Abstract
The current consensus model in mammalian olfaction is that the detection of millions of odorants requires a large number of odorant receptors (ORs) and that each OR interacts selectively with a small subset of odorants, which are typically related in structure. Here, we report the odorant response properties of an OR that deviates from this model: SR1, a mouse OR that is abundantly expressed in sensory neurons of the septal organ and also of the main olfactory epithelium. Patch-clamp recordings reveal that olfactory sensory neurons (OSNs) that express SR1 respond to many, structurally unrelated odorants, and over a wide concentration range. Most OSNs expressing a gene-targeted SR1 locus that lacks the SR1 coding sequence do not show this broad responsiveness. Gene transfer in the heterologous expression system Hana3A confirms the broad response profile of SR1. There may be other mouse ORs with such broad response profiles.
Introduction
The central tenet in mammalian olfaction is that the full spectrum of distinct molecular structures perceived by the olfactory system requires a large number of odorant receptors (ORs), each capable of interacting with a small number of odorous ligands (Axel, 2005). Olfactory perception initiates with the binding of odorants to ORs, which triggers a cAMP-dependent signal transduction cascade leading to action potential firing in the olfactory sensory neurons (OSNs) (Firestein, 2001). Each OSN is thought to express one functional OR, which defines both its response profile and its projection site in the olfactory bulb (Mombaerts, 2006). Since the cloning of mammalian ORs (Buck and Axel, 1991), the receptor–ligand relationships have been under extensive investigation. Progress has been made in the identification of putative chaperone molecules and other factors (Saito et al., 2004; Shirokova et al., 2005; Von Dannecker et al., 2006; Zhuang and Matsunami, 2007), which facilitate OR expression in heterologous cell lines. The consensus view is that ORs are used combinatorially to encode odor identities (Buck, 2005). Indeed, the best-studied mammalian ORs respond preferably to a small number of structurally related compounds from a list of up to a few hundred, such as rat I7 to octanal (Araneda et al., 2000), M71 to acetophenone (Bozza et al., 2002), mOR–EG to eugenol (Katada et al., 2005), and MOR23 to lyral (Touhara et al., 1999). The available data support the central tenet that mammalian ORs are relatively selective in their responses to odorants. In contrast, odorant receptors in the fruitfly Drosophila melanogaster display a diversity of response spectra, ranging from narrow to broad (Hallem and Carlson, 2006).
Here, we report that a mouse OR, SR1 (also known as MOR256-3 and Olfr124) has an unusually broad response profile. This OR is expressed ventrally in the main olfactory epithelium (MOE) and most notably at high density in the septal organ (SO) (Tian and Ma, 2004). The SO is a small island of olfactory epithelium at the base of the nasal septum (Rodolfo-Masera, 1943) and contains ∼10,000 OSNs. Nine ORs are expressed in >90% of SO–OSNs, with SR1 expressed in ∼50% of SO–OSNs (Tian and Ma, 2004). A single SO–OSN expresses one OR, with some exceptions (Tian and Ma, 2008). Using patch-clamp recordings in a novel strain of gene-targeted mice, we demonstrate that OSNs expressing SR1 are broadly responsive to various odorants and over a wide concentration range. A knockout of the coding region of SR1 disrupts this pattern, indicating that SR1 in and of itself—instead of the cells that choose to express SR1—is responsible for the broad responsiveness. We further confirm the broad response profile of SR1 in a heterologous expression system. The existence of broadly responsive ORs and broadly responsive OSNs has implications for the strategies of olfactory coding in mammals.
Materials and Methods
Gene targeting at the SR1 locus.
Similarly to a previous report (Cutforth et al., 2003), we generated two targeted mutations in the SR1 (MOR256-3, Olfr124) locus. A 13 kb BamHI fragment comprising the SR1 coding sequence was subcloned from bacterial artificial chromosome clone CT-350K7 (CITB-CJ7-B library, male CJ7/129SV; Research Genetics, Invitrogen) into pBluescript-SK(−), from which the internal HindIII restriction site had been eliminated. A 3.3 kb 5′ fragment, containing the outside probe, was removed by SpeI digestion resulting in a 9.9 kb SpeI/BamHI fragment with 6.3 kb and 3.6 kb homology arms for homologous recombination in embryonic stem cells. To generate the SR1–internal ribosome entry site (IRES)–tauGFP targeting vector, a 5 kb HindIII shuttle was subcloned, an AscI restriction site was engineered 3 nt downstream of the TAG stop codon, and an IRES–tauGFP–ACNF cassette was ligated to the AscI site (see Fig. 4A). To generate the ΔSR1–red fluorescent protein (RFP) targeting vector, the SR1 coding sequence was neatly replaced with a sequence coding for monomeric RFP1 by overlap PCR using PacI and AscI restriction sites, and a cassette containing ACNF was introduced at the AscI site (see Fig. 4A). Homologous recombination in E14 embryonic cells and blastocyst injection were performed as described previously (Mombaerts et al., 1996). Mice are in a mixed 129 × C57BL/6 background. These strains are publicly available from The Jackson Laboratory: SR1–IRES–tauGFP, strain name B6;129P2–Olfr124tm1Mom/MomJ, JAX #6717; ΔSR1–RFP, also referred to as mRFP → SR1, strain name B6;129P2–Olfr124tm4Mom/MomJ, JAX #6720.
Figure 4.
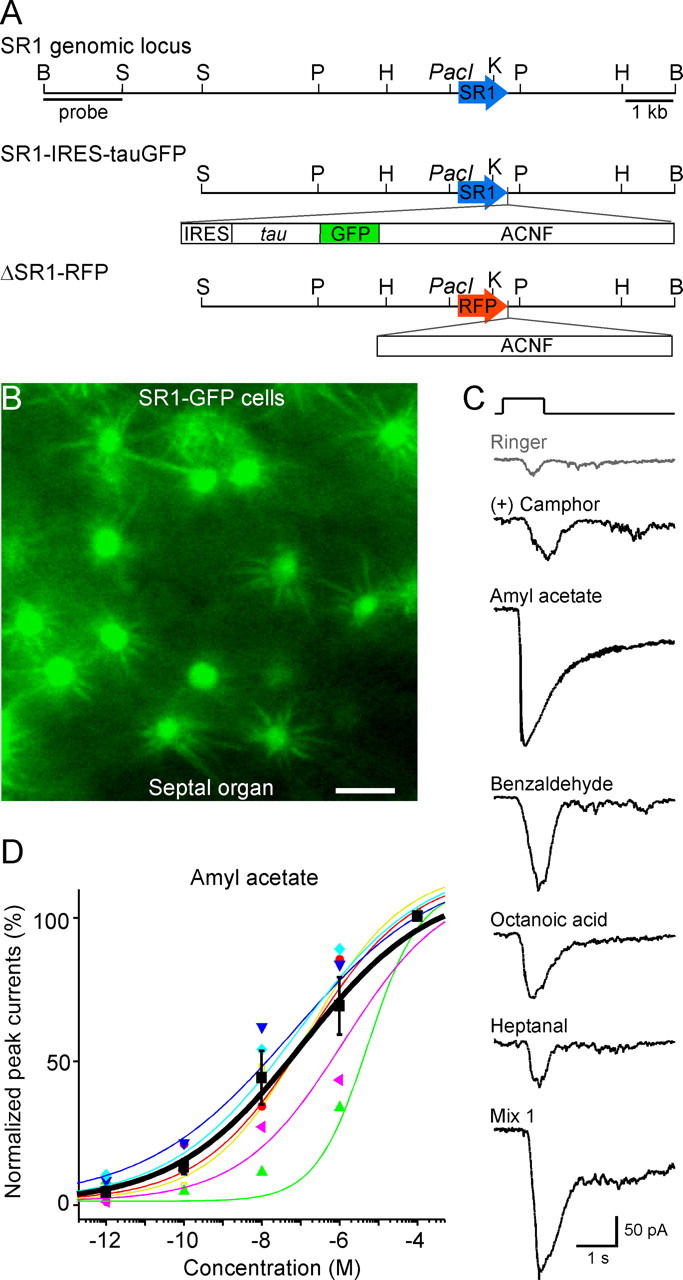
SR1–GFP cells in the SO respond broadly and over a wide concentration range. A, The SR1 genomic locus and the targeting vectors for generating strains with SR1–IRES–tauGFP and ΔSR1–RFP mutations. B, Image taken under fluorescent illumination of the SO from an SR1–IRES–tauGFP mouse. Scale bar, 5 μm. C, A single SR1–GFP cell responded to all five odorants at 1 μm, and to Mix 1 at 1 μm, under voltage-clamp mode. The gray trace (Ringer) was the mechanical response induced by a Ringer puff that was delivered with the same pressure as the odorant puffs. D, The dose–response curve for amyl acetate was averaged from six SR1–GFP cells. The holding potential was −65 mV for all neurons. Each colored line (and the corresponding symbols) represents the data from a single cell, and the thick black line represents the averaged curve from all six neurons. Error bars indicate SEM. Note that the averaged curve has K1/2 = 0.11 μm and n = 0.3, which are slightly different from the averaged values from individual curves reported in the text.
Patch clamp.
Intact epithelial preparations were as in our published procedures (Ma and Shepherd, 2000). Wild-type (C57BL/6) or homozygous gene-targeted mice (male or female, 3 to 12 weeks) were deeply anesthetized by injection of ketamine HCl and xylazine (200 mg/kg and 20 mg/kg body weight, respectively) and then decapitated. The head was immediately put into ice-cold Ringer's solution, which contained (in mm) 124 NaCl, 3 KCl, 1.3 MgSO4, 2 CaCl2, 26 NaHCO3, 1.25 NaH2PO4, and 15 glucose (osmolarity, 305 mOsm). The pH was kept at 7.4 by bubbling with 95% O2 and 5% CO2. The nose was dissected out en bloc; the olfactory mucosa attached to the nasal septum and the dorsal recess was removed and kept in oxygenated Ringer. Before use, the entire mucosa was peeled away from the underlying bone and transferred to a recording chamber with the mucus layer facing up. Oxygenated Ringer was continuously perfused at 25 ± 2°C.
The dendritic knobs of OSNs were visualized through an upright differential interference contrast microscope (Olympus; BX51WI) equipped with a charge-coupled device camera (Dage-MTI) and a 40× water-immersion objective. An extra 4× magnification was achieved by an accessory lens in the light path. The GFP- and RFP-tagged cells were visualized under fluorescent illumination. Superimposition of the fluorescent and bright-field images allowed identification of the fluorescent cells under bright field, which directed the recording pipettes (Grosmaitre et al., 2006). Electrophysiological recordings were controlled by an EPC-10 amplifier combined with Pulse software (HEKA Electronic). Perforated patch-clamp was performed on the dendritic knobs by including 260 μm nystatin in the recording pipette, which was filled with the following solution (in mm): 70 KCl, 53 KOH, 30 methanesulfonic acid, 5.0 EGTA, 10 HEPES, 70 sucrose; pH 7.2 (KOH), and 310 mOsm. The junction potential was ∼9 mV and was corrected in all experiments off-line. Under voltage-clamp mode, the signals were initially filtered at 10 kHz and then at 2.9 kHz. For voltage-gated ionic currents, the signals were sampled at 50 kHz. For odorant-induced transduction currents (which are slow and long lasting), the signals were sampled at 333 Hz. Further filtering off-line at 100 Hz (to remove noise) did not change the response kinetics or amplitudes, indicating that the sampling rate was sufficient and signal aliasing was not a concern. Similarly, under current-clamp mode, the signals were filtered at 2.9 kHz and sampled at 5 kHz. Further filtering off-line at 1.5 kHz did not change the response kinetics or amplitudes.
A seven-barrel pipette was used to deliver stimuli by pressure ejection through a picospritzer (Pressure System IIe; Toohey Company). The stimulus electrode was placed ∼20 μm downstream from the recording site, and both mechanical and odorant responses could be elicited in some neurons (Grosmaitre et al., 2007). All stimuli were delivered by 138 kPa (20 psi) on the picospritzer. The pulse length was kept at 1 s to ensure that the neurons were stimulated by the intrapipette concentration (Grosmaitre et al., 2006). Odorants were prepared in 0.5 m solution in dimethylsulfoxide (DMSO) and kept at −20°C. Final solutions were prepared before each experiment by adding Ringer. Odorant mixture Mix 1 contains 19 compounds in equal molar concentration: heptanol, octanol, hexanal, heptanal, octanal, heptanoic acid, octanoic acid, cineole, amyl acetate, (+) limonene, (−) limonene, (+) carvone, (−) carvone, 2-heptanone, anisaldehyde, benzaldehyde, acetophenone, 3-heptanone, and ethyl vanilline. The odorant mixture Mix 2 contained 19 compounds in equal molar concentration: hexanol, nonyl alcohol, nonanal, hexanoic acid, nonanoic acid, valeric acid, ethyl caproate, 1-octen-3-ol, phenethyl alcohol, γ-octanoic lactone, pyrrolidine, methyl anthranilate, benzenethiol (thiophenol), pyridine, pyrazine, geraniol, eugenol, benzyl salicylate, isoamyl acetate. The final concentration of DMSO in the odorant solutions ranges from 0.0002 (v/v) (for a single odorant at 1 μm) to 0.4% (for Mix 1 and Mix 2 at 100 μm). To rule out potential effects of DMSO on electrical responses, we compared the transduction currents in SO–OSNs (n = 6) induced by puffs of Ringer without DMSO or with 0.2–0.5% DMSO; we did not observe any differences. We, therefore, used the Ringer solution as controls in most of the experiments. All compounds and chemicals were obtained from Sigma-Aldrich, unless otherwise stated. Lyral was provided as a generous gift from International Fragrances and Flavors (New York, NY). The peptides AAPDNRETF and SYFPEITHI were synthesized by Genemed Synthesis, and α-phellandren and β-phenethylamine were purchased from Fluka.
Heterologous expression of SR1 in Hana3A cells.
The coding sequence of SR1 was amplified from genomic DNA of C57BL/6 mice and subcloned into the pcDNA3.1/TOPO vector (Invitrogen) with an N-terminal tag of the first 20 aa of rhodopsin. The SR1 amino acid sequence is identical between 129 and C56BL/6 mice. The Dual-Glo Luciferase Assay (Promega) was used to determine the activities of firefly and Renilla luciferase. Firefly luciferase, driven by a cAMP response element promoter (CRE-Luc; Stratagene), was used to determine OR activation levels. Renilla luciferase, driven by a constitutively active SV40 promoter (pRL-SV40; Promega), functioned as an internal control for transfection efficiency and cell viability. Hana3A cells stably expressing RTP1L, RTP2, REEP1, and Gαolf were plated on poly-d-lysine-coated 96-well plates (Nalge Nunc) and incubated overnight in minimum essential medium eagle (Sigma) with 10% FBS at 37°C and 5% CO2. The following day, cells were transfected using Lipofectamine 2000 (Invitrogen). For each 96-well plate, we transfected 1 μg pRL-SV40, 1 μg CRE-Luc, 1 μg mouse RTP1S, and 6 μg of SR1 plasmid DNA. After transfection (24 h), medium was replaced with 25 μl of odorant solution diluted in CD293 chemically defined medium (Invitrogen), and cells were further incubated for 4 h at 37°C and 5% CO2. The manufacturer's protocols were followed to measure firefly luciferase and Renilla luciferase activities. A Wallac Victor 1420 plate reader (Perkin-Elmer) was used to measure luminescence. Data were analyzed using Microsoft Excel and GraphPad Prism 4. Normalized activity was further calculated using the following formula: [Luc/RLuc(N)−Luc/RLuc(lowest)]/[Luc/RLuc(highest)−Luc/RLuc(lowest)], where Luc/RLuc(N) = luminescence of firefly luciferase divided by luminescence of Renilla luciferase in a certain well; Luc/RLuc(lowest) = lowest firefly luminescence divided by Renilla luminescence of a plate or set of plates; Luc/RLuc(highest) = highest firefly luminescence divided by Renilla luminescence of a plate or set of plates.
Results
Many septal organ neurons are broadly responsive
We first investigated the odorant response properties of SO–OSNs in intact epithelial preparations from wild-type C57BL/6 mice. Using perforated patch-clamp conditions, odorant-induced transduction currents (under voltage-clamp mode) or membrane potential changes (under current-clamp mode) were recorded from the dendritic knobs of individual OSNs. Odorant stimulation was delivered by pressure ejection (puffing) of odorant solutions through a seven-barrel pipette, in which one barrel is filled with pure Ringer to serve as a control for mechanical responses. A positive odorant response is defined as a response amplitude that is >160% of the Ringer response under voltage-clamp mode (Grosmaitre et al., 2007). Odorant puffs elicited excitatory responses in 70% (228 of 328) of SO–OSNs. The nonresponsive cells were healthy: they displayed normal voltage-gated ionic currents, action potentials, and an intact cAMP cascade (Grosmaitre et al., 2007).
Among 43 individually tested compounds which cover a variety of chemical classes, 19 induced responses in ≥50% of SO–OSNs and are thus highly likely ligands for SR1-expressing SO–OSNs, in view of the expression of SR1 in 50% of SO–OSNs (Fig. 1). The effective ligands include alcohols, aldehydes, acids, esters, and ketones. We did, however, observe some selectivity among SO–OSNs. For example, cineole (10 or 300 μm) elicited responses in 17% of neurons, and 2,5-dimethyl pyrazine did not induce responses in 20 cells examined. Not effective for SO–OSNs were selected amines that stimulate MOE neurons expressing trace amine-associated receptors (TAARs) (Liberles and Buck, 2006) and two major histocompatibility complex (MHC)-related peptides AAPDNRETF and SYFPEITHI, which elicit responses in very small subsets of neurons in the vomeronasal organ (Leinders-Zufall et al., 2004) and the MOE (Spehr et al., 2006) (Fig. 1).
Figure 1.
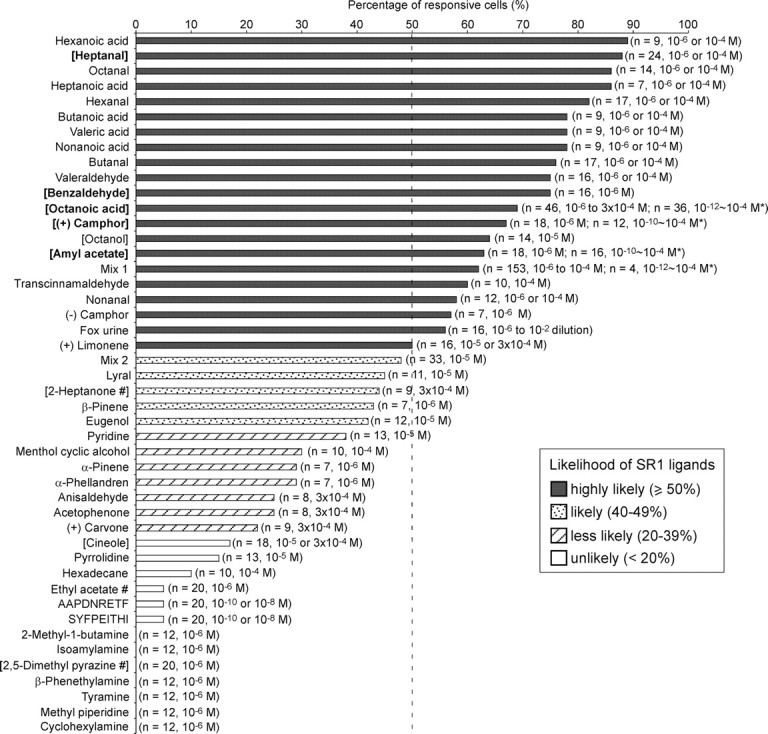
SO–OSNs respond to a variety of compounds and mixtures. The results from 328 SO–OSNs of wild-type C57BL/6 mice are summarized. The compounds are listed according to the percentage of responsive cells (high to low). The cell numbers are included in parentheses. A concentration range marked with * indicates that multiple concentrations were tested in individual cells. We did not observe a systematic increase in the number of responsive neurons at 1 μm or higher, suggesting that the response threshold to many effective odorants is <1 μm. Therefore, we combined the percentage of responsive neurons tested at concentrations ≥1 μm. Odorants in BOLD were also tested in SR1–GFP neurons and those in square brackets were also tested in Hana3A cells. Chemicals marked with # are previously reported as pheromones. The dashed line marks 50%, which corresponds to the percentage of SO–OSNs that express SR1.
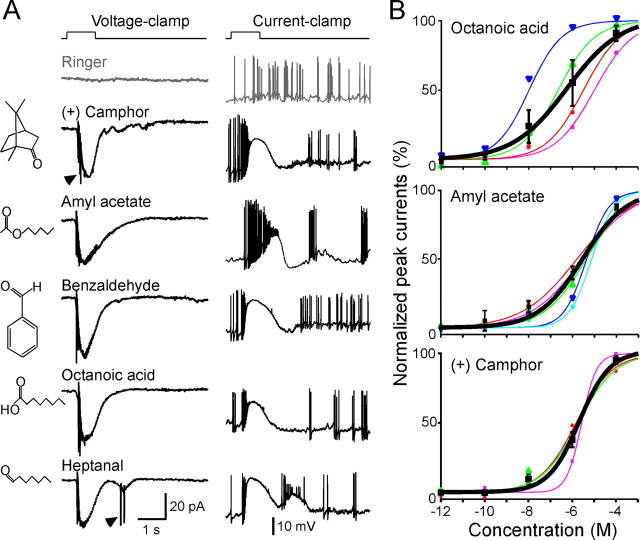
A subset of SO–OSNs was tested using five odorants with distinct structural features and human odor qualities: heptanal (strong fruity), octanoic acid (rancid), amylacetate (banana), benzaldehyde (almond), and (+) camphor (strong aromatic). Octanoic acid and (+) camphor are among the most dissimilar pair of odorants according to an odor metric (Haddad et al., 2008). Because an extensive set of odorants cannot be tested on individual OSNs due to practical constraints, we define operationally an OSN as “broadly responsive” if it responds to these five odorants, and an OSN as “selectively responsive” if it responds to none or only one of these five odorants. Figure 2A shows a SO–OSN that responded to all five compounds individually, each at 1 μm. When tested with homologous compounds carrying the same functional group (acid or aldehyde), most of the responsive SO–OSNs responded to all compounds (Fig. 3). We determined dose–response curves for octanoic acid, amyl acetate, and (+) camphor between 10−12 and 10−4 m, sequentially applied from the lowest to the highest with an interval >1 min to minimize odorant adaptation (Fig. 2B). A nonzero baseline mechanical response (Ibaseline) was elicited by Ringer puffs (Grosmaitre et al., 2007). After subtracting Ibaseline, the normalized peak currents were fitted with the modified Hill equation, Inorm = (I − Ibaseline)/Imax = 1/(1 + (K1/2/C)n), where I represents the peak current, Imax the maximum response at saturating concentrations, K1/2 the concentration at which half of the maximum response was reached, C the concentration of odorant, and n the Hill coefficient. For octanoic acid, the threshold ranged between 10−12 and 10−8 m (18 cells), n = 0.5 ± 0.0, and K1/2 = 3.2 ± 2.3 μm (four cells). For amyl acetate, the threshold ranged from 10−10 to 10−8 m (nine cells), n = 0.6 ± 0.1 and K1/2 = 4.0 ± 1.0 μm (five cells). For (+) camphor, the threshold ranged from 10−8 to 10−6 m (eight cells), n = 0.8 ± 0.3, and K1/2 = 2.0 ± 0.3 μm (three cells). Thus, common features are a subnanomolar to submicromolar threshold and a wide concentration range.
Figure 2.
SO–OSNs respond to structurally diverse odorants over a wide concentration range. A, A single neuron responded to five odorants at 1 μm each under voltage-clamp conditions (left column, holding potential is −65 mV) and current-clamp conditions (right column, resting membrane potential is near −65 mV). The arrowheads mark action potentials generated under voltage-clamp mode due to imperfect space clamp. The gray trace (Ringer) was the mechanical response induced by a Ringer puff that was delivered with the same pressure as the odorant puffs. B, Dose–response curves for octanoic acid, amyl acetate, and (+) camphor between 10−12 and 10−4 m. Each colored line (and the corresponding symbols) represents the data from a single cell, and the thick black line represents the averaged curve from all neurons shown in the same graph. Error bars indicate SEM. The holding potential was −65 mV for all neurons.
Figure 3.
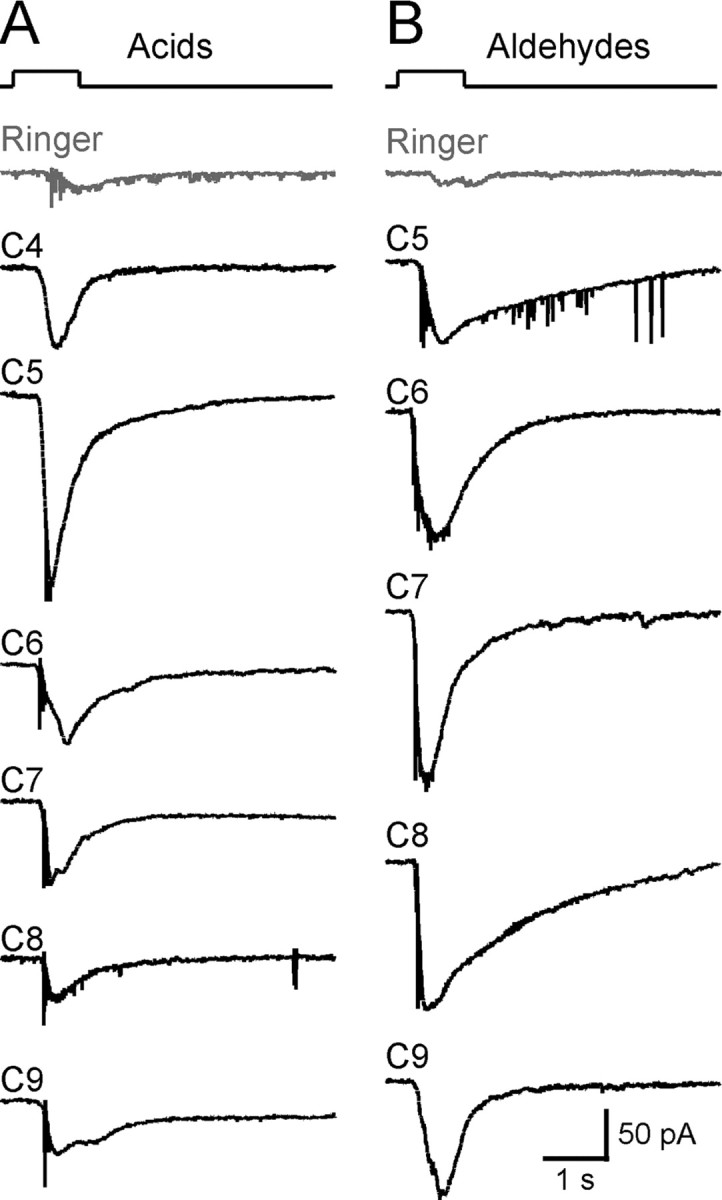
SO–OSNs respond to homologous compounds with the same functional group. We tested the responses of 26 SO neurons to acids or aldehydes with different carbon chain length (C4 to C9). Most of the responsive neurons (19 of 26; 73%) responded to all acids or aldehydes at concentrations of 1 or 100 μm. A, A single neuron responded to aliphatic acids (all at 100 μm). B, Another neuron responded to aliphatic aldehydes (all at 100 μm). The holding potential was −65 mV for both neurons. The gray trace (Ringer) in each cell was the mechanical response induced by a Ringer puff that was delivered with the same pressure as the odorant puffs.
SR1-expressing neurons are broadly responsive
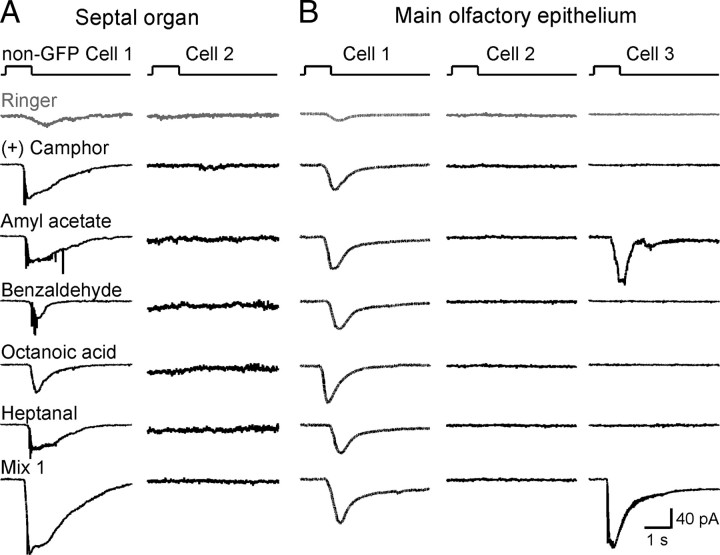
These experiments on wild-type mice do not identify the ORs that underlie the broad responsiveness of SO–OSNs and do not reveal if the SO–OSNs themselves exhibit unusual response properties, instead of the particular ORs that they express. We, therefore, generated a mouse strain with a gene-targeted mutation in the SR1 locus, SR1–IRES–tauGFP (Fig. 4A). In these mice, SR1-expressing (SR1+) neurons coexpress SR1 and tauGFP by virtue of an IRES. The cells that appear green under fluorescent illumination are referred to as SR1–GFP cells. Such cells are densely packed in the SO, with an average density of 1.66 ± 0.17 per 100 μm2 surface area (n = 8 frames taken from different preparations) (Fig. 4B). To determine the response properties of SR1–GFP cells, we applied the same set of five odorants in random order. We used 1 μm, because this concentration likely falls in the middle of the dose–response curves (Fig. 2B). We find that 100% of SR1–GFP cells (n = 8) responded to all five compounds at 1 μm and to a mixture of 19 compounds (Mix 1) at 1 μm (Fig. 4C). There were no statistically significant differences in the response amplitudes induced by these five odorants (pairwise t tests). We obtained dose–response curves to amyl acetate for another set of SR1–GFP cells (n = 6). These cells display a wide dynamic range with a Hill coefficient n = 0.4 ± 0.1 and K1/2 = 1.2 ± 0.8 μm (n = 6) (Fig. 4D), comparable with values seen in SO–OSNs of wild-type mice (Fig. 2B). Thus, SR1+ OSNs in the SO respond to a variety of compounds including two aldehydes, an acid, an ester, and a ketone, and exhibit wide dynamic ranges.
Consistent with the recordings in wild-type mice (Fig. 1), SR1–GFP neurons in the SO showed some selectivity. A set of 10 SR1–GFP neurons failed to respond to two MHC peptides (AAPDNRETF and SYFPEITHI at 0.1 or 10 nm), 2,5-dimethyl pyrazine (1 μm), and ethyl acetate (1 μm). Another set of six SR1–GFP neurons failed to respond to amines that stimulate TAARs (β-phenethylamine, tyramine, methyl piperidine, cyclohexylamine, 2-methyl-1-butamine, and isoamylamine at 1 μm) (data not shown). These data indicate that SR1–GFP neurons did not respond to every compound from our collection.
Are the unusual response properties of SR1+ OSNs due to their anatomical location in the SO? There are also SR1–GFP cells in the MOE but at a much lower density (0.09 ± 0.01 per 100 μm2 surface area, n = 6) than in the SO (Fig. 5A). We find that 100% of SR1–GFP cells in the MOE (n = 7) responded to all five odorants and to Mix 1 at 1 μm (Fig. 5B). Dose–response curves to amyl acetate show that SR1–GFP neurons in the MOE resemble their counterparts in the SO in terms of the wide dynamic range, with a Hill coefficient n = 0.4 ± 0.1 and K1/2 = 0.13 ± 0.08 μm (n = 7) (Fig. 5C). Thus, SR1+ neurons respond broadly and over a wide concentration range, regardless of their location in the SO versus the MOE. The broad response profile of SR1+ OSNs is in sharp contrast with those of other genetically labeled OSNs of the MOE, such as MOR23+ OSNs, which all respond to lyral (Grosmaitre et al., 2006) but not to Mix 1, even at high concentrations (up to 300 μm; data not shown).
Figure 5.
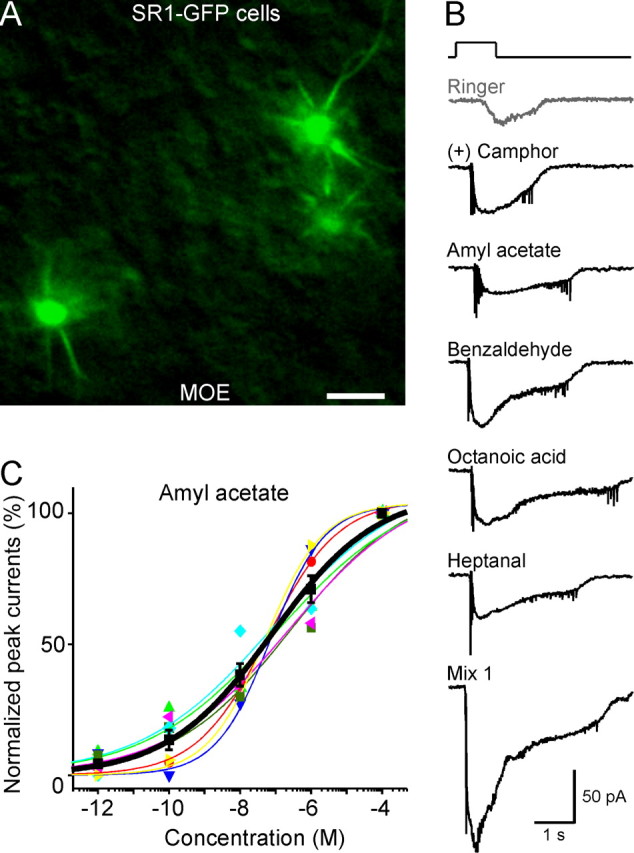
SR1–GFP cells in the MOE respond broadly and over a wide concentration range. A, Image taken under fluorescent illumination of the most ventral region of the MOE from an SR1–IRES–tauGFP mouse. Scale bar, 5 μm. B, A single SR1–GFP cell responded to all five distinct odorants at 1 μm each, and to Mix 1 at 1 μm, under voltage-clamp mode. The gray trace (Ringer) was the mechanical response induced by a Ringer puff that was delivered with the same pressure as the odorant puffs. C, The dose–response curve for amyl acetate was averaged from seven neurons. The holding potential was −65 mV for all neurons. Each colored line (and the corresponding symbols) represents the data from a single cell; the thick black line represents the averaged curve from all seven neurons. Error bars indicate SEM. Note that the averaged curve has K1/2 = 0.08 μm and n = 0.3, which are slightly different from the averaged values from individual curves reported in the text.
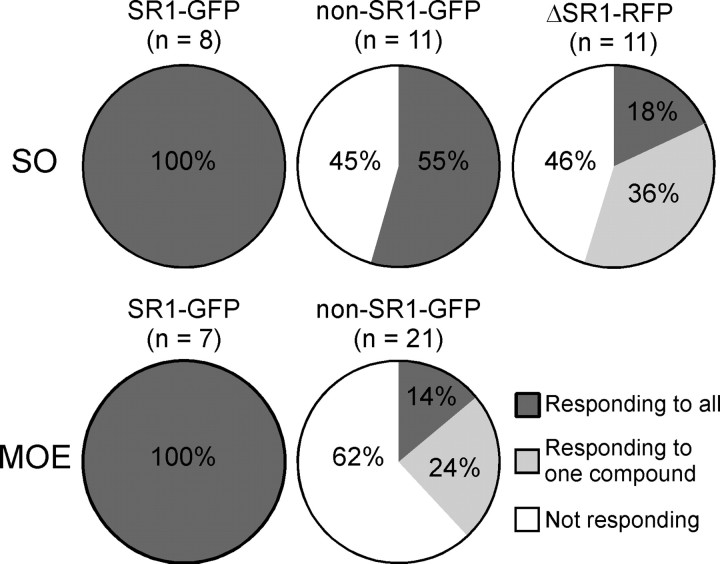
Is SR1 the only broadly responsive OR expressed by SO–OSNs? We found that among non-GFP cells (n = 11) in the SO of SR1–GFP mice, 55% responded to the same set of five odorants, and 45% did not respond to any one of them (Fig. 6A). Thus, in addition to SR1+ OSNs, SO–OSNs expressing some other ORs are also broadly responsive. Furthermore, among non-GFP cells in the MOE of SR1–GFP mice (n = 22), 14% responded to all five odorants, 24% responded to one odorant, and 62% did not respond to any odorant (Fig. 6B). Thus, there are other broadly responsive OSNs in the MOE, but to a lesser extent than in the SO, consistent with the greater diversity of OR gene expression in the MOE.
Figure 6.
Non-SR1–GFP cells in the SO and MOE display diverse response profiles. Non-GFP (=SR1-negative) cells from SR1–IRES–tauGFP mice were stimulated by the same set of five odorants (at 1 μm each) and Mix 1 (1 μm) as tested in the SR1–GFP cells. A, Non-GFP Cell 1 in the SO responded to all five odorants, and Cell 2 did not respond to any odorants including Mix 1. B, Non-GFP Cell 1 responded to all five odorants, Cell 2 did not respond to any odorants including Mix 1, and Cell 3 responded to one of the five odorants. The holding potential was −65 mV for all neurons. The gray trace (Ringer) was the mechanical response induced by a Ringer puff that was delivered with the same pressure as the odorant puffs.
SR1 itself underlies the broad responsiveness
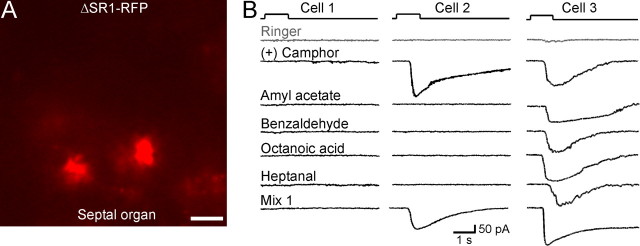
Is the unusually broad response profile of SR1+ neurons attributable to SR1 in and of itself or to unusual features of these cells? As a first approach to this question, we generated by gene targeting a strain with a knock-out mutation in the SR1 locus, ΔSR1–RFP. The intron-less coding region of SR1 is here replaced with the sequence encoding a monomeric RFP, which is an improved, less toxic form of DsRed (Discosoma RFP). The red-fluorescent cells in this strain are referred to as ΔSR1–RFP cells. The density of ΔSR1–RFP cells in the SO (Fig. 7A) is 14× lower (0.12 ± 0.02 per 100 μm2 surface area, n = 8) than that of SR1–GFP cells in the SO of SR1–GFP mice, presumably due to cell death or OR gene switching in the absence of a functional OR produced from the mutant locus (Wang et al., 1998; Serizawa et al., 2003; Lewcock and Reed, 2004; Shykind et al., 2004; Bozza et al., 2009). The recorded ΔSR1 neurons were nonetheless healthy: they displayed similar membrane properties (leakage currents, voltage-gated ionic currents, and firing patterns) as SR1–GFP neurons (data not shown). Among ΔSR1–RFP neurons in the SO (n = 11), 18% responded to all five odorants, 36% responded to only one, and 45% did not respond to any (Figs. 7B, 8). This response profile is very different from that of SR1–GFP cells (Fig. 8) and implicates SR1 itself in the broad responsiveness.
Figure 7.
SO–OSNs expressing the ΔSR1 allele show diverse patterns of odorant responses. A, Image under fluorescent illumination of the SO from a ΔSR1–RFP mouse. Scale bar, 5 μm. Because RFP is not fused to the tau protein, OSN cilia are not visible. B, Under voltage-clamp mode, ΔSR1–RFP neurons were stimulated by the same set of five odorants tested in the SR1–GFP cells. Cell 1 (left column) did not respond to any odorant including Mix 1. Cell 2 (middle column) responded to one of the five odorants. Note that this neuron also responded to Mix 1, which does not contain (+) camphor. Cell 3 (right column) responded to all five odorants. The holding potential was −65 mV for all neurons. The gray trace (Ringer) in each cell was the mechanical response induced by a Ringer puff that was delivered with the same pressure as the odorant puffs.
Figure 8.
Summary of the response profiles of the five cell groups in response to the same set of five distinct odorants. The n number in parentheses indicates the total number of cells tested in each group.
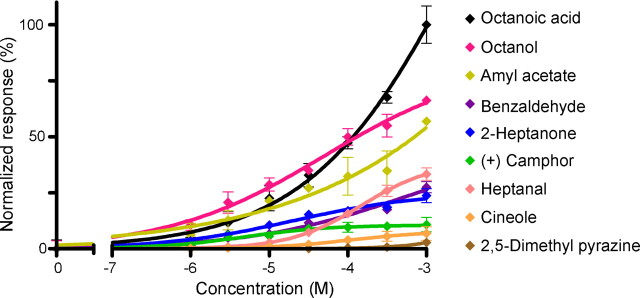
A second approach to the role of SR1 was taken by heterologous gene transfer. We expressed SR1 in Hana3A cells (Saito et al., 2004, 2009; Zhuang and Matsunami, 2007), which are HEK293T cells that are stably transfected to express Gαolf and the OR accessory molecules RTP1L, RTP2, and REEP1. Using a luciferase assay, we measured the cAMP level that results from SR1 activation by odorants. Dose–response curves for nine odorants (including the five odorants tested above) are shown in Figure 9. Consistent with the patch-clamp recordings, SR1 conferred responsiveness to seven of nine odorants but not to cineole and 2,5-dimethyl pyrazine, which also stimulate SO–OSNs poorly (Fig. 1). Thus, the results from the in vitro assay with Hana3A cells confirm and extend the observations made in intact epithelial preparations.
Figure 9.
Hana3A cells transfected with SR1 respond to multiple odorants in a dose-dependent manner. Data were normalized. Error bars indicate SEM, and n = 4 for each point. Dose–response curves were fitted by setting the baseline (no odorant stimulation) to 0 and the maximum response to 100. For the effective ligands, the K1/2 (or EC50) value ranged from 83.2 μm (for octanoic acid) to several millimolars, and the Hill coefficient ranged from 0.2 to 0.7.
Discussion
Here, we report the unusually broad response profile of a mouse OR, SR1, which is abundantly expressed in neurons of the septal organ and to a lesser extent in the main olfactory epithelium. Using patch-clamp recordings, we report that a large fraction of SO neurons responds to diverse odorants over a wide concentration range. We show that SR1 underlies this broad responsiveness, by analyzing genetically tagged SR1–GFP and ΔSR1–RFP neurons. We then independently confirm the broad response profile of SR1 in a heterologous system. Our results indicate that the mouse nose contains broadly responsive OSNs, and we speculate that these OSNs provide special sensory information to the olfactory system.
SO–OSNs respond to diverse chemicals with different size, shape, and functional groups. We identified 19 single compounds that stimulated at least 50% of SO–OSNs from wild-type mice and that are thus highly likely ligands for SR1+ SO–OSNs in view of the expression of SR1 in 50% of SO–OSNs. The set of five odorants that we tested subsequently in detail on SR1–GFP neurons does not provide a full characterization of the response profiles. We expect to identify additional ligands for SR1+ neurons with an expanded list of odorants. Since these five odorants represent five different chemical classes and SR1+ neurons respond to all of them, the response profiles to these five compounds provide a reasonable description for the various groups of cells under investigation (Fig. 8). Notably, we rarely observed OSNs with an intermediate response profile (responding to two to four odorants), presumably because the five odorants we chose are chemically distinct.
The response profile of SR1 is unusual given the current models of ligand binding for G-protein-coupled receptors (GPCRs). Most nonolfactory GPCRs use ionic or hydrogen bonding interactions to ensure high selectivity and sensitivity in binding their agonists or antagonists (Goddard and Abrol, 2007). In contrast, OR-ligand binding (e.g., the mOR–EG receptor to 22 compounds with a common motif) is likely achieved by hydrophobic and van der Waals interactions, which result in selective binding with relatively low affinities (Katada et al., 2005). Broadly responsive olfactory receptors have been reported in Drosophila (Hallem and Carlson, 2006), but these receptors likely function as ligand-gated ion channels (Sato et al., 2008; Wicher et al., 2008). It is plausible to postulate that mammalian ORs and their host OSNs also show diverse response properties, ranging from highly selective to broadly responsive. SR1 may be toward one extreme of the spectrum. Our findings have implications for the overall strategy for olfactory processing, specifically as to whether odor information is densely or sparsely coded in the olfactory system. The identification of a broadly responsive OR provides evidence at the receptor level that olfactory codes can be densely distributed, consistent with functional analyses in the olfactory bulb (Xu et al., 2000).
It would be interesting to study olfactory performance in ΔSR1 mice. However, there are other broadly responsive ORs, based on the analysis of non-GFP cells in SR1–GFP mice. Studies by electrophysiological recordings and calcium imaging have shown that some rodent OSNs can respond to diverse odorants (Duchamp-Viret et al., 1999; Malnic et al., 1999; Araneda et al., 2004). Because SR1 is unlikely to be the sole OR with such broad response profile in the repertoire of ∼1200 mouse ORs, the effect of a knockout of a single broadly responsive OR may be relatively minor and experimentally undetectable in behavioral assays. Another complication of interpreting olfactory performance of ΔSR1 mice is the complexity of a ΔOR phenotype in terms of OR gene regulation. We show that 18% (2 of 11) of ΔSR1–RFP cells in the SO respond to all five odorants tested, and 36% (4 of 11) to one of the five odorants tested. Thus, at least half of the SO–OSNs that express the ΔSR1 locus appear to express another OR locus that encodes a functional OR. There are various explanations for this phenomenon: concomitant expression of a ΔOR locus with an OR locus encoding a functional OR, lack of negative feedback from a functional OR, or gene switching. Coexpression of an OR gene that encodes a functional OR has been well characterized in a fraction of OSNs that express a ΔOR locus or ΔOR transgene (Serizawa et al., 2003; Feinstein et al., 2004; Lewcock and Reed, 2004; Shykind et al., 2004; Bozza et al., 2009). We have previously shown that OSNs that express a ΔOR locus show a diversity of odorant response profiles (Feinstein et al., 2004).
Curiously, (+) camphor induced relatively small responses in the in vitro system, although it induced similar responses as the other four odorants tested in SR1–GFP neurons (Figs. 4, 5). In addition, the sensitivity of Hana3A cells is 100–1000-fold lower than the most sensitive SR1+ neurons in the intact epithelium. This discrepancy may result from biological differences between heterologous cells and endogenous OSNs or technical differences between the measuring methods. Depending on the physicochemical properties of the odorants, the mucus (which contains odorant-binding proteins and other proteins) covering the OSNs may differentially influence the effective concentrations in the patch-clamp assay. Furthermore, the odorants are puffed for 1 s in the patch-clamp assay but incubated with Hana3A cells for 4 h. Although the in vitro data do not match the patch-clamp results in every detail, they confirmed that SR1 responds to multiple odorants including two aldehydes, an alcohol, an acid, an ester, and a ketone.
SO–OSNs respond to diverse odorants over a wide concentration range: four log units from threshold to saturation. This finding sheds light on a mismatch between the dynamic ranges of concentration codes at the OSN level (narrow range) and the glomerular level (wide range). The current study suggests that the wider range observed at the glomerular level is not necessarily an emerging feature from axonal convergence but instead may be a built-in feature at the level of ORs or OSNs. The properties of SR1-expressing OSNs make them suited to serve as general odorant detectors and/or sensors for the total odorant concentration.
Both in wild-type mice and in gene-targeted mice, there is a close correlation between broad responsiveness and mechanosensitivity (Grosmaitre et al., 2007). Of the 58 neurons summarized in Figure 8, all broadly responsive OSNs (n = 26) also showed mechanical responses to Ringer puffs delivered at the same pressure as the odorant puffs, regardless of OR identity. The remaining cells (nine responding to one odorant and 23 not responding to any of the five odorants) did not display mechanosensitivity. These findings suggest the possible involvement of the SR1 protein in mechanosensitivity. We speculate that broadly responsive OSNs can thus also sense the airflow (Carey et al., 2009). The SO contains a higher percentage of broadly responsive OSNs than the MOE, supporting the hypothesis that the SO, as a strategically placed outpost of olfactory epithelium, could quickly alert the olfactory system for odorant and airflow changes and prime it for receiving new odorant information. The SO may also monitor the total concentration of odorants in the external environment, such that breathing patterns and sniffing frequency can be regulated and the overall sensitivity of the olfactory system can be adjusted.
Footnotes
This work was supported by grants from the National Institute on Deafness and Other Communication Disorders–National Institutes of Health (R01s to H.M., P.M., and M.M.; R03s to X.G. and S.H.F.; and a training fellowship to A.C.L.).
References
- Araneda RC, Kini AD, Firestein S. The molecular receptive range of an odorant receptor. Nat Neurosci. 2000;3:1248–1255. doi: 10.1038/81774. [DOI] [PubMed] [Google Scholar]
- Araneda RC, Peterlin Z, Zhang X, Chesler A, Firestein S. A pharmacological profile of the aldehyde receptor repertoire in rat olfactory epithelium. J Physiol. 2004;555:743–756. doi: 10.1113/jphysiol.2003.058040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axel R. Scents and sensibility: a molecular logic of olfactory perception (Nobel lecture) Angew Chem Int Ed Engl. 2005;44:6110–6127. doi: 10.1002/anie.200501726. [DOI] [PubMed] [Google Scholar]
- Bozza T, Feinstein P, Zheng C, Mombaerts P. Odorant receptor expression defines functional units in the mouse olfactory system. J Neurosci. 2002;22:3033–3043. doi: 10.1523/JNEUROSCI.22-08-03033.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozza T, Vassalli A, Fuss S, Zhang JJ, Weiland B, Pacifico R, Feinstein P, Mombaerts P. Mapping of class I and class II odorant receptors to glomerular domains by two distinct types of olfactory sensory neurons in the mouse. Neuron. 2009;61:220–233. doi: 10.1016/j.neuron.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- Buck LB. Unraveling the sense of smell (Nobel lecture) Angew Chem Int Ed Engl. 2005;44:6128–6140. doi: 10.1002/anie.200501120. [DOI] [PubMed] [Google Scholar]
- Carey RM, Verhagen JV, Wesson DW, Pírez N, Wachowiak M. Temporal structure of receptor neuron input to the olfactory bulb imaged in behaving rats. J Neurophysiol. 2009;101:1073–1088. doi: 10.1152/jn.90902.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutforth T, Moring L, Mendelsohn M, Nemes A, Shah NM, Kim MM, Frisén J, Axel R. Axonal ephrin-As and odorant receptors: coordinate determination of the olfactory sensory map. Cell. 2003;114:311–322. doi: 10.1016/s0092-8674(03)00568-3. [DOI] [PubMed] [Google Scholar]
- Duchamp-Viret P, Chaput MA, Duchamp A. Odor response properties of rat olfactory receptor neurons. Science. 1999;284:2171–2174. doi: 10.1126/science.284.5423.2171. [DOI] [PubMed] [Google Scholar]
- Feinstein P, Bozza T, Rodriguez I, Vassalli A, Mombaerts P. Axon guidance of mouse olfactory sensory neurons by odorant receptors and the β2 adrenergic receptor. Cell. 2004;117:833–846. doi: 10.1016/j.cell.2004.05.013. [DOI] [PubMed] [Google Scholar]
- Firestein S. How the olfactory system makes sense of scents. Nature. 2001;413:211–218. doi: 10.1038/35093026. [DOI] [PubMed] [Google Scholar]
- Goddard WA, 3rd, Abrol R. 3-Dimensional structures of G protein-coupled receptors and binding sites of agonists and antagonists. J Nutr. 2007;137:1528S–1538S. doi: 10.1093/jn/137.6.1528S. discussion 1548S. [DOI] [PubMed] [Google Scholar]
- Grosmaitre X, Vassalli A, Mombaerts P, Shepherd GM, Ma M. Odorant responses of olfactory sensory neurons expressing the odorant receptor MOR23: a patch clamp analysis in gene-targeted mice. Proc Natl Acad Sci U S A. 2006;103:1970–1975. doi: 10.1073/pnas.0508491103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosmaitre X, Santarelli LC, Tan J, Luo M, Ma M. Dual functions of mammalian olfactory sensory neurons as odor detectors and mechanical sensors. Nat Neurosci. 2007;10:348–354. doi: 10.1038/nn1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddad R, Khan R, Takahashi YK, Mori K, Harel D, Sobel N. A metric for odorant comparison. Nat Methods. 2008;5:425–429. doi: 10.1038/nmeth.1197. [DOI] [PubMed] [Google Scholar]
- Hallem EA, Carlson JR. Coding of odors by a receptor repertoire. Cell. 2006;125:143–160. doi: 10.1016/j.cell.2006.01.050. [DOI] [PubMed] [Google Scholar]
- Katada S, Hirokawa T, Oka Y, Suwa M, Touhara K. Structural basis for a broad but selective ligand spectrum of a mouse olfactory receptor: mapping the odorant-binding site. J Neurosci. 2005;25:1806–1815. doi: 10.1523/JNEUROSCI.4723-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinders-Zufall T, Brennan P, Widmayer P, Chandramani SP, Maul-Pavicic A, Jäger M, Li XH, Breer H, Zufall F, Boehm T. MHC class I peptides as chemosensory signals in the vomeronasal organ. Science. 2004;306:1033–1037. doi: 10.1126/science.1102818. [DOI] [PubMed] [Google Scholar]
- Lewcock JW, Reed RR. A feedback mechanism regulates monoallelic odorant receptor expression. Proc Natl Acad Sci U S A. 2004;101:1069–1074. doi: 10.1073/pnas.0307986100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberles SD, Buck LB. A second class of chemosensory receptors in the olfactory epithelium. Nature. 2006;442:645–650. doi: 10.1038/nature05066. [DOI] [PubMed] [Google Scholar]
- Ma M, Shepherd GM. Functional mosaic organization of mouse olfactory receptor neurons. Proc Natl Acad Sci U S A. 2000;97:12869–12874. doi: 10.1073/pnas.220301797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malnic B, Hirono J, Sato T, Buck LB. Combinatorial receptor codes for odors. Cell. 1999;96:713–723. doi: 10.1016/s0092-8674(00)80581-4. [DOI] [PubMed] [Google Scholar]
- Mombaerts P. Axonal wiring in the mouse olfactory system. Annu Rev Cell Dev Biol. 2006;22:713–737. doi: 10.1146/annurev.cellbio.21.012804.093915. [DOI] [PubMed] [Google Scholar]
- Mombaerts P, Wang F, Dulac C, Chao SK, Nemes A, Mendelsohn M, Edmondson J, Axel R. Visualizing an olfactory sensory map. Cell. 1996;87:675–686. doi: 10.1016/s0092-8674(00)81387-2. [DOI] [PubMed] [Google Scholar]
- Rodolfo-Masera T. Su 1′esistenza di un particolare organo olfacttivo nel setto nasale della cavia e di altri roditori. Arch Ital Anat Embryol. 1943;48:157–212. [Google Scholar]
- Sato K, Pellegrino M, Nakagawa T, Nakagawa T, Vosshall LB, Touhara K. Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature. 2008;452:1002–1006. doi: 10.1038/nature06850. [DOI] [PubMed] [Google Scholar]
- Saito H, Kubota M, Roberts RW, Chi Q, Matsunami H. RTP family members induce functional expression of mammalian odorant receptors. Cell. 2004;119:679–691. doi: 10.1016/j.cell.2004.11.021. [DOI] [PubMed] [Google Scholar]
- Saito H, Chi Q, Zhuang H, Matsunami H, Mainland JD. Odor coding by a mammalian receptor repertoire. Sci Signal. 2009;2:ra9. doi: 10.1126/scisignal.2000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serizawa S, Miyamichi K, Nakatani H, Suzuki M, Saito M, Yoshihara Y, Sakano H. Negative feedback regulation ensures the one receptor-one olfactory neuron rule in mouse. Science. 2003;302:2088–2094. doi: 10.1126/science.1089122. [DOI] [PubMed] [Google Scholar]
- Shirokova E, Schmiedeberg K, Bedner P, Niessen H, Willecke K, Raguse JD, Meyerhof W, Krautwurst D. Identification of specific ligands for orphan olfactory receptors. G protein-dependent agonism and antagonism of odorants. J Biol Chem. 2005;280:11807–11815. doi: 10.1074/jbc.M411508200. [DOI] [PubMed] [Google Scholar]
- Shykind BM, Rohani SC, O'Donnell S, Nemes A, Mendelsohn M, Sun Y, Axel R, Barnea G. Gene switching and the stability of odorant receptor gene choice. Cell. 2004;117:801–815. doi: 10.1016/j.cell.2004.05.015. [DOI] [PubMed] [Google Scholar]
- Spehr M, Kelliher KR, Li XH, Boehm T, Leinders-Zufall T, Zufall F. Essential role of the main olfactory system in social recognition of major histocompatibility complex peptide ligands. J Neurosci. 2006;26:1961–1970. doi: 10.1523/JNEUROSCI.4939-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H, Ma M. Molecular organization of the olfactory septal organ. J Neurosci. 2004;24:8383–8390. doi: 10.1523/JNEUROSCI.2222-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H, Ma M. Activity plays a role in eliminating olfactory sensory neurons expressing multiple odorant receptors in the mouse septal organ. Mol Cell Neurosci. 2008;38:484–488. doi: 10.1016/j.mcn.2008.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touhara K, Sengoku S, Inaki K, Tsuboi A, Hirono J, Sato T, Sakano H, Haga T. Functional identification and reconstitution of an odorant receptor in single olfactory neurons. Proc Natl Acad Sci U S A. 1999;96:4040–4045. doi: 10.1073/pnas.96.7.4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Dannecker LE, Mercadante AF, Malnic B. Ric-8B promotes functional expression of odorant receptors. Proc Natl Acad Sci U S A. 2006;103:9310–9314. doi: 10.1073/pnas.0600697103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Nemes A, Mendelsohn M, Axel R. Odorant receptors govern the formation of a precise topographic map. Cell. 1998;93:47–60. doi: 10.1016/s0092-8674(00)81145-9. [DOI] [PubMed] [Google Scholar]
- Wicher D, Schäfer R, Bauernfeind R, Stensmyr MC, Heller R, Heinemann SH, Hansson BS. Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature. 2008;452:1007–1011. doi: 10.1038/nature06861. [DOI] [PubMed] [Google Scholar]
- Xu F, Greer CA, Shepherd GM. Odor maps in the olfactory bulb. J Comp Neurol. 2000;422:489–495. doi: 10.1002/1096-9861(20000710)422:4<489::aid-cne1>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Zhuang H, Matsunami H. Synergism of accessory factors in functional expression of mammalian odorant receptors. J Biol Chem. 2007;282:15284–15293. doi: 10.1074/jbc.M700386200. [DOI] [PubMed] [Google Scholar]