogen bonds between the stacked bases are quasi-statistical unions that continuously break apart and form again; such a situation poses the risk that the nucleotide bases may gyrate away from the double-helix axis and become exposed. DNA naturally solves this structural-stress problem by further coiling upon its own axis, thus avoiding the exposure of the nucleotide bases, but becoming negatively supercoiled in a similar fashion to a pulled house-telephone cord.35,36 Thus, the naked DNA loops display a gradient of supercoiling that goes from lower to higher from tip to base of the loop,36 save for the fact that the structural properties of MARs are such that they also function as buffers or sinks of negative supercoiling37,38 thus avoiding maximal supercoiling at the base of the loops. The NM plus the naked DNA loops anchored to it constitute a nucleoid. Under the conditions of lysis employed to generate nucleoids, the DNA remains essentially intact, although it lacks the nucleosome structure because of the dissociation of histones and most other nuclear proteins usually associated with DNA; yet, the DNA loops remain topologically constrained and supercoiled as depicted in Fig. 1. Indeed, nucleoids are also known as nuclear halos since the exposure of such structures to DNA-intercalating agents like ethidium bromide leads to unwinding of the DNA loops that form a DNA halo around the NM periphery (Fig. 1C). A typical DNA loop can be divided into four topological zones according to their relative proximity to the NM. Each of these zones would manifest an identifiable behaviour when exposed to non-specific nucleases that are sensitive to the local DNA topology (Fig. 1A). We have previously shown that in nucleoid preparations, the relative resistance of a given loop-DNA sequence to a limited concentration of DNase I is directly proportional to its proximity to the NM anchoring point.31,39 Two main factors determine this property. (i) Steric hindrance resulting from the proteinaceous NM that acts as a physical barrier relatively protecting the naked loop DNA that is closer to the NM from endonuclease action. (ii) The local degree of loop DNA supercoiling that is lower in the distal portions of the loop and higher in the regions proximal to the NM. Supercoiling is a structural barrier against the action of non-specific endonucleases, such as DNase I, that hydrolyze the DNA backbone by a single-strand cleavage (nicking) mechanism.40 Both factors only confer relative but not absolute DNase I resistance to loop DNA. However, in a large sample of nucleoids exposed to a limited concentration of DNase I, there is a consistent trend in which the loop-DNA sensitivity to the enzyme is inversely proportional to its distance relative to the NM and so distal regions of the loop are digested first whereas the regions closer to the NM are digested later. Indeed, it is known that the DNA embedded within the NM is very resistant to DNase I action, and there is a fraction corresponding to some 2% the total DNA that is basically non-digestible even when exposed to high concentrations of the enzyme. This fraction corresponds to fragments with an average length of 1.6 kb in rat hepatocytes,12 likely to represent the regions that include the actual MARs (LARs) anchored to the NM. This pattern of sensitivity to DNase I holds provided that the DNA is basically devoid of histones and most other proteins that form chromatin. Indeed, whole chromatin attached to the NM shows an inverse pattern of nuclease sensitivity compared with that of naked loop DNA. In looped chromatin, those sequences closer to the matrix attachment point are preferentially cleaved by nucleases.41
Figure 1.
Properties of naked DNA loops attached to the NM. (A) Drawing illustrating the local topology along a typical supercoiled DNA loop that correlates with both distance relative to the NM and sensitivity to DNase I. (B) Phase contrast micrograph showing the NM of a hepatocyte nucleoid. (C) Fluorescence micrograph showing the DNA halo around the NM of a hepatocyte nucleoid caused by the unwinding of the supercoiled DNA loops by treatment of the nucleoid with the DNA-intercalating agent ethidium bromide (80 µg/ml). Scale bars 10 µm.
3.2. Differential kinetics of loop DNA digestion as a function of proximity to the NM
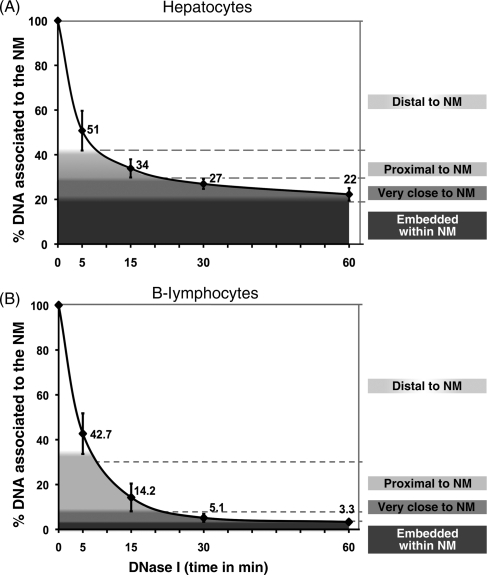
Treatment of nucleoids from quiescent, freshly isolated rat hepatocytes, with a limited concentration of DNase I (0.5 U/ml) produces a highly reproducible kinetics of digestion of loop DNA (Fig. 2A). It is possible to identify three different phases in the digestion curve: the first corresponds to a very fast kinetics of digestion that removes almost 60% the total DNA associated with the NM within the first 5 min. Such a DNA corresponds to the loop fraction distal to the NM; in such a fraction, relatively minor DNA supercoiling is the only barrier to the endonuclease action. Moreover, as the DNase I nicks the loop DNA, each nick becomes a point of further DNA unwinding, thus increasing the reduction in loop supercoiling in time and so making more accessible the loop DNA to the action of the endonuclease. The second phase that lasts ∼10 min shows a reduced kinetics of digestion in which some further 10% loop DNA is removed. This slower kinetics is the result of the slower rate of supercoiling loss in the loop DNA proximal to the NM (originally highly supercoiled) that slows down the action of the enzyme. The third and slowest phase, lasting 45 min, shows the removal of some further 10% total DNA associated with the NM. This very slow kinetics results from two factors: the effect of any residual DNA supercoiling and principally of the proximity between the loop DNA and the NM proteins that act as physical barriers against the action of DNase I on the loop DNA that is very close to the NM. Finally, there is about 20% total DNA that remains bound to the NM even after 60 min of treatment with DNase I. This fraction corresponds to the DNA that is actually embedded within the NM, and so it is rather inaccessible to the limited concentration of DNase I used.
Figure 2.
Kinetics of nucleoid DNA digestion. Nucleoids were digested with DNase-I (0.5 U/ml). Each time-point value is the average of separate experiments with samples obtained from separate animals (n = 9 for hepatocytes, n = 4 for naïve B-lymphocytes). Bars indicate the corresponding SD. The topological zones relative to the NM correspond to decreasing percentages of total DNA bound to the NM. (A) For hepatocytes: distal to NM (100–42% total DNA), proximal to NM (42–30% total DNA), very close to NM (30–19% total DNA) and embedded within NM (19–0% total DNA). The corresponding slopes are the following: 0–5 min = −9.80; 5–15 min = −1.70; 15–30 min = −0.47 and 30–60 min = −0.17. Hence, the slope of the digestion curve became close to zero by 60 min of DNase I treatment and remained like that even after 120 min of incubation with the enzyme (slope 60–120 min = −0.17). (B) For naïve B-lymphocytes: distal to NM (100–33% total DNA), proximal to NM (33–7% total DNA), very close to NM (7–4% total DNA) and embedded within NM (4–0% total DNA). The corresponding slopes are the following: 0–5 min = −11.46; 5–15 min = −2.84; 15–30 min = −0.61 and 30–60 min = −0.06 (slope 60–120 min = −0.03). Hence, the slope of the digestion curve also became close to zero by 60 min of DNase I treatment.
Nucleoids from naïve B-lymphocyte treated with the same limited concentration of DNase I display a faster kinetics of loop-DNA digestion, and so after 60 min of treatment with the enzyme, the amount of total DNA embedded within the NM is very small (≤4.0%), yet the digestion of further loop DNA stalls and becomes negligible afterwards (Fig. 2B). The average DNA loop size in naïve B-lymphocytes is rather similar to that in hepatocytes, yet the DNA–NM interactions are more fragile in B-lymphocytes when compared with hepatocytes, and comparative PAGE analysis shows that the relative amount of typical NM proteins is significantly reduced in naïve B-lymphocytes, in particular the three nuclear lamins (A, B and C) that are major components of standard NM preparations (our unpublished results). Hence, the presence of a less dense NM in the nucleoids from naïve B-lymphocytes explains why the fraction of total DNA that is relatively protected from DNase I action is significantly smaller than that in nucleoids from hepatocytes. Depending on the cell type of the nucleoids studied, preliminary experiments must be carried out so as to define the concentration of DNase I suitable to produce consistent tri-phasic digestion curves (Fig. 2A and B). On the basis of our experience working with several cell lines31 and different types of primary cells, the range of DNase I concentrations to be considered is between 1.0 and 0.05 U/ml. Indeed, extensive damage to DNA due to incorrect nucleoid preparation and handling, or the natural presence of nicked DNA in some terminally differentiated cell types (such as non-naïve, activated lymphocytes) produce bi-phasic nucleoid-DNA digestion curves since the loop-DNA supercoiling is either absent or severely reduced. Nevertheless, in all nucleoid preparations treated with a limited amount of DNase I, there is a fraction of total DNA that is resistant to the enzyme by being embedded within the NM, such a DNA is easily identifiable in the digestion curves as the fraction that is digested very slowly so that the local slope becomes close to zero and remains like that even after very long incubation times (Fig. 2A and B).
3.3. Mapping the relative position to the NM of DNA sequences along the genomic region containing the rat albumin locus
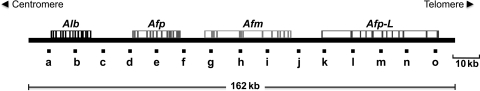
On the basis of the above considerations and results, we used the general strategy depicted in Fig. 3 in order to map the position relative to the NM of 15 small DNA sequences located along a region corresponding to 162 kb of chromosome 14 in the rat, which includes the loci of the following gene members of the albumin gene family: albumin (Alb), alpha-fetoprotein (Afp), afamin (Afm) and the pseudo-gene similar to Afp (Afp-L). Such small DNA sequences were spaced some 10 kb apart (Fig. 4). Therefore, nucleoids prepared from freshly isolated hepatocytes or naïve B-lymphocytes (Fig. 3A) were incubated with DNase I (0.5 U/ml) for different times so as to obtain nucleoid samples with differential amounts of DNA associated with the NM (Fig. 3B and C), and such samples with partially digested NM-bound DNA were used for PCR amplification of the chosen target sequences located along the 162-kb genomic region under study (Fig. 3A). By correlating the amplification data (Fig. 3A) with the kinetics of the nucleoid-DNA digestion (Fig. 3B), it is possible to locate each target sequence within a topological zone relative to the NM (Fig. 2). Previous studies have shown that the average size of the nuclear DNA fragments liberated by non-specific nucleases in rat hepatocytes is 0.8 kb.12 Thus, the DNA sequences to be mapped are <550 bp in length (Table 1), and so likely to be cut as whole units by the endonuclease instead of being progressively eroded by partial digestions. Therefore, in our mapping protocol, we score the specific templates as either present (amplifiable) or absent (non-amplifiable) as a function of endonuclease-digestion time (Fig. 5 and Supplementary Fig. S1). This was scored without considering the intensity of the amplicon signals, but just whether such signals are detected or not by an image-analysis program (Kodak 1D Image Analysis Software 3.5), using the default settings. We established these criteria because in our topological-mapping approach, it is the average relative position to the NM anchoring point and not the actual template length the critical parameter that determines the average sensitivity to DNase I of each sequence mapped. Nevertheless, the chosen amplicons were also selected on the basis that all of them may be amplified with similar efficiency using the same amplification program (see Section 2). Thus, the absence of amplified product at a given digestion time-point indicates that the relative abundance of the target template has fallen to a non-amplifiable level within the large nucleoid population analysed in each sample.6
Figure 3.
Procedure for mapping the position of DNA sequences relative to the NM. (A) Nucleoids prepared from freshly isolated rat primary cells were incubated with DNase I so as to progressively digest the loop DNA, obtaining a tri-phasic kinetics of digestion (B). Nucleoid samples with partially digested NM-bound DNA, as shown in the drawing and fluorescent micrographs, (C) were used for PCR amplification of target DNA sequences located along the genomic region containing members of the rat albumin gene family. The amplicons were run in agarose gels and scored as present (+) or absent (−) by an image-analysis software (A) as a function of nucleoid-sample digestion time (C).
Figure 4.
The 162-kb genomic region containing members of the rat albumin gene family (chromosome 14): Alb (albumin), Afp (alpha-fetoprotein), Afm (afamin), Afp-L (pseudo-gene similar to Afp). The letters indicate the location of the target DNA sequences to be positionally mapped relative to the NM.
Figure 5.
Positional mapping relative to the NM of specific target DNA sequences by PCR. Rat-hepatocyte nucleoids were treated with DNase-I (0.5 U/ml) for different times. The residual NM-bound DNA was directly used as template for PCR amplification of the target sequences (a–o). The specific amplicons were resolved in 2% agarose gels stained with ethidium bromide (0.5 µl/ml). C, control. Topological zones with respect to NM: D, distal; P, proximal; VC, very close; E, embedded within NM. (−), negative control (no template); (+), positive control (pure genomic DNA as template). The amplification patterns were consistently reproduced in separate experiments with samples from independent animals (n = 4).
It is important to emphasize that the method described here is not intended for identification and characterization of actual MARs (LARs) in a given genomic region. Indeed, some general properties of MARs such as the presence of A-T tracts and repetitive sequences make them unsuitable for efficient PCR amplification under stringent conditions for achieving highly specific amplification of most other non-repetitive genomic sequences. Thus, primer-design programs will grant a low score to any set of primers targeting amplicons containing potential MARs, even more if such amplicons are meant to be highly specific and small-sized (≤500 bp), as those targeted in the present protocol.
3.4. Determination of the structural DNA loop organization in the region of the rat albumin locus
By coupling the highly reproducible amplification experiments (Fig. 5 and Supplementary Fig. S1), together with the consistent kinetics of loop DNA digestion by DNase I (Fig. 2), we were able to locate each mapped sequence within a given topological zone relative to the NM (Tables 2 and 3). Such data were used to draw the most likely DNA loop organization relative to the NM of the 162-kb region studied, by considering the distance (in kb) between the separate sequences mapped and the topological positions of such sequences relative to the NM. The data suggest that in the hepatocytes, the region is organized in five loops anchored to the NM (Fig. 6A). The resulting size from tip to base of such loops is not bigger than 20 kb; thus, the largest loop of the set, containing both Afp and Afm sequences, would be some 40 kb in length. The genomic region studied is a high-gene-density region. Through evolution, a series of duplications formed the genes within the Alb family that includes the vitamin D-binding protein (DBP) gene located almost 1 Mb upstream of the Alb gene.42 Nevertheless, such a gene family has been travelling as a cluster within the mammalian genomes. The fact that the region studied is organized into five rather compact structural DNA loops whose size (40–20 kb) is smaller than the estimated average rat-hepatocyte DNA loop size (80 kb) may be relevant to the functionality of such a genomic region, since fundamental processes of nuclear physiology occur at macromolecular complexes organized on the NM.17–19 It remains to be explored whether high-gene-density regions are usually organized into relatively small-sized structural DNA loops; at least another report suggests that such may be the case.26 However, despite the fact that the average DNA loop size in naïve B-lymphocytes is quite similar to that in hepatocytes (our unpublished results), in naïve B-lymphocytes the corresponding 162 kb region is very close to the NM as most sequences map within the VC topological region. Moreover, the target sequence h consistently maps to the DNase-resistant fraction of NM-bound DNA, and so it must be embedded within the NM. Only the l sequence remains proximal to the NM (Table 3 and Supplementary Fig. S1). This means that in naïve B-lymphocytes the 162-kb region is organized in numerous but tiny (<14 kb) DNA loops (Table 3 and Fig. 6B). This is quite interesting since the genes within the region studied are not known to be expressed in B-lymphocytes as it was confirmed by RT–PCR (data not shown), suggesting that packaging of genes in truly small DNA loops may be correlated with the stable transcriptional silencing of such genes (see below).
Table 2.
Location of the target sequences (amplicons) in the specific topological zones relative to the NM from rat hepatocytes
| Amplicon | Topological zones relative to the NM |
|||
|---|---|---|---|---|
| Distal to NM | Proximal to NM | Very close to NM | Embedded within NM | |
| a | + | + | − | − |
| b | + | − | − | − |
| c | + | − | − | − |
| d | + | + | − | − |
| e | + | − | − | − |
| f | + | − | − | − |
| g | + | − | − | − |
| h | + | + | − | − |
| i | + | − | − | − |
| j | + | − | − | − |
| k | + | + | − | − |
| l | + | − | − | − |
| m | + | + | − | − |
| n | + | − | − | − |
| o | + | + | − | − |
The amplicons were scored either as positive or as negative as a function of nuclease digestion time and for each topological zone relative to the NM, depending on whether or not they were detected by a digital image-analysis system (Kodak 1D Image Analysis Software 3.5) using the default settings.
Table 3.
Location of the target sequences (amplicons) in the specific topological zones relative to the NM in naïve B-lymphocytes
| Amplicon | Topological zones relative to NM in B-lymphocytes |
|||
|---|---|---|---|---|
| Distal to NM | Proximal to NM | Very close to NM | Embedded within NM | |
| a | + | + | + | − |
| b | + | + | + | − |
| c | + | + | + | − |
| d | + | + | + | − |
| e | + | + | + | − |
| f | + | + | + | − |
| g | + | + | + | − |
| h | + | + | + | + |
| i | + | + | + | − |
| j | + | + | + | − |
| k | + | + | + | − |
| l | + | + | − | − |
| m | + | + | + | − |
| n | + | + | + | − |
| o | + | + | + | − |
The amplicons were scored either as positive or as negative as a function of nuclease digestion time and for each topological zone relative to the NM, depending on whether or not they were detected by a digital image-analysis system (Kodak 1D Image Analysis Software 3.5) using the default settings.
Figure 6.
Experimentally determined structural DNA loop organization of the 162-kb region containing members of the rat albumin gene family in hepatocytes (A) and naïve B-lymphocytes (B). The letters indicate the position of the target sequences within the DNA loops. The bold lines indicate the position of the whole named genes. The dashed lines indicate projected loop regions outside of the region studied. The left bar indicates the relative DNA loop size from tip to base; hence the whole DNA loop length is approximately the double of such value. The right bar indicates the topological zones relative to the NM according to the kinetics of nucleoid DNA digestion. The illustration is according to scale in kb along the x and y axis.
3.5. Predicted MARs in silico do not fully correspond to actual MARs in vivo
We used four different available computer programs for predicting the location of potential MARs along the 162-kb region studied. All these programs have been quoted as MAR-prediction or even as MAR-identifying devices in several published papers.27,28 However, each program uses different criteria so as to define the respective MAR-searching algorithms. This can be appreciated as not all programs coincide in their predictions (Table 4). Moreover, the mapping results in hepatocytes indicate that loop-DNA regions containing actual MARs (LARs) are located next to the following sequences mapped: a, d, h, k, m, o. However, only the region next to the d sequence is consistently identified as a MAR-containing region by the four programs, whereas the other experimentally defined LAR-containing regions are only partially identified by some of the four MAR-searching programs used and all programs predict rather inconsistently the presence of MARs in regions that according to the experimental results are unlikely to be bound to the NM in hepatocytes (Supplementary Fig. S2). The programs detect potential MARs close to most of the sequences mapped along the region studied, but even sequences such as b and c that lack any program-identified MAR nearby (Supplementary Fig. S2) are located very close to the NM in naïve B-lymphocytes, suggesting that there are actual MARs (LARs) that cannot be detected by any of the MAR-identifying programs. Only the sequence l that also lacks any program-identified MAR nearby is consistently far from the NM in naïve B-lymphocytes. These results suggest that MAR-finding programs are highly unreliable tools for actual MAR identification, a fact that has already been shown by others.28
Table 4.
Location of potential MARs in the 162-kb genomic region studied as predicted by four different MAR-searching computer programs
| MarFinder |
MARScan |
ChrClass |
SMARtest |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Start | End | Length (bp) | Start | End | Length (bp) | Start | End | Length (bp) | Start | End | Length (bp) |
| 36 380 | 36 580 | 201 | 1762 | 1824 | 63 | 5035 | 5595 | 561 | 36 686 | 37 120 | 435 |
| 62 080 | 62 580 | 501 | 2980 | 3002 | 23 | 35 884 | 36 384 | 501 | 47 786 | 48 090 | 305 |
| 76 480 | 76 880 | 401 | 36 939 | 37 003 | 65 | 51 284 | 51 584 | 301 | 51 306 | 51 640 | 335 |
| 88 480 | 89 580 | 1101 | 55 573 | 55 589 | 17 | 59 884 | 60 284 | 401 | 78 281 | 78 580 | 300 |
| 102 380 | 102 980 | 601 | 69 392 | 69 513 | 122 | 88 984 | 89 884 | 901 | 88 811 | 89 300 | 490 |
| 141 901 | 142 601 | 701 | 84 791 | 84 806 | 16 | 90 784 | 91 084 | 301 | 108 916 | 109 295 | 380 |
| 88 179 | 88 232 | 54 | 102 561 | 102 861 | 301 | 138 866 | 139 280 | 415 | |||
| 102 643 | 102 669 | 27 | 141 301 | 141 801 | 501 | 141 341 | 141 650 | 310 | |||
| 131 451 | 131 487 | 37 | 142 101 | 142 601 | 501 | 142 106 | 142 570 | 465 | |||
| 142 475 | 142 491 | 17 | 153 801 | 154 601 | 801 | ||||||
| 142 773 | 142 788 | 16 | |||||||||
The numbers indicate the nucleotide start and ending position number along the genomic region studied. The length of the predicted MARs is indicated in bp.
3.6. Conclusions
The method described in this paper makes it possible to determine the organization in structural DNA loops of any long segment of genomic DNA with known sequence, provided that such a DNA is attached to the NM as is the case in nucleoid preparations of freshly isolated hepatocytes and naïve B-lymphocytes. The procedure used for isolating the nucleoids only preserves DNA–NM interactions that are resistant to high-salt extraction, therefore, facultative non-structural attachments to the NM that may result from some transient functional process such as active transcription are eliminated.43 Our method exploits the topological properties of naked DNA attached to the NM as well as elementary topological principles such as that points in a deformable string (DNA) can be positionally mapped relative to a position-reference invariant (NM), and from such mapping, it can be deduced the configuration of the string in third dimension. Moreover, both the intrinsic persistence length of mixed-sequence DNA, estimated at some 240 bp, and negative supercoiling confer relative rigidity to the naked DNA loops,36 thus limiting their deformability in such a way that the determined positions of loop-DNA sequences relative to the NM are highly reproducible among experiments, indicating limited fluctuation of the DNA loop morphology over time. Nevertheless, some sequences that happen to lie close to the boundary between two topological zones may display some variability in their relative position to the NM, such is the case of sequence e in hepatocytes that in one of four experiments it was detected in the proximal zone, whereas in three experiments it was only detectable in the distal zone relative to the NM (Fig. 5). It must be stressed that the presence of the corresponding amplicons in a given topological zone was determined by an image-analysis system using its default settings independently of the apparent intensity of the amplicon signal in the corresponding gel photograph. Thus, for example, in one of four experiments with hepatocytes, the k amplicon was barely apparent in the proximal region; nevertheless, it was consistently detected in such a region by the image-analysis software in the four separate experiments (Fig. 5).
The present protocol works with NM-bound DNA templates and so the actual MARs (LARs) present in such preparations are deeply embedded within the NM, thus being poorly available to the action of any externally added enzyme. Most LAR-identifying protocols depend on the proteolytic destruction of the NM so as to liberate the NM-bound DNA fragments that may be further characterized by a number of techniques.22,24,26,30,44 Thus, our protocol addresses the problem of determining the configuration of structural DNA loops in a given, large enough genomic region without the need of mapping or characterizing the actual MARs (LARs) present in such a region as a precondition for determining the possible loop configuration. Nevertheless, our protocol delimits the genomic regions where such LARs are more likely to be found. Yet, pinpointing the actual extension of such LARs depends on applying further techniques beyond the present protocol. So far, there is only one published protocol that directly maps LARs on NM-bound DNA; this protocol depends on the activation of endogenous, NM-bound topoisomerase II that releases DNA loops by cutting along MARs embedded within the NM.23,45
The in vivo, local spatial configuration of DNA into structural loops, as determined by the present protocol, constitutes the starting point for any further refinement or modification of such a configuration by chromatin proteins and epigenetic mechanisms affecting chromatin structure. However, chromatin as such plays no role in determining the structural DNA loop organization since chromatin proteins are completely eliminated by the high-salt extraction, and yet stable DNA–NM interactions persist. In any case, the organization of the genome into structural DNA loops must be highly relevant for the nuclear physiology. Indeed, current evidence suggest that the structural DNA loops may correspond to the actual replicons,5,6,19,39 yet the pattern of such structural loops may also determine the limits of further local structural changes that may be related to transient chromatin modifications resulting in functional interactions (related to transcription) between loop DNA and protein complexes located upon the NM.18,46,47 The genomic region studied contains classical examples of developmentally regulated genes. Indeed, Alb, Afp and Afm are primarily expressed in the liver, yet while Afp and Alb are highly expressed in the fetal liver, the transcription of Afp is rapidly reduced after birth, but Alb and Afm continue to be expressed in the adult liver.42,48 There are three enhancers in the intergenic region between Alb and Afp whose action is required for Afp and Alb activation early in liver development and for Afp reactivation during liver regeneration, yet later during fetal development Alb expression becomes independent of such enhancers due to the action of an Alb-specific enhancer that lies between 8.5 and 10 kb upstream the Alb transcription-start site.42,49 Interestingly, the intergenic enhancers are located in the region between the sequences corresponding to amplicons c and d48 which is clearly detached from the NM in the adult rat (Fig. 6A). We actually mapped the position relative to the NM of enhancer 1 (E1) which is located 2 kb upstream of sequence d that belongs to the Afp promoter region (Fig. 4). E1 mapped within the region proximal to the NM (data not shown) the same as d (Table 2). So far the Alb-specific enhancer has not been formally characterized in the rat. However, comparison of the mouse Alb-enhancer sequence indicates that a closely related sequence is present in the rat in a 1.3-kb region located 8.3 kb upstream of the Alb transcription-start site. Our results suggest the presence of a LAR some 5 kb upstream of sequence a that belongs to the Alb promoter (Figs 4 and 6A and Supplementary Fig. S2). Hence the putative rat Alb-enhancer is located some 2 kb upstream of the possible LAR and so the enhancer could be very close to the NM, although this needs to be experimentally verified.
It is known that replication and transcription occur at factories organized upon the NM.18,50 However, replication and transcription factories never co-localize since they occupy distinct but rather fixed sites that are conserved throughout several cell generations.17,51,52 In mammalian lymphocytes it is known that genes separated by tens of mega-bases in cis and genes that belong to separate chromosomes somehow migrate to preassembled transcription factories for their actual transcription.50,53 Such a process requires that the genes undergoing active transcription loop out away from their respective chromosome territories so that distant genomic regions are brought together within one nuclear complex for transcription.54 Hence the fact that in naïve B-lymphocytes the genomic region studied is very close to the NM, suggests that the genes present in the region are prevented from looping out towards a transcription factory, since the genes are included into numerous tiny loops tightly bound to the NM (Fig. 6B).
The high-salt-resistant LARs that determine structural attachments to the NM constitute long-term interactions between DNA and the NM. Nevertheless, as predicted by some models, there is evidence that under the influence of a combination of mechanical, biochemical and thermodynamical factors the pattern of such attachments may be modified during development and aging leading to stable changes in cell differentiation.10,13,55,56 Varied and important evidence suggests that the high-salt resistant structural DNA loops correspond to the actual replicons in vivo.5,19 Thus it is remarkable that in totipotent blastomeres the cell cycle consists of overlapping S and M phases with no G1 or G2, the rapid DNA replication observed involves the use of high numbers of closely spaced attachments between DNA and the NM and so the average DNA loops are very small, yet this is compatible with very active DNA replication but not with active transcription as genes in the chromosomes of blastomeres are largely inactive during cleavage.57 Therefore in naïve B-lymphocytes the presence of tiny loops in the region studied may contribute to the transcriptional silencing of the corresponding genes without preventing such a region from being replicated after B-lymphocyte activation.
The nucleotype has been defined as those characters of nuclear DNA that may affect the phenotype independently of the information encoded in such a DNA,58 and it has been suggested that besides species-specific nucleotypic characters such as karyotype, and the C-value for nuclear DNA that correlates with overall cell-cycle length, there could be tissue-specific nucleotypic characters, such as the pattern of structural DNA loops, that correlate with cell differentiation.10 Our results support the existence of such tissue specific nucleotypic characters.
Supplementary Data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Funding
This work was sponsored by CONACYT, Mexico, grant 48447-Q (25506) and UAEM, Mexico, grant 2212/2006, both awarded to A.A.-A. J.C.R.-M. is a CONACYT Research Scholar within the Graduate Program in Biomedical Sciences at IFC-UNAM, México.
Supplementary Material
Footnotes
Edited by Mitsuo Oshimura
References
- 1.Nickerson J.A. Experimental observations of a nuclear matrix. J. Cell Sci. 2001;114:463–74. doi: 10.1242/jcs.114.3.463. [DOI] [PubMed] [Google Scholar]
- 2.Tsutsui K.M., Sano K., Tsutsui K. Dynamic view of the nuclear matrix. Acta Med. Okayama. 2005;59:113–20. doi: 10.18926/AMO/31953. [DOI] [PubMed] [Google Scholar]
- 3.Mika S., Rost B. NMPdb: database of nuclear matrix proteins. Nucleic Acids Res. 2005;33:D160–3. doi: 10.1093/nar/gki132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ottaviani D., Lever E., Takousis P., Sheer D. Anchoring the genome. Genome Biol. 2008;9:2011. doi: 10.1186/gb-2008-9-1-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Razin S.V. The nuclear matrix and chromosomal DNA loops: is there any correlation between partitioning of the genome into loops and functional domains? Cell Mol. Biol. Lett. 2001;6:59–69. [PubMed] [Google Scholar]
- 6.Maya-Mendoza A., Hernández-Muñoz R., Gariglio P., Aranda-Anzaldo A. Gene positional changes relative to the nuclear substructure correlate with the proliferating status of hepatocytes during liver regeneration. Nucleic Acids Res. 2003;31:6168–79. doi: 10.1093/nar/gkg825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heng H.H.Q., Goetze S., Ye J.C., et al. Chromatin loops are selectively anchored using scaffold/matrix-attachment regions. J. Cell Sci. 2004;117:999–1008. doi: 10.1242/jcs.00976. [DOI] [PubMed] [Google Scholar]
- 8.Petrova N.V., Iarovaia O.V., Verbovoy V.A., Razin S.V. Specific radial positions of human chromosomes X, 1 and 19 remain unchanged in chromatin depleted nuclei of primary human fibroblasts: evidence for the organizing role of the nuclear matrix. J. Cell. Biochem. 2005;96:850–7. doi: 10.1002/jcb.20592. [DOI] [PubMed] [Google Scholar]
- 9.Berezney R. Dynamic properties of the nuclear matrix. In: Busch H., editor. The Cell Nucleus. Orlando: Academic Press; 1979. pp. 413–56. [Google Scholar]
- 10.Aranda-Anzaldo A. On the role of chromatin higher-order structure and mechanical interactions in the regulation of gene expression. Speculations Sci. Technol. 1989;12:163–76. [Google Scholar]
- 11.Hakes D.J., Berezney R. DNA binding properties of the nuclear matrix and individual nuclear matrix proteins. J. Biol. Chem. 1991;266:11131–40. [PubMed] [Google Scholar]
- 12.Berezney R., Buchholtz L.A. Dynamic association of replicating DNA fragments with the nuclear matrix of regenerating liver. Exp. Cell Res. 1981;132:1–13. doi: 10.1016/0014-4827(81)90076-8. [DOI] [PubMed] [Google Scholar]
- 13.Maya-Mendoza A., Hernández-Muñoz R., Gariglio P., Aranda-Anzaldo A. Natural ageing in the rat liver correlates with progressive stabilisation of DNA-nuclear matrix interactions and withdrawal of genes from the nuclear substructure. Mech. Ageing Dev. 2005;126:767–82. doi: 10.1016/j.mad.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 14.Bode J., Ríos-Ramírez M., Mielke C., Stengert M., Kay V., Klehr-Wirth D. Scaffold/matrix-attached regions: structural properties creating transcriptionally active loci. Int. Rev. Cytol. 1995;162A:389–454. doi: 10.1016/s0074-7696(08)61235-8. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y., Xi Z., Hedge R.S., Shakked Z., Crothers D.M. Predicting indirect readout effects in protein-DNA interactions. Proc. Natl Acad. Sci. USA. 2004;101:8337–41. doi: 10.1073/pnas.0402319101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsutsui K., Tsutsui K., Muller M.T. The nuclear scaffold exhibits DNA-binding sites selective for supercoiled DNA. J. Biol. Chem. 1988;263:7235–41. [PubMed] [Google Scholar]
- 17.Wei X., Samarabandu J., Devdhar R.S., Siegel A.J., Acharya R., Berezney R. Segregation of transcription and replication sites into higher order domains. Science. 1998;281:1502–5. doi: 10.1126/science.281.5382.1502. [DOI] [PubMed] [Google Scholar]
- 18.Cook P.R. The organization of replication and transcription. Science. 1999;282:1790–5. doi: 10.1126/science.284.5421.1790. [DOI] [PubMed] [Google Scholar]
- 19.Anachkova B., Djeliova V., Russev G. Nuclear matrix support of DNA replication. J. Cell Biochem. 2005;96:951–61. doi: 10.1002/jcb.20610. [DOI] [PubMed] [Google Scholar]
- 20.Aranda-Anzaldo A., Dent M.A.R. Loss of DNA-loop supercoiling and organization in cells infected by herpes simplex virus type 1. Res. Virol. 1997;148:397–408. doi: 10.1016/s0923-2516(97)83629-1. [DOI] [PubMed] [Google Scholar]
- 21.Aranda-Anzaldo A., Orozco-Velasco F., García-Villa E., Gariglio P. p53 is a rate-limiting factor in the repair of higher-order DNA structure. Biochim. Biophys. Acta. 1999;1446:181–92. doi: 10.1016/s0167-4781(99)00086-x. [DOI] [PubMed] [Google Scholar]
- 22.Gombert W.M., Farris S.D., Rubio E.D., Morey-Rosler K.M., Schubach W.H., Krumm A. The c-myc insulator element and matrix attachment region define the c-myc chromosomal domain. Mol. Cell Biol. 2003;23:9338–48. doi: 10.1128/MCB.23.24.9338-9348.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iarovaia O.V., Bystritsky A., Ravcheev D., Hancock R., Razin S.V. Visualization of individual DNA loops and a map of loop domains in the human dystrophin gene. Nucleic Acids Res. 2004;32:2079–86. doi: 10.1093/nar/gkh532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petrov A., Pirozhkova I., Carnac G., Laoudj D., Lipinski M., Vassetzky Y.S. Chromatin loop domain organization within the 4q35 locus in facioscapulohumeral dystrophy patients versus normal human myoblasts. Proc. Natl Acad. Sci. USA. 2006;103:6982–7. doi: 10.1073/pnas.0511235103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hair A., Vassetzky Y. Determination of the chromatin domain structure in arrayed repeated regions: organization of the somatic 5S RNA domain during embryogenesis in Xenopus laevis. J. Cell Biochem. 2007;102:1140–8. doi: 10.1002/jcb.21413. [DOI] [PubMed] [Google Scholar]
- 26.Kumar P., Bischof O., Purbey P., et al. Functional interaction between PML and SATB1 regulates chromatin-loop architecture and transcription of the MHC class 1 locus. Nat. Cell Biol. 2007;9:45–56. doi: 10.1038/ncb1516. [DOI] [PubMed] [Google Scholar]
- 27.Frisch M., Frech K., Klingenhoff A., Cartharius K., Liebich I., Werner T. In silico prediction of scaffold/matrix attachment regions in large genomic sequences. Genome Res. 2002;12:349–54. doi: 10.1101/gr.206602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Evans K., Ott S., Hansen A., Koentges G., Wernisch L. A comparative study of S/MAR prediction tools. BMC Bioinformatics. 2007;8:71. doi: 10.1186/1471-2105-8-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ottaviani D., Lever E., Mitter R., et al. Reconfiguration of genomic anchors upon transcriptional activation of the human major histocompatibility complex. Genome Res. 2008;18:1778–1786. doi: 10.1101/gr.082313.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Linnemann A.K., Platts A.E., Krawetz S.A. Differential nuclear scaffold/matrix attachment marks expressed genes. Hum. Mol. Genet. 2009;18:645–54. doi: 10.1093/hmg/ddn394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maya-Mendoza A., Aranda-Anzaldo A. Positional mapping of specific DNA sequences relative to the nuclear substructure by direct polymerase chain reaction on nuclear matrix-bound templates. Anal. Biochem. 2003;313:196–207. doi: 10.1016/s0003-2697(02)00611-5. [DOI] [PubMed] [Google Scholar]
- 32.Singh G.B., Kramer J.A., Krawetz S.A. Mathematical model to predict regions of chromatin attachment to the nuclear matrix. Nucleic Acids Res. 1997;25:1419–25. doi: 10.1093/nar/25.7.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Drunen C.M., Sewalt R.G.A.B., Oosterling R.W., Weisbeek P.J., Smeekens S.C.M., van Driel R. A bipartite sequence element associated with matrix/scaffold attachment regions. Nucleic Acids Res. 1999;27:2924–30. doi: 10.1093/nar/27.14.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Glazko G.V., Rogozin I.B., Glazkov M.V. Comparative study and prediction of DNA fragments associated with various elements of the nuclear matrix. Biochim. Biophys. Acta. 2001;1517:351–64. doi: 10.1016/s0167-4781(00)00297-9. [DOI] [PubMed] [Google Scholar]
- 35.Nelson D.L., Cox M.M. Lehninger Principles of Biochemistry. 3rd edition. New York: Worth Publishers; 2000. pp. 915–8. [Google Scholar]
- 36.Calladine C.R., Drew H.R., Luisi B.F., Travers A.A. Understanding DNA. 3rd edition. London: Elsevier-Academic Press; 2004. pp. 94–138. [Google Scholar]
- 37.Bode J., Kowhi Y., Dickinson L., et al. Biological significance of unwinding capability of nuclear matrix-associating DNAs. Science. 1992;255:195–7. doi: 10.1126/science.1553545. [DOI] [PubMed] [Google Scholar]
- 38.Benham C., Kohwi-Shigematsu T., Bode J. Stress-induced duplex destabilization in scaffold/matrix attachment regions. J. Mol. Biol. 1997;2774:181–96. doi: 10.1006/jmbi.1997.1385. [DOI] [PubMed] [Google Scholar]
- 39.Maya-Mendoza A., Hernández-Muñoz R., Gariglio P., Aranda-Anzaldo A. Gene positional changes relative to the nuclear substructure during carbon tetrachloride-induced hepatic fibrosis in rats. J. Cell Biochem. 2004;93:1084–98. doi: 10.1002/jcb.20264. [DOI] [PubMed] [Google Scholar]
- 40.Lewin B. Gene Expression 2. 2nd edition. New York: John Wiley and Sons; 1980. pp. 360–2. [Google Scholar]
- 41.Gromova I.I., Nielsen O.F., Razin S.V. Long-range fragmentation of the eukaryotic genome by exogenous and endogenous nucleases proceeds in a specific fashion via preferential DNA cleavage at matrix attachment sites. J. Biol. Chem. 1995;270:18685–90. doi: 10.1074/jbc.270.31.18685. [DOI] [PubMed] [Google Scholar]
- 42.Jin L., Long L., Green M.A., Spear B.T. The alpha-fetoprotein enhancer region activates the albumin and alpha-fetoprotein promoters during liver development. Dev. Biol. 2009 doi: 10.1016/j.ydbio.2009.09.026. doi:10.1016/j.dbio.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Razin S.V. DNA interaction with the nuclear matrix and spatial organization of replication and transcription. BioEssays. 1987;6:19–23. doi: 10.1002/bies.950060106. [DOI] [PubMed] [Google Scholar]
- 44.Rollini P., Namciu S.J., Marsden M.D., Fournier R.E.K. Identification and characterization of nuclear matrix-attachment regions in the human serpin gene cluster at 14q32.1. Nucleic Acids Res. 1999;27:3779–91. doi: 10.1093/nar/27.19.3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gromova I.I., Thomsen B., Razin S.V. Different topoisomerase II antitumor drugs direct similar specific long-range fragmentation of an amplified c-myc gene locus in living cells and in high-salt-extracted nuclei. Proc. Natl Acad. Sci. USA. 1995;92:102–6. doi: 10.1073/pnas.92.1.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davie J.R. Histone modifications, chromatin structure and the nuclear matrix. J. Cell Biochem. 1996;62:149–57. doi: 10.1002/(sici)1097-4644(199608)62:2<149::aid-jcb2>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 47.Kantidze O.L., Razin S.V. Chromatin loops, illegitimate recombination and genome evolution. BioEssays. 2009;31:278–86. doi: 10.1002/bies.200800165. [DOI] [PubMed] [Google Scholar]
- 48.Lazarevich N.L. Molecular mechanisms of alpha-fetoprotein gene expression. Biochemistry (Mosc.) 2000;65:117–33. [PubMed] [Google Scholar]
- 49.Pinkert C.A., Ornitz D.M., Brinster R.L., Palmiter R.D. An albumin enhancer located 10 kb upstream functions along with its promoter to direct efficient, liver-specific expression in transgenic mice. Genes Dev. 1987;1:268–76. doi: 10.1101/gad.1.3.268. [DOI] [PubMed] [Google Scholar]
- 50.Mitchell J.A., Fraser P. Transcription factories are nuclear subcompartments that remain in the absence of transcription. Genes Dev. 2008;22:20–5. doi: 10.1101/gad.454008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jackson D.A., Pombo A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J. Cell Biol. 1998;140:1285–95. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dimitrova D.S., Berezney R. The spatio-temporal organization of DNA replication sites is identical in primary, immortalized and transformed mammalian cells. J. Cell Sci. 2002;115:4037–51. doi: 10.1242/jcs.00087. [DOI] [PubMed] [Google Scholar]
- 53.Osborne C.S., Chakalova L., Mitchell J.A., et al. Myc dynamically and preferentially relocates to a transcription factory occupied by Igh. PLoS Biol. 2007;5:e192. doi: 10.1371/journal.pbio.0050192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Linnemann A.K., Krawetz S.A. Maintenance of a functional higher order chromatin structure: the role of the nuclear matrix in normal and disease states. Gene Ther. Mol. Biol. 2009;13:231–43. [PMC free article] [PubMed] [Google Scholar]
- 55.Martínez-Ramos I., Maya-Mendoza A., Gariglio P., Aranda-Anzaldo A. A global but stable change in HeLa cell morphology induces reorganization of DNA structural loop domains within the cell nucleus. J. Cell Biochem. 2005;96:79–88. doi: 10.1002/jcb.20428. [DOI] [PubMed] [Google Scholar]
- 56.Aranda-Anzaldo A. A structural basis for cellular senescence. Aging. 2009;1:598–607. doi: 10.18632/aging.100074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lemaitre J-M., Danis E., Pasero P., Vassetzky Y., Mechali M. Mitotic remodelling of the replicon and chromosome structure. Cell. 2005;123:787–801. doi: 10.1016/j.cell.2005.08.045. [DOI] [PubMed] [Google Scholar]
- 58.Bennett M.D. The time and duration of meiosis. Philos. Trans. R. Soc. Lond. B, Biol. Sci. 1977;277:201–26. doi: 10.1098/rstb.1977.0012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.