Abstract
The volume of biomedical literature has experienced explosive growth in recent years. This is reflected in the corresponding increase in the size of MEDLINE®, the largest bibliographic database of biomedical citations. Indexers at the U.S. National Library of Medicine (NLM) need efficient tools to help them accommodate the ensuing workload. After reviewing issues in the automatic assignment of Medical Subject Headings (MeSH® terms) to biomedical text, we focus more specifically on the new subheading attachment feature for NLM’s Medical Text Indexer (MTI). Natural Language Processing, statistical, and machine learning methods of producing automatic MeSH main heading/subheading pair recommendations were assessed independently and combined. The best combination achieves 48% precision and 30% recall. After validation by NLM indexers, a suitable combination of the methods presented in this paper was integrated into MTI as a subheading attachment feature producing MeSH indexing recommendations compliant with current state-of-the-art indexing practice.
Keywords: Abstracting and Indexing as Topic/*methods/statistics & numerical data; Artificial Intelligence; Dictionaries, Medical; *Evaluation Studies as Topic; *MEDLINE; Medical Subject Headings; Natural Language Processing
Background
Reflecting the latest developments in biomedical research, a significant increase in the indexing load is anticipated by the U.S. National Library of Medicine (NLM) in order to keep the MEDLINE® database up to date in the next decade. As many as 1 million journal articles are expected to be indexed each year by 2015 compared to 670,943 1 in 2007. To accommodate this 45% increase in the indexing load, tools must be developed in order to assist indexers in their daily task. In this paper, we report on the subheading attachment project conducted at NLM over the past two years. This effort investigated fine-grained indexing methods for the biomedical literature and led to the integration of a subheading attachment feature in the Medical Text Indexer (MTI) [1], a tool that automatically recommends Medical Subject Headings (MeSH®) main headings to NLM indexers. At the end of this section, we provide a summary of earlier stages of the project that have been described in [2] and [3].
A definition of “Indexing”
As the size of the biomedical literature grows, it becomes more diverse in terms of format and content. The methods to process and archive these documents become equally diverse and increasingly sophisticated, so that the notion of “indexing” has become ubiquitous. It is used in different, yet related, domains to generally denote the act of assigning descriptors to a document. However, the specific nature and purpose of these descriptors, and the rules or methods used to assign them can vary significantly among communities such as information retrieval, information science, computer science, image processing and so on.
In this paper, we will refer to indexing as the task of assigning to a document a limited number of terms denoting concepts that are substantively discussed in the document. This type of indexing is useful for retrieval purposes, but it also has a strong semantic descriptive value, in that the set of terms chosen to describe a document will serve as a synopsis of the subject matter discussed in the document. As a result, each indexing term must reflect an important aspect of the document, and its selection constitutes a difficult cognitive task implying a thorough understanding of the content of the document. Although free-text indexing is not necessarily incompatible with this type of indexing, automatic free-text indexing such as performed by SMART [4] or Latent Semantic Indexing (LSI) systems [5] does not conform to our definition. In the remainder of this paper, we will focus on the particular controlled indexing task of assigning indexing terms from the MeSH thesaurus to biomedical text referenced in MEDLINE, also known as citations.
MeSH indexing
MeSH is a thesaurus that has been developed at NLM since 19602. A new version is released every year, supplying controlled terms representing biomedical concepts to be used for the indexing of publications included in the MEDLINE database. MeSH contains two different types of term of concern in this paper: main headings (also known as descriptors) that denote biomedical concepts such as Diabetes Mellitus and Foot and subheadings (also known as qualifiers) that may be attached to a main heading in order to denote a more specific aspect of the concept such as metabolism and surgery. For each main heading, MeSH defines a set of subheadings that can be combined with it. These are also known as “allowable qualifiers”. As a result, certain pairs are permitted (for example, Diabetes Mellitus/metabolism and Foot/surgery are possible pairs) while others are not (for example, Foot/metabolism cannot be used because metabolism is not an allowable qualifier for the main heading Foot).
In the remainder of this paper, by “MeSH indexing terms” we refer to either main headings or main heading/subheading pairs. The task of MeSH indexing for MEDLINE requires indexers to assign MeSH indexing terms to biomedical articles in the following way: (1) select main headings to represent all concepts that are substantively discussed in the article (an average of approximately a dozen headings are selected, but the number may vary depending upon the article’s length and complexity), (2) attach the appropriate subheadings to the main headings selected, (3) mark the most substantively discussed concepts as “major” and (4) make sure appropriate “checktags”3 are selected, all the while (5) complying with instructions detailed in the indexing manual.
Issues in MeSH automatic indexing
Although the automatic assignment of MeSH indexing terms to a body of biomedical text has been extensively studied in the literature (see for example [1], [5], [6], [7], [8], [9], [10]), several major aspects of the task are often misunderstood or understated. Most issues pertain to the following topics:
multi-label assignment
scalability
compliance with indexing policies
Multi-label assignment
Controlled indexing is often viewed as a categorization problem because indexers must essentially decide whether a document substantively discusses the concept denoted by a given indexing term, i.e. whether the document is relevant to the category defined by this indexing term or label. In this respect, MeSH indexing is a multi-label categorization task because more than one term should be assigned to a document. Furthermore, it is referred to as a fuzzy classification problem [6] because the combinations between the indexing terms are numerous and varied. Some work addressing MeSH indexing attempts to elude the complexity due to the high number of indexing terms and variety of their combinations by selecting test collections where the documents are assigned a limited number of MeSH descriptors and therefore a limited number of combinations [6], as remarked by Rak et al. [10]. Although MeSH indexing could be viewed as a series of binary classification problems (for each descriptor, build a classifier to decide whether the document should be assigned the corresponding label), a recent experiment investigating Support Vector Machine (SVM) classifiers for the assignment of ICD-9 codes to clinical text found that better results were obtained when the categories given to the system were all the possible combinations of labels rather than the labels themselves [11]. If this were also the case for MeSH indexing, the scalability issues faced by machine learning approaches would be several orders of magnitude above what we describe below.
Scalability
With an average of 2,500 MEDLINE citations completed daily, the processing load of a practical, functional MeSH indexing system faces scalability issues. But more importantly, the exact number of indexing terms needs to be taken into account when discussing the MeSH indexing task; Table 1 gives counts of MeSH main headings, pairs and indexing terms4. Note that in [6], MeSH main headings are referred to as “MeSH categories”. Most efforts addressing MeSH indexing attempt to tackle indexing by solely using main headings which involves about 24,000 categories [1], [7]. However, in practice, MeSH indexing terms also include main heading/subheading pairs. The actual scale of the MeSH indexing problem is in fact in the range of 550,000 categories (and even more if one were to consider all possible combinations of indexing terms). Although Rak et al. [10] acknowledged the importance of multi-label assignment, the authors generalized main headings to the second level of the hierarchy (or tree5), i.e. scaling down to about 108 categories. Cai and Hoffman [8] opted for a similar use of the MeSH hierarchy, and Yang [9] acknowledged that most otherwise high-performance machine learning methods failed to accommodate the large-scale problem posed by MeSH indexing, with a k-Nearest Neighbors classifier being the most viable and robust approach. It is the underlying principle of the probabilistic, topic-based model for content similarity described by Lin and Wilbur [12] that was implemented in the “Related Articles” feature of PubMed® (NLM’s access point to MEDLINE). This feature is also a component of MTI [1]. The most recent work on main heading assignment revisits machine learning methods for MeSH indexing and more specifically Naïve Bayes and least square classifiers [13]. The authors address the issue of the size of training sets required by machine learning algorithms and introduce a method to obtain optimal training sets. They present an evaluation on a set of 20 main headings. Although the method could conceivably be applied to all MeSH indexing terms, including main heading/subheading pairs, it is hard to tell how well it would scale up from this study.
Table 1.
Number of MeSH indexing terms from 2004 to 2008
| MeSH version | Main Headings | Allowable pairs | MeSH Indexing terms |
|---|---|---|---|
| 2008 | 24,767 | 556,793 | 581,560 |
| 2006 | 23,883 | 534,981 | 558,864 |
| 2004 | 22,568 | 500,495 | 523,063 |
Compliance with indexing policies
MeSH indexing requires adherence to NLM’s indexing policies described in the Indexing Manual (e.g. choose a dozen main headings representing concepts substantively discussed in the document, combine with subheadings where appropriate, etc.) as well as more specific rules. An example of a specific indexing rule is the sample “coordination rule” shown in Figure 1. When machine learning approaches to MeSH indexing are used, the underlying assumption is that all the indexing rules will be derived from the set of labelled documents and be seamlessly “learned” by the system. However, the number of MeSH indexing term combinations may make it difficult to find a proper training corpus containing at least one sample application of each indexing rule.
Figure 1.
A sample coordination rule
1 DISEASE refers to a MeSH main heading belonging to the Diseases category or Mental Disorders subcategory (in the Psychiatry and Psychology category).
2 DRUG refers to a MeSH main heading belonging to the Chemicals and Drugs category.
Prior work on Subheading Attachment
As reported in [2], our work on subheading attachment first focused on the Genetics domain, which covered almost 20% of articles indexed for MEDLINE in 2005. Preliminary work was conducted on three genetics-related subheadings (genetics, metabolism and immunology). Based on encouraging results obtained in the context of MeSH indexing of French health resources [14], dictionary and rule-based approaches were developed and evaluated on a genetics corpus. This evaluation showed that both methods translated well to MeSH indexing of the biomedical literature in English. Therefore, we decided to extend the work beyond the genetics domain. We also refined the methods used (e.g. devised a semi-automatic scheme to increase the size of the dictionary) and investigated additional subheading attachment methods. Progress on the project was reported in [3], which presents an evaluation on a random MEDLINE corpus (a subset of the large test corpus described in this paper). The present paper reports on the overall subheading attachment project, including previously unpublished material.
Methods
Training and Test Corpora
Throughout the study, we used two large training and test corpora for quantitative evaluations and two smaller test corpora for qualitative evaluations.
Large training and test corpora
A large training set was built using 100,000 citations randomly selected from MEDLINE 20066. A same-size test corpus7 was built in the same way, with the additional constraint of selecting only citations that were not in the training corpus, so that the corpora are disjoint8.
Small test corpora
Two smaller test corpora of MEDLINE citations were used to carry out qualitative evaluations of the pair recommendations. These corpora each consisted of three journals selected by staff in the Index Section at NLM. The journals were chosen because of the anticipated high topical relevance of subheading recommendations to the subject matter they covered:
A genetics corpus: Hum Hered. 2006;62(2), Vet Immunol Immunopathol. 2006 Nov 15;114(1–2) and Genet Test. 2006 Fall;10(3)
A surgery corpus: Ann Plast Surg. 2007 May;58(5), Ann Transplant. 2006;11(3) and J Laparoendosc Adv Surg Tech A. 2007 Apr;17(2)
Four NLM indexers were shown the main heading/subheading pair recommendations for citations in these corpora before the MEDLINE indexing was available, in order to avoid bias in the indexers’ relevance judgments. The indexers were asked to look at the pair recommendations, and to determine whether a recommendation was useful and/or appropriate. Indexers were also asked to point out which were the worst recommendations according to them and to explain why a recommendation was not useful or appropriate. Finally, various ways of presenting the recommendations were also discussed (e.g. showing 2-letter codes vs. full names for subheadings). The indexers reviewed the recommendations on their own, and commented on them informally during project meetings – this means that there was no specific count of the number of “useful” or “appropriate” recommendations for a given citation or journal in these corpora. The purpose of this aspect of the study was to obtain trends of indexers’ opinion on the recommendations as a whole in order to make sure the resulting tool would meet their expectations in terms of usefulness and usability.
Automatically producing MeSH main heading/subheading recommendations
In the context of the Subheading Attachment Project, several methods were investigated to produce MeSH main heading/subheading recommendations. All of them aim at completing existing MTI main heading recommendations obtained with default filtering9 by attaching subheadings to the main headings.
“Jigsaw puzzle” methods
The “jigsaw puzzle” methods were intended as a simple type of approach to subheading attachment relying on the idea that the whole (MeSH pairs) could be created out of assembling its elements (main headings and subheadings). They work by separately extracting MeSH main headings and subheadings relevant to an article, and then attaching the subheadings to main headings when allowable. A dictionary method (DIC) introduced in [2] uses MTI-retrieved main headings. Subheadings are then extracted based on the presence of certain dictionary words or expressions in the title or abstract of the article. For example, the subheading genetics will be retrieved if words such as “gene”, “genes”, “genetic”, “heredity”, “DNA”, “RNA”, etc. are found. At first, the dictionary was composed of words that could be related to the subheadings based on the indexing manual chapter on assigning subheadings10. It was then expanded based on statistical fingerprinting of the subheadings over the entire MEDLINE collection using a technique similar to that described in [15]. For each subheading, the citations that used the subheading at least once were collected to form a subheading-specific corpus. After stop words were removed, a score S was computed for each word w in the subheading corpus SHi as follows:
| (1) |
The score of a word is based on its frequency (number of occurrences) in the subheading corpus vs. the MEDLINE collection and its frequency in the subheading corpus vs. the frequency of all content words in this corpus. The top 100 words according to this ranking were considered for addition in the dictionary. They were added to the dictionary if they improved the performance of the dictionary method on two training corpora11. Bigram statistics obtained from the subheading corpora were also used. Specifically, a score Sb,SHi was computed for bigrams b (two-word sets) according to Equation 1, so that bigrams were also considered for inclusion in the dictionary.
A JDI method was derived from Journal Descriptor Indexing (JDI), described in [16] and [17]. JDI automatically indexes text according to journal descriptors (JDs) which are a set of about 120 MeSH terms representing biomedical disciplines (e.g., Cardiology; Genetics, Medical; Surgery). For each journal, a set of JDs is manually assigned and recorded in NLM’s List of Serials Indexed for Online Users12 (LSIOU). JDI uses statistical associations between JDs and words or between JDs and MeSH indexing terms from a training set of MEDLINE citations, the JDs corresponding to the journals in the citations based on the contents of the LSIOU. For example, words and indexing terms in citations in the training set from the journal Foot and Ankle Clinics become statistically associated with the JD Orthopedics, because this is the JD for this journal in the serials file. The result of JDI of a word is a vector, consisting of JDs with their scores (between 0 – 1) for that word. Computation of word-JD vectors and MeSH indexing term JD vectors is described in [16]. JDI can also be performed on a subheading (SH), resulting in a JD vector for that SH.
Using a vector cosine similarity measure, the JD vector of a word can be compared to the JD vectors of each of the subheadings. As a result, a word-SH vector for the word can be created, where the score for each SH in the SH vector is the similarity between the word-JD vector and the JD vector for that SH. The ordering of SHs by score for a word gives a picture of the best to worst SHs for that word. For this study, word-SH vectors have been computed for words in a three-year MEDLINE training set (1999–2001).
To create a ranked list of SHs for a text outside the training set, the SH vectors for matching words in the training set are used. The scores for each SH are averaged across the words, forming a text-SH vector, where we use the top-five ranked SHs. For example, Table 2 shows the results of applying the JDI method to the title of MEDLINE citation # 15165580, “The role of surgical decompression for diabetic neuropathy”.
Table 2.
The text-SH vector showing the top-five SHs returned by the JDI method (sorted alphabetically) for citation #15165580.
| Words in Title | ||||||
|---|---|---|---|---|---|---|
| SHs | surgical | decompression | diabetic | neuropathy | average | rank |
| blood supply | 0.8075 | 0.5518 | 0.3348 | 0.5495 | 0.5609 | 5 |
| complications | 0.7400 | 0.4903 | 0.4499 | 0.6413 | 0.5804 | 3 |
| etiology | 0.7777 | 0.5140 | 0.4226 | 0.6200 | 0.5836 | 2 |
| physiopathology | 0.6009 | 0.4256 | 0.5364 | 0.7034 | 0.5666 | 4 |
| surgery | 0.9613 | 0.7455 | 0.1963 | 0.4339 | 0.5842 | 1 |
The final jigsaw puzzle method, the MTI method works by inferring relevant subheadings based on the main headings themselves. For example, if any main heading in the MeSH subcategory G13 (Genetic Phenomena) were retrieved by MTI, the method infers that the subheading genetics might be relevant for indexing the article. It would then be attached to the main headings also retrieved by MTI, when allowable. There is at least one such rule for all subheadings except drug effects.
Rule-based methods
Rule-based methods reflect the indexers’ practice of finding the best indexing terms by looking for indicator snippets of text in the articles and building on terms they have already selected to make the indexing set coherent and comprehensive. Post-processing (PP) rules infer pair recommendations from a pre-existing set of indexing terms - in our case, MTI main heading recommendations. A sample rule is shown in Figure 2. These rules were developed in the same spirit as the subheadings inferred in the MTI method above – in fact, Mutation is a G13 subcategory term. However, they are much more specific as they define which type of main heading the subheading should be attached to. Furthermore, before a new rule is added to the set, it is evaluated on the training corpora used for the dictionary method.
Figure 2.
A sample post-processing rule
1 DISEASE refers to a MeSH main heading belonging to the Diseases category or Mental Disorders subcategory (in the Psychiatry and Psychology category).
Natural Language Processing (NLP) rules use cues from the title or abstract of an article to infer pair recommendations. More specifically, interactions between medical entities are retrieved from the text in the form of Unified Medical Language System® (UMLS®) triplets using SemRep [18]. UMLS triplets are composed of two concepts from the UMLS Metathesaurus® together with their respective UMLS Semantic Types (STs) and the relation between them, according to the UMLS Semantic Network. The knowledge expressed in these triplets is then translated into MeSH pairs using rules and a restrict-to-MeSH algorithm [19]. A sample rule is that the triplet (Enzyme AFFECTS Disease or Syndrome) translates into MeSH by attaching the subheading enzymology to the corresponding <DISEASE> term. However, some rules are more complicated and must be tailored to several term categories. For example, the triplet (Therapeutic or Preventive Procedure TREATS Disease or Syndrome) translates into MeSH by attaching the subheading surgery if the procedure is surgical (MeSH subcategory E04) or the subheading radiotherapy if the procedure involves radiation (MeSH tree node E02.815), etc. The PP and NLP rules are described in more detail in [2].
Statistical method
Statistical methods build on an existing set of indexed articles by postulating that similar articles should be indexed in a similar way. The PubMed Related Citations (PRC) method that we used was first introduced in [20] and is further described in [12]. It uses a k-Nearest Neighbors approach to find citations in the MEDLINE database that are similar to the new article to be indexed. MeSH pair recommendations are then inferred from the existing indexing of the ten nearest neighbors. Pairs used in the indexing of more than one of the ten nearest neighbors are recommended by this method.
Combining recommendations
Several previous indexing experiments [1], [11] showed that when multiple automatic methods are used, the best overall results are obtained by combining the methods. For this reason, we investigated several ways of combining the methods described in the previous section.
Pooling
We assessed the performance of the recommendations when they came from a pool of at least N methods, for N between 1 and 5.
Filtering
Based on the feedback received from the indexers on the small test corpora (see second subsection in the Results section), three methods of filtering were enforced. The first method is based on the frequency of occurrence (the number of occurrences, or nb_occurrences) of a given pair in the entire MEDLINE collection. For a given MeSH pair MH/SH, we defined the relative frequency FRel as follows:
| (2) |
where Q(MH) represents the set of allowable qualifiers for the main heading MH.
Pairs with a relative frequency beneath a certain threshold (determined using the training corpus) were filtered out of the recommendation list. The second method uses the stand-alone subheading list obtained from the PRC method, which was found to have a recall of 86% [3]. Pairs involving subheadings that are not in this list are filtered out. Finally, a third filtering method uses a list of main headings specifically prepared by the indexers while performing the qualitative evaluations on the small test corpora (see section below for additional details). This final list currently contains 92 main headings (e.g. Humans; Hybrid Cells and Mice, Inbred A) as well as all main headings in MeSH subcategories G05 (Genetic Processes), G13 (Genetic Phenomena) and G14 (Genetic Structures). Pairs involving main headings that are in this indexer-supplied list are filtered out.
Coordination rules (COORD)
A specific module was built in order to enforce the coordination rules explicitly stated in the indexing manual (such as the one shown in Figure 1) based on the set of pair recommendations obtained after pooling and filtering have been applied. A total of 38 coordination rules were included in the module.
Number of subheadings attached per main heading
After assessing recommendations made on the small test corpora, indexers decided that a maximum of three subheadings per main heading should be recommended. In order to select the best three subheadings when more than three subheadings are attached to a given main heading, we considered two approaches. One was based on the hierarchical relationships existing between subheadings; for example, therapy is an ancestor for the subheadings diet therapy, drug therapy, surgery, etc. The rule was to select only the most specific subheadings, so that if both therapy and surgery were attached to the same main heading, surgery would be selected over its ancestor therapy. However, experiments on the large training corpus proved this method of selection to be flawed as cases where more than three subheadings were attached to a main heading remained. Besides, it seemed to have a small adverse impact on performance. For these reasons, we finally decided to use a second method based on the precision obtained by each method on the training corpus. When more than three subheadings were recommended for a given main heading, we computed a score for each subheading based on which methods recommended it. The score consisted of the sum of the precisions obtained by each method on the training corpus. The three subheadings with the highest scores were selected.
Stand-alone subheading recommendations
In addition to pair recommendations, which are our primary objective, we found that stand-alone subheading recommendations obtained from the PRC method could also be useful to indexers if displayed separately from the pairs in the subheading tab of the Data Creation and Maintenance System (DCMS) indexing interface.
Evaluation measures
As reported by Lancaster [22], it is difficult to adequately evaluate the quality of indexing because even in the case of controlled indexing, there is no unique correct indexing set to use as a reference. However, as in previous studies mentioned in the background section, we used existing MEDLINE indexing as the “gold standard” indexing for a citation. Throughout the study, we used precision, recall and F-measure to perform quantitative evaluations of the results. At the beginning of the study, we expected that the pair recommendations produced automatically by our methods would be presented to NLM indexers as they work on creating MEDLINE indexing. Specifically, pair recommendations would be shown along with stand-alone main heading recommendations. In compliance with indexing rules, indexers would look at the recommendations, select appropriate main headings first, and then consider the subheadings that should be attached to them. For this reason, we evaluate subheading attachment performance after filtering out pair recommendations involving main headings not selected by indexers. Precision corresponds to the number of pairs recommended that were also in the MEDLINE indexing divided by the total number of pairs recommended (for which the main heading was in the MEDLINE indexing). Recall corresponds to the number of pairs recommended that were also in the MEDLINE indexing divided by the total number of correct pairs according to the MEDLINE indexing. The F-measure is computed as shown in Equation (3):
| (3) |
where P is precision and R is recall. A sample computation of these measures is shown below in the section Indexing of a sample citation.
Results
In this section, we present the performance of the methods that needed parameter adjustment on the training corpus. Based on these results, optimal parameters are selected and used as final settings when running the methods on the test corpora. The qualitative feedback received from the indexers after recommendations were produced for the small corpora were also used to make further adjustments before the large test corpus was processed. Finally, we illustrate the results by showing the final set of recommendations and corresponding performance scores obtained for a specific citation in the large test corpus.
Performance on the large training corpus
In this section, we present representative results obtained on the large training corpus in order to illustrate how we set the parameters used for the test corpus. Table 3 presents the performance of the JDI method when the 5, 10 and 15 top subheadings retrieved are attached to applicable main headings. The best precision (P) recall (R) and F-measure (F) are bolded.
Table 3.
Performance of the JDI method on the large training corpus when parameters vary.
| JDI – number of SH considered | P | R | F |
|---|---|---|---|
| top 5 | 26 | 33 | 29 |
| top 10 | 19 | 47 | 27 |
| top 15 | 16 | 54 | 24 |
Table 4 illustrates the method used for building the dictionary for the dictionary method. Bold figures indicate an increase in performance over the previous best results when the candidate term is added to the dictionary. Terms are included in the dictionary when they result in a positive contribution to the method performance, i.e. an increase in F-measure or an increase in precision if the F-measure remains stable. For example, the addition of the term “dogs” improves the F-measure (+9 points) so, even though it decreases the precision (− 21 points) it is included in the dictionary. On the other hand, the addition of the term “pigs” decreases the precision (− 7 points) and has no positive impact on F-measure. Therefore, it is not included in the dictionary. After preliminary investigations were made manually, the “hill climbing” process was partly automated.
Table 4.
Performance obtained on the large training corpus while adding veterinary-related terms in the dictionary.
| Terms considered for Veterinary | Performance | Decision | ||
|---|---|---|---|---|
| P | R | F | ||
| Initial set of terms: “veterinary” and “veterinarian” |
58 | 3 | 6 | - |
| Horses | 68 | 7 | 13 | Include |
| Dogs | 47 | 16 | 24 | Include |
| Horse | 47 | 17 | 25 | Include |
| Dog | 46 | 18 | 26 | Include |
| Cattle | 50 | 25 | 34 | Include |
| Calves | 49 | 27 | 35 | Include |
| Cows | 51 | 30 | 38 | Include |
| Cats | 52 | 33 | 40 | Include |
| Pigs | 45 | 36 | 40 | Discard |
| Pig | 45 | 35 | 39 | Discard |
| Sheep | 49 | 36 | 42 | Include |
Table 5 illustrates the pooling of methods. The best precision (P), recall (R), and F-measure (F) are bolded. As expected, recall is higher when few methods are required to produce the recommendations, and precision is higher when more methods are required to produce the recommendations. However, the best precision/recall balance (F=41) is obtained when N=2.
Table 5.
Pooling of N methods on the large training corpus.
| N | P | R | F |
|---|---|---|---|
| 1 | 22 | 68 | 33 |
| 2 | 36 | 46 | 41 |
| 3 | 51 | 26 | 35 |
| 4 | 63 | 8 | 15 |
| 5 | 78 | 2 | 4 |
For the purpose of scoring subheadings in cases where more than three subheadings were attached to a given main heading (see section Combining recommendations above) we used the precision obtained for JDI (26%; see Table 2 for top 5), MTI (24%), DIC (26%), PP (58%), PRC (35%), NLP (38%) and COORD (23%).
Feedback obtained from indexers on the small test corpora
Citations in the small test corpora were automatically indexed before MEDLINE MeSH indexing was available in order to obtain feedback on the automatic recommendations in the form of a critical review. The remarks made by the indexers addressed the following issues:
Missing recommendations: according to coordination rules, several recommendations were missing and caused the automatic indexing as a whole to look inconsistent and inadequate.
Erroneous recommendations: a pattern was identified where subheadings were attached erroneously albeit consistently to certain specific main headings (e.g. checktag Mice). Furthermore, pairs that looked very unlikely because they rarely occurred in MEDLINE were also recommended. Although some of these recommendations were correct, indexers thought these cases required special attention at the time of indexing, and that having them recommended automatically might confuse junior indexers. The indexers also thought it best to limit to three the number of subheadings attached to a particular main heading.
Performance on the large test corpus
Table 6 presents the overall results obtained on the large test corpus for each of the methods separately and then combined. It shows the total number of pair recommendations yielded (N) for main headings that were in the MEDLINE indexing as well as the number of subheadings covered by each method (Scope)13. It also presents the overall results obtained in terms of precision (P), recall (R), and F-measure (F). The best precision, recall, and F-measure are bolded. The performance obtained by each method on the large corpus using the parameters that were established from tests on the training corpus is reported in the top section of the table. The performance of the additional recommendations yielded through the application of coordination rules on previous recommendations is shown in the middle section of the table. Finally, the bottom section of the table presents the results obtained when combining the recommendations obtained from the various methods. First, when at least two of the six methods produced the recommendation and both main heading and subheading filtering is applied; second, when the full combination strategy14 is applied on all six methods; and finally when the full combination strategy is applied on four of the six methods (excluding NLP and JDI) as will be the case when the subheading attachment results are first integrated into the production environment. When filtering is applied as part of the combination process, about 79% of the recommendations that are filtered are removed because of the number of methods they came from, 18% are removed based on frequency, 2% are removed because they are not in the subheading list and 1% are removed because they are in the main heading exclusion list.
Table 6.
Overall results obtained on the large test corpus for all the methods and their combination (DIC= dictionary, JDI=Journal Descriptor Indexing, PP=post-processing rules, NLP=Natural Language Processing rules, PRC= PubMed Related Citations, COORD= indexing coordination rules).
| Indexing method | N | Scope | P | R | F |
|---|---|---|---|---|---|
| DIC | 865,809 | 83 | 26 | 35 | 30 |
| MTI | 344,637 | 82 | 25 | 14 | 18 |
| JDI-top5 | 726,882 | 83 | 25 | 27 | 26 |
| PP | 79,903 | 19 | 39 | 8 | 14 |
| NLP | 26,316 | 20 | 17 | 3 | 5 |
| PRC | 1,041,662 | 83 | 35 | 53 | 42 |
| COORD | 84,459 | 16 | 23 | 3 | 5 |
| At least 2 methods + MH and SH filtering |
409,227 | 83 | 48 | 28 | 36 |
| Full combination process (6 methods) |
670,097 | 83 | 48 | 30 | 36 |
| Full combination process (4 methods) |
346,475 | 83 | 49 | 25 | 33 |
More detailed data showing the performance of each method for each of the 83 subheadings as well as on main headings to which no subheading should be attached is available in supplementary files. As an example, Table 7 shows a compilation of the results obtained for two subheadings: surgery, which is one of the most frequent subheadings in MEDLINE and radionuclide imaging which is one of the least frequent. The best precision (P), recall (R), and F-measure (F) are bolded.
Table 7.
Results obtained on the large test corpus for surgery and radionuclide imaging (DIC= dictionary, JDI=Journal Descriptor Indexing, PP=post-processing rules, NLP=Natural Language Processing rules, PRC= PubMed Related Citations, COORD= indexing coordination rules).
| Surgery | Radionuclide imaging |
|||||
|---|---|---|---|---|---|---|
| Indexing method | P | R | F | P | R | F |
| DIC | 43 | 55 | 47 | 42 | 47 | 44 |
| MTI | 23 | 73 | 35 | 23 | 54 | 32 |
| JDI-top5 | 48 | 37 | 42 | 73 | 6 | 10 |
| PP | 63 | 37 | 47 | 57 | 24 | 34 |
| NLP | 64 | 7 | 13 | - | - | - |
| PRC | 47 | 67 | 55 | 26 | 56 | 36 |
| Full combination process (6 methods) |
54 | 54 | 54 | 53 | 1 | 1 |
Table 8 shows the performance of stand-alone subheading recommendations on the large test corpus for a selection of the methods.
Table 8.
Performance of stand-alone subheading recommendations
| Indexing method | P | R | F |
|---|---|---|---|
| MTI | 36 | 15 | 8 |
| PubMed Related Citations | 24 | 86 | 37 |
| Dictionary | 31 | 56 | 40 |
| Journal Descriptors Indexing-top5 | 25 | 36 | 29 |
| Journal Descriptors Indexing -top10 | 19 | 55 | 28 |
Finally, to illustrate the impact of stand-alone subheading recommendations, Table 9 shows the average number of subheadings recommended per citation by the methods as well as the average number of subheadings that are applicable to MTI-retrieved main headings or MEDLINE reference main headings.
Table 9.
Average number of allowable, recommended and used subheadings per citation in the test corpus
| Subheading Counts | |
|---|---|
| Allowable Subheadings (MTI) | 59.41 |
| Allowable Subheadings (MEDLINE) | 54.45 |
| Subheadings used by NLM indexers | 3.52 |
| Subheadings recommended by MTI | 1.18 (0.51 used) |
| Subheadings recommended by DIC | 6.13 (1.97 used) |
| Subheadings recommended by PRC | 12.48 (3.02 used) |
| Subheadings recommended by JDI5 | 4.87 (1.26 used) |
| Subheadings recommended by JDI10 | 9.74 (1.92 used) |
Indexing of a sample citation
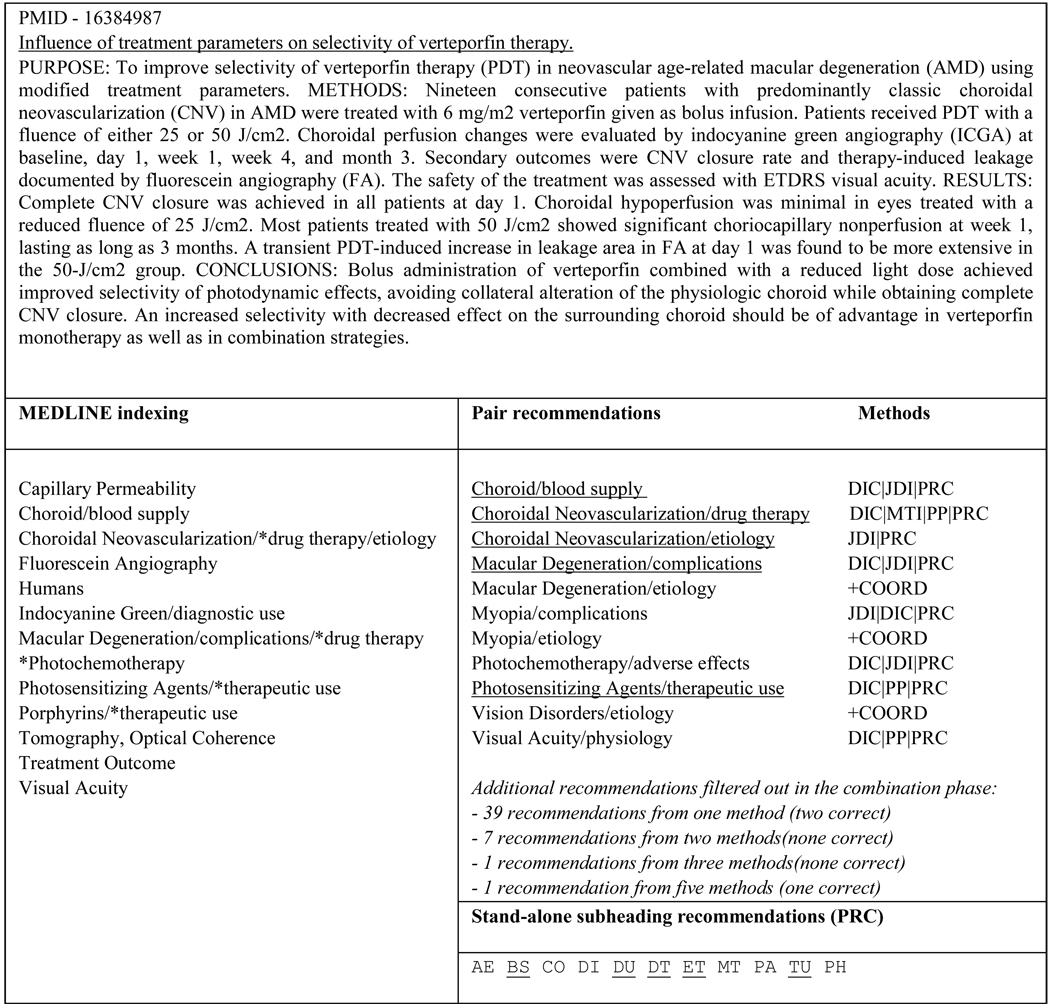
Figure 3 presents the final set of recommendations obtained for a sample citation in the large test corpus, using the full combination strategy on the six methods. We can see that the MEDLINE indexing for this citation contains eight pairs: Choroid/blood supply; Choroidal Neovascularization/drug therapy; Choroidal Neovascularization/etiology; Indocyanine Green/diagnostic use; Macular Degeneration/complications; Macular Degeneration/drug therapy; Photosensitizing Agents/therapeutic use and Porphyrins/therapeutic use. Out of the eleven pairs recommended, five were in the MEDLINE indexing (underlined). Therefore, we can compute the precision P=5/11=45%, the recall R=5/8=63% and the F-measure F=2*45*63/(45+63)=53%. Among the eleven pairs that were recommended, eight were recommended by at least two methods, and three were in fact added to the recommendation set during the combination phase through the enforcement of coordination rules (+COORD). In fact, these three pairs were triggered by the presence in the indexing set of the recommendation Photochemotherapy/adverse effects. The trigger recommendation being erroneous, the extra recommendations produced through coordination were also erroneous. Although in this case, applying coordination rules may seem detrimental to the overall quality of the recommendations, indexers insisted that having a coherent set of recommendations did make up for this inconvenience.
Figure 3.
Pair recommendations obtained for a sample citation in the Test Corpus
Discussion
Performance of the methods
The various methods exhibit complementary performance; the DIC and PRC methods tend to yield numerous recommendations and achieve high recall, while the rule-based methods (NLP and PP) tend to yield fewer recommendations but achieve high precision. The results in Table 6 average the performance of each method (and combination of methods) over the 83 subheadings as well as the 84th case where no subheading is attached to a main heading. Since the PP and NLP methods recommend fewer pairs, they achieve a very low precision (resp. 11% and 9%) for the no subheading recommendations. This explains the seemingly low overall precision (39% for PP and 17% for NLP) shown in Table 6 for these methods. In our previous evaluation [3] we reported performance averaged over the 83 subheadings only. Although the corpus used in this previous evaluation only consisted of 50,000 of the 100,000 citations used in this study, it can be noticed that the performance of the methods that cover all subheadings (MTI, DIC and PRC) is very similar to what is reported here while the precision of PP and NLP was higher (58% and 39%, respectively).
The 2% increase in recall observed between the partial combination process (at least two methods, SH and MH filtering) and the full combination process results essentially from the application of coordination rules. The overall recall of the coordination process on its own was 3%, but some of these recommendations turn out to be redundant with recommendations already provided by other methods. In addition, these figures show that the frequency filtering is efficient in weeding out mostly incorrect recommendations that do not contribute towards recall.
Table 7 illustrates more typical performance of these methods on subheadings that are within their scope.
In the case of the DIC method, it should be pointed out that the hill climbing process used to build the dictionary is dependent on the order that the terms were considered for inclusion.
The selection of stand-alone subheadings to apply to a particular citation is achieved with 86% recall with the Related Citations method (see Table 8). Although precision is only 18%, it reduces the list of applicable subheadings for a citation by about 75% (from 54 down to 12), which the indexers find useful as it may save time in deciding which subheading to use.
Combining the methods
The performance obtained for the various methods is consistent with our aim in developing them: the highest precision is obtained with the rule-based methods (NLP and PP) while the best recall is obtained with the statistical method (PRC). The other methods (JDI, DIC and MTI) have intermediate precision and recall. By applying the full combination strategy, at least one pair recommendation was made for 78% of the citations in the large test corpus (vs. 70% when only PRC, DIC, PP and MTI are combined).
In general, we observe a significant variability across methods for a given subheading and across subheadings for a given method. For example, we can see that the JDI method performs above average on surgery with 42% F-measure, whereas it performs well under average for radionuclide imaging with only 10% F-measure (see Table 7). Similarly, the NLP method yields a high precision of 64% but a low recall of 7% on surgery, whereas it produces no recommendations for radionuclide imaging which is out of its scope (see Table 7) – however, in our global evaluation (i.e. when computing the average performance data shown in Table 6) this amounts to 0% performance on this subheading. The combination of the different approaches is meant to build on the complementarities of the methods and aims at achieving the highest precision possible for a fair recall. The best recall (66%) is obtained when all the recommendations are pooled, but the corresponding precision (23%) would be unacceptable for the indexers. Table 7 shows that the combination is quite efficient with subheadings such as surgery where the F-measure is very close to that of the best method (PRC) with a significantly higher precision. However, with other subheadings such as radionuclide imaging, the good combined precision does not make up for the lack in recall. In this specific case, it is due partly to a smaller overlap in recommendations but more significantly to the fact that very few recommendations meet the frequency requirement. Future work will include efforts to improve the combination process. We anticipate that some of the work addressing the optimization of combination processes through re-ranking in the machine learning community (such as that of Ting and Witten [23]) might be difficult to adapt to our specific case for similar scalability issues as those described in the background section. Other machine learning methods aiming to mimic a curator’s decision on the relevance of indexing terms such as that described by Rodriguez-Esteban et al. [24] look more suitable for our purpose. However, they require training sets annotated by several indexers, which may be difficult to obtain.
Error Analysis
Upon careful examination, most of the main heading/subheading recommendations that do not match the gold standard fall into the pattern we first described in [2]:
Recommendation seems to be relevant
Recommendation corresponds to a concept not substantively discussed
Recommendation is incorrect
As can be seen from the indexers’ assessment presented in the next section, recommendations that are seemingly relevant or address a topic discussed in the article can be useful either because an indexer may decide to use such a recommendation even though another may not (this raises the issue of indexing consistency [25]), or because the recommendation may trigger the idea of a more suitable choice.
Errors coming from the NLP rules method, such as the recommendation of Mitotane/pharmacology in PMID “16471038” entitled “Clinical role of determination of plasma mitotane and its metabolites levels in patients with adrenal cancer”, usually fall in the first two categories because they result from a deep analysis of the text in the title and abstract of the article. Errors coming from the other methods cover all three categories. As evidenced by Figure 3, it seems there is no unique combination of methods that would help weed out truly incorrect recommendations. One recurring fault of the post-processing rules method is that a given rule may cause a subheading to be applied to several same-category terms when it should only be applied to one of these terms. For example, in PMID 16451091, the subheading drug therapy was attached to Mental Disorders, Substance-Related Disorders and Hepatitis C, Chronic when it was only relevant for the latter term. In addition, errors also occur when the term triggering the application of the rule was retrieved by MTI, but was not in the gold standard set. In the PubMed Related Citations methods, common errors result from indexing terms assigned to related articles where a different aspect of the subject matter was discussed. For example, articles related to PMID 16411348 entitled “Going smoke-free: the medical case for clean air in the home, at work and in public places.” discussed aspects of Smoking such as legislation and jurisprudence and prevention and control, which are covered in this article, but also psychology which is not. In the “jigsaw puzzle” methods, incorrect recommendations often resulted from the association of two concepts discussed in the article without relation to one another. This type of error is to be expected given the design of the method. In spite of this, jigsaw-puzzle methods contribute to enhance the overall recall.
NLM indexers’ assessment of results
The indexers’ primary concern is that the automatic recommendations not impede the indexing process. Therefore, avoiding obviously erroneous recommendations should be as important a priority as providing correct recommendations. In this respect, the performance of 32% precision (82% recall and 46% F-measure as can be seen in the “ALLresults” supplementary file) obtained on main headings to which our automatic feature did not attach any subheadings can be considered a positive result. Moreover, the F-measure obtained by combining all the methods and applying full post-processing (36%) is comparable to the inter-indexer agreement reported in [25] for main heading/subheading pairs.
The recommendation of relevant or near correct indexing terms is deemed useful even if these terms are not selected in the final indexing set. Their value lies in that they trigger the selection of a final indexing term. However, the downside of almost-correct recommendations is that they might confuse junior indexers who may not have sufficient training to distinguish between almost-correct and correct recommendations.
Based on the assessment by the NLM indexers the pair recommendations obtained with our methods will be added to the MTI display in the DCMS system (See Figure 4). In practice, the presentation of clickable attached subheadings with the MTI recommendations in DCMS led to integrating a similar feature for other tools used daily by most indexers, such as the “Neighbor” tool that shows related citations that have been previously indexed in MEDLINE. Pair recommendations obtained from four of the six methods presented (MTI, DIC, PP and PRC) are submitted to the post-processing protocol and are expected to appear in DCMS in Fall 2008. After technical issues are resolved, the two remaining methods (JDI and NLP) may be added to the production process at a later time. Stand-alone recommendations obtained from PRC are also expected to appear in the “subheading” tab of DCMS at a later date. In addition, pair recommendations will also be added as a feature of the MTI version freely available to UMLS licensees through NLM’s Semantic Knowledge Representation scheduler facility15.
Figure 4.
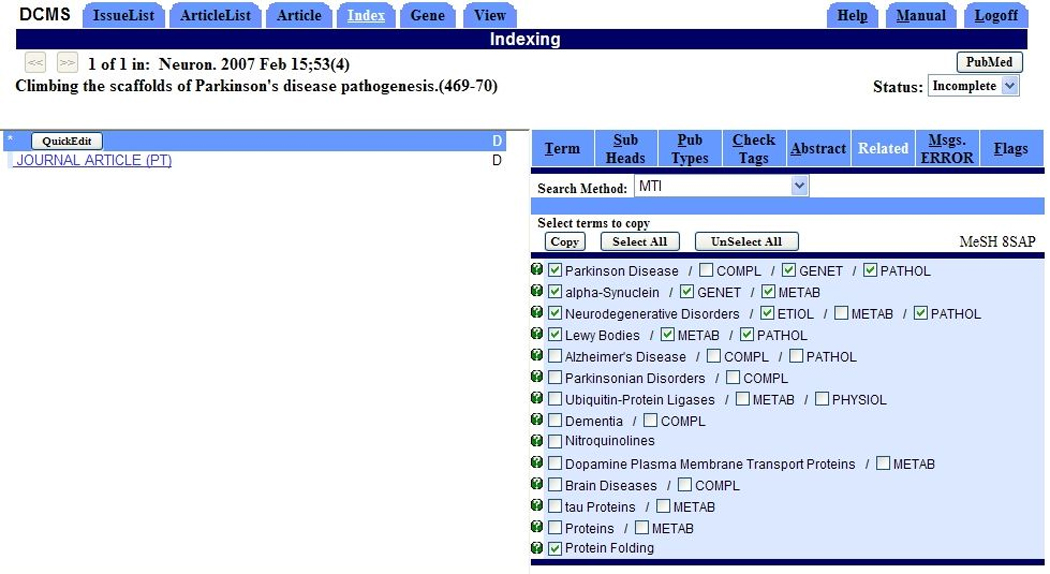
Screen Capture of the MTI tab in the DCMS system showing the automatic MeSH main heading and pair recommendations provided to NLM indexers by MTI for a sample citation. The indexing terms selected by an indexer are checked in green.
Conclusions
In this paper we have described the complexity of MeSH indexing for MEDLINE citations and reported on the latest efforts of NLM’s Subheading Attachment Project to develop advanced tools producing automatic indexing recommendations compliant with current NLM indexing policies. As a result, NLM’s Medical Text Indexer will be enhanced with a subheading attachment feature that produces main heading/subheading recommendations in addition to isolated main heading recommendations. This new feature will be used to display automatic MeSH indexing recommendations in DCMS, the interface used by indexers to create MEDLINE citations. The results of this work may also be used in the future for NLM cataloguing. Further improvements to the subheading attachment feature are still expected with the investigation of Inductive Logic Programming (ILP) as a method of automatically producing indexing rules.
Supplementary Material
Acknowledgements
This work was supported in part by the Intramural Research Program of the NIH, National Library of Medicine and by an appointment of A. Névéol to the NLM Research Participation Program sponsored by the National Library of Medicine and administered by the Oak Ridge Institute for Science and Education. The authors would like to thank James Marcetich and Joe Thomas of NLM’s Indexing Section for their interest and feedback on this work. The authors also acknowledge Willie J. Rogers for his technical help with implementing and running the JDI method.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
According to http://www.nlm.nih.gov/bsd/index_stats_comp.html. Additional data on key MEDLINE indicators may be found at http://www.nlm.nih.gov/bsd/bsd_key.html (retrieved on May 20, 2008).
NLM’s MeSH factsheet was retrieved from http://www.nlm.nih.gov/pubs/factsheets/mesh.html on May 20, 2008. It contains additional information about the MeSH thesaurus.
Checktags are a set of frequently assigned main headings such as those designating gender, age groups and animals. (e.g. Female; Child, Preschool; Mice).
This data is derived from the MeSH ASCII files retrieved from http://www.nlm.nih.gov/mesh/filelist.html on March 27, 2008.
There are 11 MeSH tree levels for 2008.
See http://mbr.nlm.nih.gov/Reference/MEDLINE_Baseline_Repository_Detail.pdf for additional details on the MEDLINE 2006 Baseline.
This corpus includes the 50,000 citations used for the evaluation reported at AMIA 2007 [3] and an additional 50,000 citations.
The list of PMIDs for each corpus is provided as supplementary material.
MTI produces main heading recommendations using a Natural Language Processing path and a Statistical path. After these approaches are merged, the results can be displayed using different levels of filtering, including the “default filtering” used here. Additional details on MTI and filtering can be found in [[1]].
http://www.nlm.nih.gov/mesh/indman/chapter_19.html (March 12, 2007)
A preliminary training corpus composed of about 17,000 citations randomly extracted from MEDLINE 2004 and our large training corpus.
http://www.nlm.nih.gov/tsd/serials/lsiou.html (April 4, 2008)
Note that the sets of indexing rules used in these experiments comprised 61 NLP rules and 778 PP rules.
i.e. when all the combination strategies described in section Combining recommendations above are applied: use of coordination rules, pooling of at least two methods, filtering based on main heading and subheading lists, limitation of the number of subheadings attached per main heading.
http://skr.nlm.nih.gov/ (retrieved on September 5, 2008)
Contributor Information
Aurélie Névéol, Email: neveola@nlm.nih.gov.
Sonya E. Shooshan, Email: sonya@nlm.nih.gov.
Susanne M. Humphrey, Email: humphrey@nlm.nih.gov.
James G. Mork, Email: mork@nlm.nih.gov.
Alan R. Aronson, Email: alan@nlm.nih.gov.
References
- 1.Aronson AR, Mork JG, Gay CW, Humphrey SM, Rogers WJ. The NLM Indexing Initiative's Medical Text Indexer. Stud Health Technol Inform. 2004;107(Pt 1):268–272. [PubMed] [Google Scholar]
- 2.Névéol A, Shooshan SE, Humphrey SM, Rindflesch TC, Aronson AR. Multiple approaches to fine-grained indexing of the biomedical literature. Pac Symp Biocomput. 2007:292–303. [PubMed] [Google Scholar]
- 3.Névéol A, Shooshan SE, Mork JG, Aronson AR. Fine-Grained Indexing of the Biomedical Literature: MeSH Subheading Attachment for a MEDLINE Indexing Tool. AMIA Annu Symp Proc. 2007 Oct 11;:553–557. [PMC free article] [PubMed] [Google Scholar]
- 4.Salton G, editor. The SMART retrieval system; experiments in automatic document processing. Englewood Cliffs, NJ: Prentice-Hall; 1983. [Google Scholar]
- 5.Deerwester S, Dumais S, Furnas G, Landauer T, Harshman R. Indexing by latent semantic analysis. J Am Soc Inf Sci. 1990 Sep;41(6):391–407. [Google Scholar]
- 6.Ruiz ME, Srinivasan P. Hierarchical text categorization using neural networks. Inf Retr. 2002;5(1):87–118. [Google Scholar]
- 7.Ruch P, Baud R, Geissbühler A. Learning-free Text Categorization. Proc AIME LNAI. 2003;2780:199–204. [Google Scholar]
- 8.Cai L, Hofmann T. Hierarchical document categorization with support vector machines. Proc CIKM. 2004:396–402. [Google Scholar]
- 9.Yang Y. An Evaluation of Statistical Approaches to Text Categorization. School of Computer Science, Carnegie Mellon University; Technical report. 1997 Retrieved on 01/20/2008 at http://reports-archive.adm.cs.cmu.edu/anon/1997/CMU-CS-97-127.ps.
- 10.Rak R, Kurgan LA, Reformat M. Multilabel associative classification categorization of MEDLINE articles into MeSH keywords. IEEE Eng Med Biol Mag. 2007 Mar–Apr;26(2):47–55. doi: 10.1109/memb.2007.335581. [DOI] [PubMed] [Google Scholar]
- 11.Aronson AR, Bodenreider O, Demner-Fushman D, Fung KW, Lee VK, Mork JG, Névéol A, Peters L, Rogers WJ. ACL. 2007. From Indexing the Biomedical Literature to Coding Clinical Text: Experience with MTI and Machine Learning Approaches. Workshop BioNLP. [Google Scholar]
- 12.Lin J, Wilbur WJ. PubMed related articles: a probabilistic topic-based model for content similarity. BMC Bioinformatics. 2007 Oct 30;8:423. doi: 10.1186/1471-2105-8-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sohn S, Kim W, Comeau DC, Wilbur WJ. Optimal training sets for bayesian prediction of MeSH assignment. J Am Med Inform Assoc. 2008 Jul–Aug;15(4):546–553. doi: 10.1197/jamia.M2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Névéol A, Rogozan A, Darmoni SJ. Automatic indexing of online health resources for a French quality controlled gateway. Inf Process Manage. 2006;42:695–709. [Google Scholar]
- 15.Liu Y, Brandon M, Navathe S, Dingledine R, Ciliax BJ. Text mining functional keywords associated with genes. Stud Health Technol Inform. 2004;107(Pt 1):292–296. [PubMed] [Google Scholar]
- 16.Humphrey SM. Automatic indexing of documents from journal descriptors: a preliminary investigation. J Am Soc Inf Sci Technol. 1999 Jun;50(8):661–674. doi: 10.1002/(SICI)1097-4571(1999)50:8<661::AID-ASI4>3.0.CO;2-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Humphrey SM, Lu CJ, Rogers WJ, Browne AC. Journal descriptor indexing tool for categorizing text according to discipline or semantic type. AMIA Annu Symp Proc. 2006:960. [PMC free article] [PubMed] [Google Scholar]
- 18.Rindflesch TC, Fiszman M. The interaction of domain knowledge and linguistic structure in natural language processing: interpreting hypernymic propositions in biomedical text. J Biomed Inform. 2003 Dec;36(6):462–477. doi: 10.1016/j.jbi.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 19.Bodenreider O, Nelson SJ, Hole WT, Chang HF. Beyond synonymy: exploiting the UMLS semantics in mapping vocabularies. Proc AMIA Symp. 1998:815–819. [PMC free article] [PubMed] [Google Scholar]
- 20.Kim W, Aronson AR, Wilbur WJ. Automatic MeSH term assignment and quality assessment. Proc AMIA Symp. 2001:319–323. [PMC free article] [PubMed] [Google Scholar]
- 21.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proc AMIA Symp. 2001:17–21. [PMC free article] [PubMed] [Google Scholar]
- 22.Lancaster FW. Indexing and abstracting in theory and practice. Champaign, IL: University of Illinois; 1991. [Google Scholar]
- 23.Ting WK, Witten I. Stacking bagged and dagged models. Proc 14th International Conference on Machine Learning; 1997. pp. 367–375. [Google Scholar]
- 24.Rodriguez-Esteban R, Iossifov I, Rzhetsky A. Imitating Manual Curation of Text-Mined Facts in Biomedicine. PLoS Comput. Biol. 2006 Sep 8;2(9):e118. doi: 10.1371/journal.pcbi.0020118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Funk ME, Reid CA. Indexing consistency in MEDLINE. Bull Med Libr Assoc. 1983 Apr;71(2):176–183. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.