Abstract
The ability to bioengineer three-dimensional (3D) tissues is a potentially powerful approach to treat diverse diseases such as cancer, loss of tissue function, or organ failure. Traditional tissue engineering methods, however, face challenges in fabricating 3D tissue constructs that resemble the native tissue microvasculature and microarchitectures. We have developed a bioprinter that can be used to print 3D patches of smooth muscle cells (5 mm × 5 mm × 81 μm) encapsulated within collagen. Current inkjet printing systems suffer from loss of cell viability and clogging. To overcome these limitations, we developed a system that uses mechanical valves to print high viscosity hydrogel precursors containing cells. The bioprinting platform that we developed enables (i) printing of multilayered 3D cell-laden hydrogel structures (16.2 μm thick per layer) with controlled spatial resolution (proximal axis: 18.0 ± 7.0 μm and distal axis: 0.5 ± 4.9 μm), (ii) high-throughput droplet generation (1 s per layer, 160 droplets/s), (iii) cell seeding uniformity (26 ± 2 cells/mm2 at 1 million cells/mL, 122 ± 20 cells/mm2 at 5 million cells/mL, and 216 ± 38 cells/mm2 at 10 million cells/mL), and (iv) long-term viability in culture (>90%, 14 days). This platform to print 3D tissue constructs may be beneficial for regenerative medicine applications by enabling the fabrication of printed replacement tissues.
Introduction
Recent breakthroughs in regenerative medicine and tissue engineering present bioengineered three-dimensional (3D) tissues as an alternative treatment for various diseases such as loss of tissue function or organ failure.1–5 Often in tissue engineering, two-dimensional (2D) or 3D scaffolds are employed to generate tissues in vitro.6,7 However, engineered tissues generated in 2D cultures do not mimic the complex microarchitecture of native tissues. Also, current 3D polymer scaffolding approaches are not suitable for fabricating complex tissue structures due to lack of spatial and temporal control during cell seeding.8–10 In the past decade, deposition of polymers/metals/cells by printing has gained momentum in electronic circuit board printing, printing of transistors, and tissue printing.11,12 Printing technology shows promise in overcoming the limitations associated with seeding cells on scaffolds. For example, bioprinting methods, such as inkjet13–15 and laser printing16–19 techniques, have been employed to control cell placement in 2D or 3D. However, some challenges still remain in existing tissue printing systems such as low cell viability, loss of cellular functionality, and clogging.20–22 Cell printing also requires extracellular matrix (ECM) to build 3D structures for long-term culture. However, the current piezo-based inkjet printing system is not easily adapted for high viscosity solutions such as collagen ECM, since it requires high impact force to generate droplets. To overcome these limitations, alginate-based cell printing23,24 and 3D fiber deposition25 approaches were used to encapsulate cells in ECM. Alginate-based cell printing is adapted to the conventional piezo-based bioprinter to prevent the rapid clogging issues by printing a low viscosity calcium chloride as crosslinking agent. However, for gelation the calcium must diffuse into alginic acid, which limits the droplet placement resolution. During the diffusion process, a change in pH also affects cell viability.23 The other approach uses the squeezing of ECM precursors from the nozzle to eliminate clogging. This approach may be limited in terms of low resolution and throughput.
An emerging approach to enhance bioprinting is to use a nozzle-free acoustic ejector, which prevents clogging during droplet generation.26–28 Another approach features a mechanical valve ejector that uses a pressure source to overcome the surface tension of high viscosity liquids.29–31 This mechanical ejector was applied for cryopreservation of cells in droplets and for cell printing. In this article, we built on the system by creating a cell-laden hydrogel droplet deposition system that can create 3D structures made of collagen, a temperature-sensitive gel. We adopted the system to evaluate a model structure using bladder smooth muscle cells (SMCs) to engineer tissues. We demonstrate that this bioprinting system can be used to (i) pattern cell-laden hydrogel droplets with microscale resolution, (ii) print hydrogel droplets containing cells in a rapid and uniform manner, and (iii) maintain long-term cell viability.
Materials and Methods
SMC collagen encapsulation
Primary bladder SMCs from Sprague Dawley rat were harvested according to a previously established protocol.32 SMC culture medium was prepared by mixing 445 mL Dulbecco's modified Eagle's medium (Gibco, Carlsbad, CA, 11965-092), 50 mL fetal bovine serum (Gibco, 10439-024), and 5 mL Pen/Strep (Sigma, St. Louis, MO, P4333) through a sterile filter (500 mL, Express Plus 0.22 μm membrane, SCGPU05RE). SMCs were cultured under standard conditions (37°C, 5% CO2) in a humidified incubator (Forma Scientific, Waltham, MA, CO2 water jacketed incubator). After the culture reached 80% confluency, cells were trypsinized (10 ×, 0.5 trypsin–EDTA; Gibco, 15400), washed, and resuspended in SMC medium to be mixed with collagen. Collagen solution was prepared by mixing 250 μL type I bovine collagen (MP Biomedicals, Solon, OH) with 50 μL sterile H2O, 50 μL 10 × phosphate-buffered saline (PBS) (DPBS, Carlsbad, CA, 14190), 50 μL fetal bovine serum, 50 μL SMC medium, and 50 μL NaOH (0.1 M, Sigma, 55881) and kept at 4°C before being mixed with SMCs (1:1 ratio).
3D printing using a droplet ejector
The droplet generation process was adjusted by controlling nitrogen gas pressure, valve opening duration, and cell concentration (Fig. 1). To fabricate a collagen-coated substrate, agarose (10% v/v mixture with distilled water and agarose powder; Fisher, Pittsburgh, PA, BP1360-100) was poured on the bare Petri dish (Falcon, Pittsburgh, PA, 35-3002) to enhance adhesion between the Petri dish and collagen. Collagen solution was then manually spread on the agarose surface and gelled. The cell-laden collagen droplets were printed onto the collagen-coated substrate. To maintain the droplet size, we kept the valve opening duration at 60 μs and nitrogen gas pressure at 34.4 kPa. To control the cell density in droplets, we used three different cell concentrations, 1 × 106, 5 × 106, and 10 × 106 cells/mL. The cell viability before and after printing was evaluated using a Live/Dead kit (Invitrogen, Carlsbad, CA, L3224). The staining solution was prepared with 0.5 μL of (1 mg/mL) calcein AM and 2 μL of (1 mg/mL) ethidium homodimer solution in 1 mL of PBS for 1 min. The staining solution was poured onto printed structures and incubated for 10 min at 37°C. The stained cells in the patch were manually counted under a florescent microscope (Eclipse Ti-s; Nikon, Melville, NY).
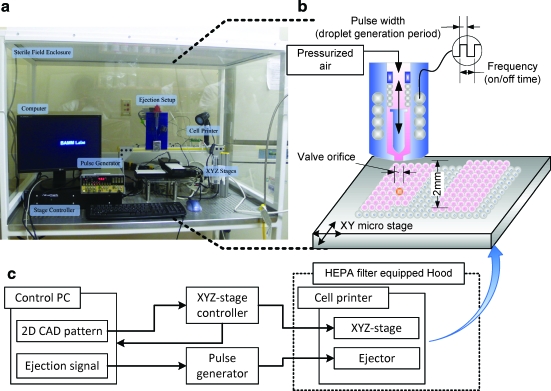
FIG. 1.
Illustration of cell encapsulating droplet printing onto a substrate. (a) Image of the cell printing setup enclosed in a sterile field (Cleanroom International, Grand Rapids, MI, 13202). (b) Schematic of droplet ejector shows cells and collagen mixture flowing into the valve driven by constant air pressure. Mixture of cells and collagen solution was loaded into a 10 mL syringe reservoir. (c) Signal flow chart shows that the xyz stage is controlled by a controller that was synchronized with a pulse generator and a control PC. With programmed sequences to build a three-dimensional (3D) structure, the apparatus can control ejection conditions, that is, stage speed, pressure, valve on/off frequency, and valve opening duration. Color images available online at www.liebertonline.com/ten.
Epitaxial layering
Using the valve-based droplet ejector setup that was previously described,29,30 cells were ejected on the prepared substrate. Using 1 × 106, 5 × 106, or 10 × 106 cells/mL, the 10 mL syringe attached to the ejector was filled with the desired cell/collagen suspension. The ejector and collagen were kept cool with liquid nitrogen (LN2, ∼5°C in gas phase) vapor to minimize viscosity changes of collagen that can solidify at room temperature. Each printed layer was gelled by incubation at 37°C for 5 min. Subsequently, another layer of collagen was printed onto the first layer. This process of layering was repeated to create 3D tissue structures.
Staining and microscopy
Printed SMC patches were gelled at 37°C for 5 min before SMC medium was added and incubated overnight. After 24 h, medium was aspirated off, and printed patches were washed three times with PBS at room temperature and fixed in 2 mL of 4% paraformaldehyde (Sigma). These patches were then rinsed with PBS three times and permeabilized with 1 mL of detergent solution (mixture of 4% bovine serum albumin and 0.1% TritonX-100 in PBS solution; Sigma). The specimens were incubated with primary antibody (actin, connexin-43, and mouse monoclonal immunoglobulin G [IgG], 1:50 dilution in PBS; Santa Cruz Biotechnology, Santa Cruz, CA) and 5 μg/mL nuclear stain 4′,6-diamidino-2-phenylindole (Invitrogen) at 37°C for 40 min. Secondary antibodies (goat anti-mouse IgG fluorescein isothiocyanate and IgG R, 1:50 dilution in PBS; Santa Cruz Biotechnology) were also incubated at 25°C for 40 min. After each incubation process, excess antibody was washed off, and stained SMC patches were imaged under the florescent microscope (Eclipse Ti-s; Nikon). The number of cells per square millimeter was plotted using SigmaPlot® that depicted cell distribution as a contour plot of an entire patch.
Results and Discussions
Uniform cell seeding density is critical for tissue engineering, since it controls the average cell-to-cell distances that influence cell-to-cell communication. The overall morphological characteristics of a tissue construct depend on this uniformity. To achieve 3D tissue structures with spatial control of cell seeding, we characterized (i) the number of cells per droplet as a function of cell loading concentration, (ii) droplet printing precision, (iii) overlapping cell-laden collagen droplets to fabricate seamless line structures, and (iv) number of cells per unit area in a printed patch.
The mechanical valve was attached to a micrometer precision xyz stage that enabled 3D spatial motion. The movement of the stage was synchronized with droplet generation signal resulting in 3D patterning capability. The platform spatially and temporally controlled the droplet placement (Fig. 1). First, we evaluated the position and density of cells in the biomaterial by printing cell-laden droplets in multiple layers. The cell-laden collagen droplets landed onto a Petri dish surface that was coated with collagen gel (Fig. 2a). This controlled placement allowed the system to deposit a cell-laden hydrogel droplet epitaxially in 2D and 3D using droplets with 650 ± 18 μm spread diameter on the surface. Uniform cell seeding was investigated by characterizing where droplets land onto a surface during droplet generation and xyz stage movement along a temporal line (distal axis, Fig. 2a). The landing locations and placement variation (δx and δy) of droplets determine the overlap between droplets when patterning lines and patches in 3D. The droplet ejection directionality was the major determinant of this variation. The system achieves 0.5 ± 4.9 and 18.0 ± 7.0 μm variation in the x (distal) and y (proximal) directions, respectively. These variations were negligible compared to the 650 ± 18 μm spread droplet diameter. To create layered structures using an intermediate collagen layer was printed between the first layer of droplets and second layer of droplets (Fig. 2b). The adjacent droplets gel together and form a single seamless layer. Further, a secondary droplet array was printed on top of the gelled layers to pattern droplets in a 3D microarchitecture (Fig. 2c). The cell-laden collagen droplet in the first layer was printed at a lower cell concentration on the substrate than the collagen droplet printed in the secondary layer to depict a layered structure.
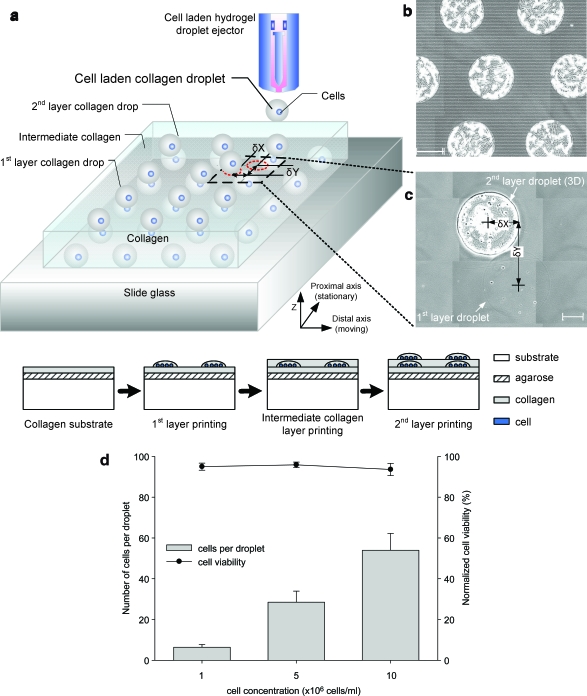
FIG. 2.
Printing platform for 3D cell-laden droplet printing. (a) Cell-laden hydrogel droplets are generated by a mechanical valve that is operated by a controlled pulse width (open period of the valve) and a frequency (on/off time of the valve) to generate required volume and timed placement of droplets onto a substrate, respectively (Fig. 1). Droplets are printed to form multiple layers of collagen; smooth muscle cell (SMC)–laden collagen droplet array (gray color sphere), intermediate collagen layer, and top SMC-laden droplet layer (blue color sphere). Image of a printed array of collagen droplets (b) and image of a multilayered array on a slide glass (c). A gray-colored droplet indicates the bottom layer of collagen shown in (c). δx and δy are measured between centers of each droplet in different layers. Mean and standard deviation values of x (distal axis) and y (proximal axis; moving axis) directional variations were 0.5 ± 4.9 and 18.0 ± 7.0 μm, respectively. (d) Number of cells per droplet and cell viability as a function of loading concentrations. Mean and standard deviation values of encapsulated cells were 6 ± 1, 29 ± 5, and 54 ± 8 cells per droplet in 1 × 106, 5 × 106, and 10 × 106 cells/mL, respectively. The cell printing platform showed 94.8 ± 0.8% average cell viability for three different concentrations compared to the culture flask. Each cell loading concentration had 94.9 ± 1.7%, 95.8 ± 1.3%, and 93.5 ± 3.0% cell viability. Scale bar: 200 μm. Color images available online at www.liebertonline.com/ten.
Second, we characterized the number of cells per droplet at three cell loading densities and the cell viability of the printing platform (Fig. 2d). It showed 6 ± 1 cells per droplet at 1 × 106 cells/mL, 29 ± 5 cells per droplet at 5 × 106 cells/mL, and 54 ± 8 cells per droplet at 10 × 106 cells/mL. The number of cells per droplet was repeatable over ejected droplets at various cell loading concentrations. Further, the number of cells per droplet increased with increasing cell loading density in the ejector reservoir. The number of cells that can be packed in a single droplet does not increase linearly with the loading density. Consequently, it is harder to pack more cells into a fixed droplet volume. To better understand cell seeding density, the mean and standard deviation for number of cells per droplet were investigated. Smaller standard deviation can be translated into a more uniform seeding density as cells are patterned to create 3D constructs. The platform also printed cells with high viability 94.8 ± 0.8% compared to the culture flask viability. The viability was calculated by the ratio of pre-ejection cell viability (96.1 ± 1.9%) and post-ejection cell viability (91.1 ± 2.3%) by counting 250 printed cells (Fig. 2d). The results showed that system precision, printing cell viability, and cells per droplet uniformity sufficed to establish controlled cell seeding density with high cell viability.
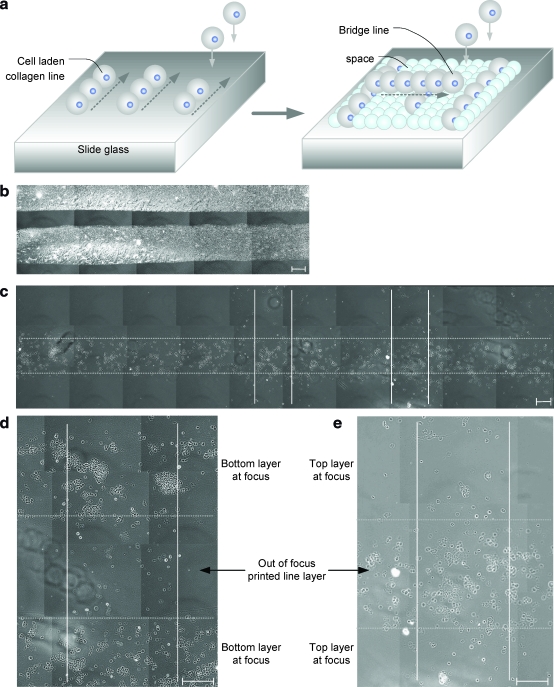
The third step was to print overlapping collagen droplets to pattern cell-laden collagen lines as we build a 3D structure. An illustration describing placement of droplets in a printed line pattern is shown by overhanging printed cell-line bridges in separate layers (Fig. 3a). The overlap between the adjacent droplets was maintained at 50% by the temporally controlled ejection. To test the system operation, two collagen lines were printed side by side in a single layer (Fig. 3b), and multiple lines were printed within separate layers of a 3D structure in a crossover pattern (Fig. 3c). These cell-laden collagen lines were placed on top of each other in the z direction by printing a cell-less collagen layer within between two layers. The magnified images of the cross-pattern bridges of printed cell lines are shown in Figures 3d and e.
FIG. 3.
Printing of cells in lines of hydrogel microstructures. (a) Illustration of printed droplets in a line pattern. Top layer of the line pattern form a 3D structure like a bridge separated by a spacing layer of hydrogel. (b, c) Dot and solid lines represent the edge of bottom and top collagen lines; dried collagen line pattern in (b) and multilayered line pattern in (c). (d, e) Magnified images show cross-patterned lines on separate layers. The top and bottom layers are shown with two focused images: bottom focused image in (d) and top focused image in (e). Scale bar: 200 μm. Color images available online at www.liebertonline.com/ten.
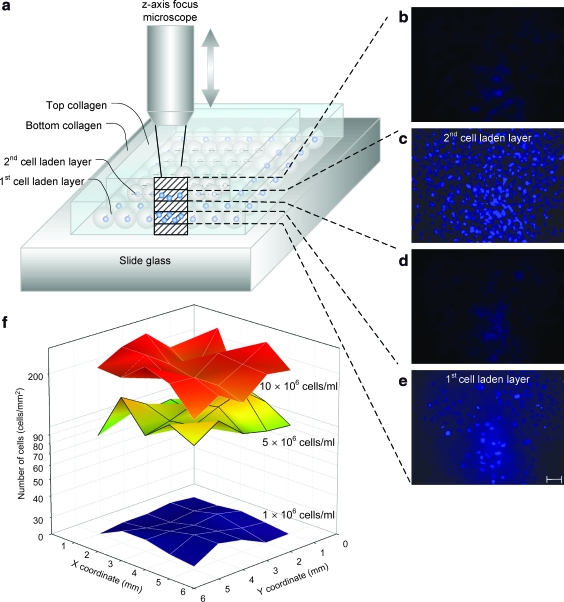
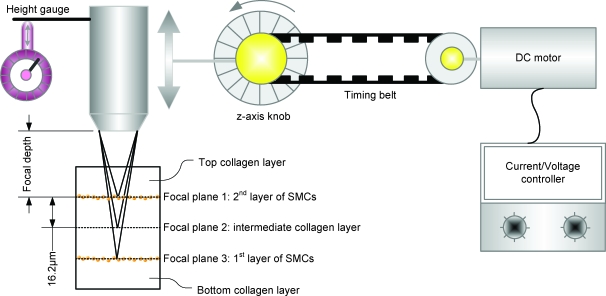
Finally, native tissue comprises multiple cell layers. To mimic such tissue architecture, the bioprinting system employs a 3D printing capability using an epitaxial method (layer by layer) (Fig. 4a). To print smooth muscle tissue constructs, cell-laden collagen droplets were patterned on top of earlier printed layers. The challenge of 3D patterning was overcome by first gelling the initial printed layer and then depositing additional cell-laden hydrogel droplets on top of the previously printed layer like in layer-by-layer epitaxy. First, a bottom cell-less collagen layer was placed in agarose. Then, on top of this layer a cell-laden collagen layer was printed. This process was repeated creating five cell-less and two cell-laden collagen layers (81 μm thick). To observe the multiple layers, a motorized system was created that steps the microscope focus (Fig. 5). Images were taken at each focus point with 16.2 μm steps (Fig. 4b–e). The printed 3D multilayer SMC-laden collagen construct was stained with 4′,6-diamidino-2-phenylindole. Focal images show printed layers with stained cells and without cells. The cell-laden layers (Fig. 4c, e) show stained circular cellular nuclei, whereas the cell-less collagen layers only show background due to staining of the gel (Fig. 4b, d). The described epitaxial method was used to observe cell seeding densities within a single printed layer at three different cell densities, 1 × 106, 5 × 106, and 10 × 106cells/mL (Fig. 4f). As shown, the cell seeding density of the printed patches was uniform right after printing: 216 ±38 cells/mm2 at 10 × 106cells/mL, 122 ± 20 cells/mm2 at 5 × 106cells/mL, and 26 ± 2cells/mm2 at 1 × 106 cells/mL.
FIG. 4.
Focal images of a printed 3D SMC tissue construct and two-dimensional cell seeding distribution. (a) Illustration of 3D patch imaging. The distance between each imaged layer is 16.2 μm which is controlled by timed imaging and moving speed of a z-axis knob (Fig. 5). (b–e) Focal images of 3D patch layers; top layer of printed collagen in (b), second layer of SMC patch in (c), intermediate collagen layer in (d), and first layer of SMC patch in (e). (f) Cell distribution of two-dimensional patch of 1, 5, and 10 million cells/mL concentration after printing (day 0). Each patch size is 5 × 5 mm. Average number and standard deviation of printed cells for each patch were 26 ± 2 cells/mm2 (average ± standard deviation) at 1 × 106 cells/mL, 122 ± 20 cells/mm2 at 5 × 106 cells/mL, and 216 ± 38 cells/mm2 at 10 × 106 cells/mL. The number of cells is represented in log scale for comparison between 1 × 106 and 10 × 106 cells/mL. Scale bar: 100 μm. Color images available online at www.liebertonline.com/ten.
FIG. 5.
Focal 3D imaging method using a motorized microscope. A direct current motor was connected to control the z-axis knob of a fluorescence microscope body by a timing belt. Each image was taken at a scheduled time by a charge-coupled device camera control software. The distance of each layer was calculated by the reference index of the microscope (65 μm/360°), motor speed (180°/s), and imaging time control (0.5 s/image). These conditions gave a resolution of 16.2 μm separation between each image for an 81-μm thick patch (five layers). Color images available online at www.liebertonline.com/ten.
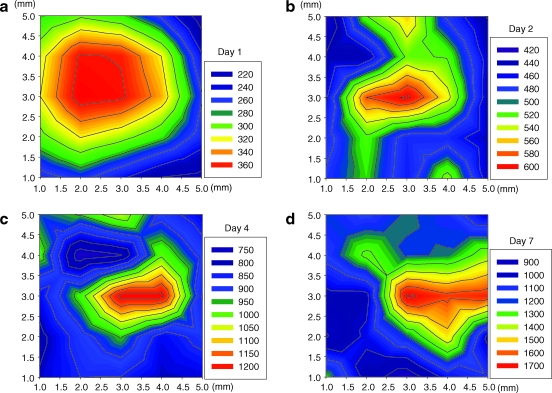
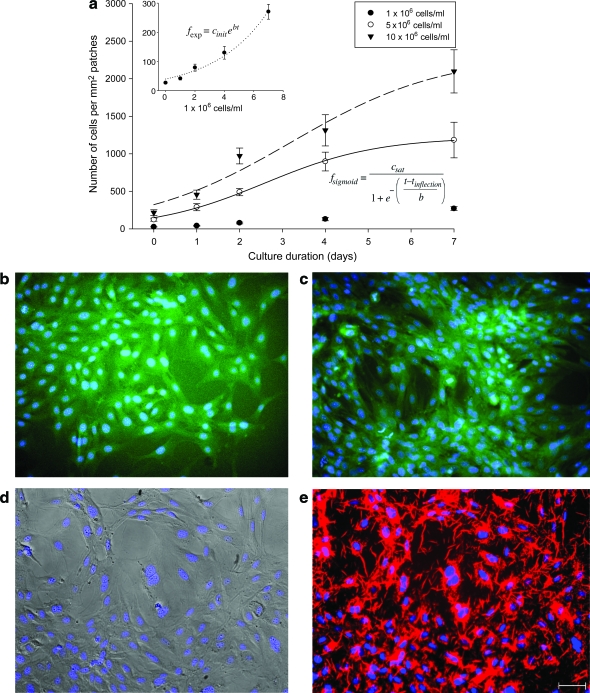
The patches were imaged after printing, and the number of cells was averaged per square millimeter in each image for an entire patch area of 25 mm2. We validated the distribution, uniformity, and variation of cell seeding density by the printing method. The topographic color coding of the top view of these patches reveals the cell distribution over 1–7 days for 5 × 106 cells/mL cell printing concentration (Fig. 6a–d). The color coding indicates the cell concentration in that area (see the legend). The increased cell seeding density correlates with the increased number of cells per droplet (Fig. 7a). This characterization is crucial, since it builds the logical tie between a cell-laden hydrogel droplet and a printed 3D tissue construct. However, the proliferation rate is not linear as a function of cell density and culture time. The rates show a sigmoid tendency as a function of culture duration, which indicates that initial high proliferation rates decrease as the number of cells per unit area increases. The inflection time, tinflection, of the sigmoid regression curves were 2.6 days for 5 × 106 cells/mL and 3.2 days 10 × 106 cells/mL. In case of 26 ±1.7 cells/mm2 initial cell loading density, the proliferation rate of cells showed an exponential increment. The exponent and the sigmoid regression functions feature unknown factor, b, which is related to cell proliferation, 0.2 for 1 × 106 cells/mL, 1.3 for 5 × 106 cells/mL, and 1.7 for 10 × 106 cells/mL. The number of cells per droplet and the precise positioning of these droplets in a 3D architecture determine the cell seeding density of the patch before the long-term culture. Such high-throughput capability and cell seeding control to create 3D tissue constructs allow potentially rapid characterization and optimization of tissues. Printing a 5 × 5 mm patch takes 10 s with 160 Hz ejection frequency. The total time becomes 10 min including the gelation time to build a secondary layer. This processing time indicates the high-throughput aspect of the system compared to the conventional scaffold methods that take 1–2 h to build a single patch. Cells are also observed to adhere and spread within the printed cell-laden collagen layer (Fig. 7b–e). In long-term culture, cells were observed to be viable as demonstrated by histological stains. During days 4 and 7, the printed cells expressed actin after the printing and culturing steps (Fig. 7b, c). Patches on the 14th day of culture expressed connexin-43 (Fig. 7d, e). This marks a positive turning point for the printed patches and indicates future possibilities for tissue engineering by this 3D bioprinting platform technology. This technology employed for tissue engineering and regenerative medicine could create avenues for functional tissues and could create a clinical impact by enhancing the quality of life for patients.
FIG. 6.
Cell distribution of printed SMC patch in culture. (a–d) Quantification of cell distribution and cell proliferation within a single layer of printed SMC patch: day(s) 1 in (a), 2 in (b), 4 in (c), and 7 in (d) for 5 × 106 cells/mL. Each patch size is 5 × 5 mm (xy-axis index). The cell distribution of printed cells for each patch was 289 ± 47 cells/mm2 (average ± standard deviation) in (a), 489 ± 48 cells/mm2 in (b), 897 ± 125 cells/mm2 in (c), and 1183 ± 236 cells/mm2 in (d). Color images available online at www.liebertonline.com/ten.
FIG. 7.
Characterization of printed SMC patch in culture. The proliferation graph shows increasing number of cells over a period of time in collagen patches for three initial cell concentrations (Cinit), that is, 1 × 106, 5 × 106, and 10 × 106 cells/mL. (a) The total number of cells per square millimeter in three different initial printing concentrations were measured from day 0 to 7. Inset represents an enlarged figure of 1 × 106 cells/mL initial cell loading density. After 7 days of culturing (Csat), 270 ± 25, 1183 ± 236, and 2097 ± 287 cells/mm2 were observed for 1 × 106, 5 × 106, and 10 × 106 cells/mL, respectively. The inflection time (tinflection) of sigmoid regression curves was 2.6 days for 5 × 106 cells/mL and 3.2 days for 10 × 106 cells/mL. In case of 26 ± 1.7 cells/mm2 initial cell loading density, proliferation rate of cells showed an exponential increment. The unknown factor for cell proliferation b is a factor of each exponent and sigmoid regression functions, 0.2 for 1 × 106 cells/mL, 1.3 for 5 × 106 cells/mL, and 1.7 for 10 × 106 cells/mL. (b–e) Stained SMC patch images for 1 × 106 cells/mL concentration after day(s) in culture: day 4 culture of SMC patch stained with 4′,6-diamidino-2-phenylindole (DAPI) (blue) and actin (green) under a light microscope (10×) in (b), day 7 SMCs stained with DAPI and actin in (c), SMCs stained with DAPI (blue) at day 14 in culture in (d), SMCs stained with DAPI and connexin-43 (red) at day 14 in culture in (e). Scale bar: 100 μm. Color images available online at www.liebertonline.com/ten.
Briefly, we presented a 3D cell patterning platform that allows efficient cell–matrix deposition with microscale spatial resolution and uniform initial cell seeding density, while maintaining cell viability over long-term culture. This high-throughput system to print tissue constructs from microdroplets has the potential to enable future therapies by providing (i) uniform cell seeding, (ii) 3D cell patterning layer by layer, and (iii) viability over long-term culture.
Acknowledgments
We would like to thank The Randolph Hearst Foundation and the department of Medicine, Brigham and Women's Hospital for the Young Investigators in Medicine Award. Y.S., F.X., and U.D. were also partially supported by R21 (EB007707). This work was performed at the BAMM Labs at the HST-Brigham and Women's Hospital Center for Bioengineering, Harvard Medical School.
Disclosure Statement
No competing financial interests exist.
References
- 1.Langer R. Vacanti J.P. Tissue engineering. Science. 1993;260:920. doi: 10.1126/science.8493529. [DOI] [PubMed] [Google Scholar]
- 2.Atala A. Bauer S.B. Soker S. Yoo J.J. Retik A.B. Tissue-engineered autologous bladders for patients needing cystoplasty. Lancet. 2006;367:1241. doi: 10.1016/S0140-6736(06)68438-9. [DOI] [PubMed] [Google Scholar]
- 3.Macchiarini P. Jungebluth P. Go T. Asnaghi M.A. Rees L.E. Cogan T.A. Dodson A. Martorell J. Bellini S. Parnigotto P.P. Dickinson S.C. Hollander A.P. Mantero S. Conconi M.T. Birchall M.A. Clinical transplantation of a tissue-engineered airway. Lancet. 2008;372:2023. doi: 10.1016/S0140-6736(08)61598-6. [DOI] [PubMed] [Google Scholar]
- 4.Khademhosseini A. Langer R. Borenstein J. Vacanti J.P. Microscale technologies for tissue engineering and biology. Proc Natl Acad Sci USA. 2006;103:2480. doi: 10.1073/pnas.0507681102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nerem R.M. Cellular engineering. Ann Biomed Eng. 1991;19:529. doi: 10.1007/BF02367396. [DOI] [PubMed] [Google Scholar]
- 6.Glicklis R. Shapiro L. Agbaria R. Merchuk J.C. Cohen S. Hepatocyte behavior within three-dimensional porous alginate scaffolds. Biotechnol Bioeng. 2000;67:344. doi: 10.1002/(sici)1097-0290(20000205)67:3<344::aid-bit11>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 7.Wang X. Yan Y. Pan Y. Xiong Z. Liu H. Cheng J. Liu F. Lin F. Wu R. Zhang R. Lu Q. Generation of three-dimensional hepatocyte/gelatin structures with rapid prototyping system. Tissue Eng. 2006;12:83. doi: 10.1089/ten.2006.12.83. [DOI] [PubMed] [Google Scholar]
- 8.Yan Y. Wang X. Pan Y. Liu H. Cheng J. Xiong Z. Lin F. Wu R. Zhang R. Lu Q. Fabrication of viable tissue-engineered constructs with 3D cell-assembly technique. Biomaterials. 2005;26:5864. doi: 10.1016/j.biomaterials.2005.02.027. [DOI] [PubMed] [Google Scholar]
- 9.Ling Y. Rubin J. Deng Y. Huang C. Demirci U. Karp J.M. Khademhosseini A. A cell-laden microfluidic hydrogel. Lab Chip. 2007;7:756. doi: 10.1039/b615486g. [DOI] [PubMed] [Google Scholar]
- 10.Jakab K. Norotte C. Damon B. Marga F. Neagu A. Besch-Williford C.L. Kachurin A. Church K.H. Park H. Mironov V. Markwald R. Vunjak-Novakovic G. Forgacs G. Tissue Engineering by self-assembly of cells printed into topologically defined structures. Tissue Eng A. 2008;14:413. doi: 10.1089/tea.2007.0173. [DOI] [PubMed] [Google Scholar]
- 11.Yan H. Chen Z. Zheng Y. Newman C. Quinn J.R. Dotz F. Kastler M. Facchetti A. A high-mobility electron-transporting polymer for printed transistors. Nature. 2009;457:679. doi: 10.1038/nature07727. [DOI] [PubMed] [Google Scholar]
- 12.Calvert P. Materials science. Printing cells. Science. 2007;318:208. doi: 10.1126/science.1144212. [DOI] [PubMed] [Google Scholar]
- 13.Nakamura M. Kobayashi A. Takagi F. Watanabe A. Hiruma Y. Ohuchi K. Iwasaki Y. Horie M. Morita I. Takatani S. Biocompatible inkjet printing technique for designed seeding of individual living cells. Tissue Eng. 2005;11:1658. doi: 10.1089/ten.2005.11.1658. [DOI] [PubMed] [Google Scholar]
- 14.Mironov V. Toward human organ printing: Charleston Bioprinting Symposium. ASAIO J. 2006;52:e27. doi: 10.1097/01.mat.0000248999.25334.6a. [DOI] [PubMed] [Google Scholar]
- 15.Boland T. Xu T. Damon B. Cui X. Application of inkjet printing to tissue engineering. Biotechnol J. 2006;1:910. doi: 10.1002/biot.200600081. [DOI] [PubMed] [Google Scholar]
- 16.Ringeisen B.R. Kim H. Barron J.A. Krizman D.B. Chrisey D.B. Jackman S. Auyeung R.Y.C. Spargo B.J. Laser printing of pluripotent embryonal carcinoma cells. Tissue Eng. 2004;10:483. doi: 10.1089/107632704323061843. [DOI] [PubMed] [Google Scholar]
- 17.Ringeisen B.R. Othon C.M. Barron J.A. Young D. Spargo B.J. Jet-based methods to print living cells. Biotechnol J. 2006;1:930. doi: 10.1002/biot.200600058. [DOI] [PubMed] [Google Scholar]
- 18.Barron J.A. Wu P. Ladouceur H.D. Ringeisen B.R. Biological laser printing: a novel technique for creating heterogeneous 3-dimensional cell patterns. Biomed Microdevices. 2004;6:139. doi: 10.1023/b:bmmd.0000031751.67267.9f. [DOI] [PubMed] [Google Scholar]
- 19.Chang R. Nam J. Sun W. Direct cell writing of 3D microorgan for in vitro pharmacokinetic model. Tissue Eng C Methods. 2008;14:157. doi: 10.1089/ten.tec.2007.0392. [DOI] [PubMed] [Google Scholar]
- 20.Sikavitsas V.I. Bancroft G.N. Holtorf H.L. Jansen J.A. Mikos A.G. Mineralized matrix deposition by marrow stromal osteoblasts in 3D perfusion culture increases with increasing fluid shear forces. Proc Natl Acad Sci USA. 2003;100:14683. doi: 10.1073/pnas.2434367100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin I. Wendt D. Heberer M. The role of bioreactors in tissue engineering. Trends Biotechnol. 2004;22:80. doi: 10.1016/j.tibtech.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 22.Xu T. Gregory C.A. Molnar P. Cui X. Jalota S. Bhaduri S.B. Boland T. Viability and electrophysiology of neural cell structures generated by the inkjet printing method. Biomaterials. 2006;27:3580. doi: 10.1016/j.biomaterials.2006.01.048. [DOI] [PubMed] [Google Scholar]
- 23.Nakamura M. Nishiyama Y. Henmi C. Iwanaga S. Nakagawa H. Yamaguchi K. Akita K. Mochizuki S. Takiura K. Ink Jet Three-dimensional digital fabrication for biological tissue manufacturing: analysis of alginate microgel beads produced by ink jet droplets for three dimensional tissue fabrication. J Imaging Sci Technol. 2008;52:1. [Google Scholar]
- 24.Yuichi N. Makoto N. Chizuka H. Kumiko Y. Shuichi M. Hidemoto N. Koki T. Development of a Three-dimensional bioprinter: construction of cell supporting structures using hydrogel and state-of-the-art inkjet technology. J Biomech Eng. 2009;131:035001. doi: 10.1115/1.3002759. [DOI] [PubMed] [Google Scholar]
- 25.Fedorovich N.E. de Wijn J.R. Verbout A.J. Alblas J. Dhert W.J.A. Three-dimensional fiber deposition of cell-laden, viable, patterned constructs for bone tissue printing. Tissue Eng A. 2008;14:127. doi: 10.1089/ten.a.2007.0158. [DOI] [PubMed] [Google Scholar]
- 26.Demirci U. Yaralioglu G.G. Haeggstrom E. Percin G. Ergun S. Khuri-Yakub B.T. Acoustically actuated flextensional SixNy and single-crystal silicon 2-D micromachined ejector arrays. IEEE Trans Semicond Manuf. 2004;17:517. [Google Scholar]
- 27.Demirci U. Acoustic picoliter droplets for emerging applications in semiconductor industry and biotechnology. J Microelectromech Syst. 2006;15:957. [Google Scholar]
- 28.Demirci U. Montesano G. Single cell epitaxy by acoustic picolitre droplets. Lab Chip. 2007;7:1139. doi: 10.1039/b704965j. [DOI] [PubMed] [Google Scholar]
- 29.Demirci U. Montesano G. Cell encapsulating droplet vitrification. Lab Chip. 2007;7:1428. doi: 10.1039/b705809h. [DOI] [PubMed] [Google Scholar]
- 30.Moon S. Lin P.A. Keles H.O. Yoo S.S. Demirci U. Cell encapsulation by droplets. J Vis Exp. 2007;8:316. doi: 10.3791/316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee W. Debasitis J.C. Lee V.K. Lee J.-H. Fischer K. Edminster K. Park J.-K. Yoo S.-S. Multi-layered culture of human skin fibroblasts and keratinocytes through three-dimensional freeform fabrication. Biomaterials. 2009;30:1587. doi: 10.1016/j.biomaterials.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 32.Roby T. Olsen S. Nagatomi J. Effect of sustained tension on bladder smooth muscle cells in three-dimensional culture. Ann Biomed Eng. 2008;36:1744. doi: 10.1007/s10439-008-9545-5. [DOI] [PubMed] [Google Scholar]