Abstract
Regulator of G-protein signaling (RGS) proteins are strong modulators of G-protein-mediated pathways in the nervous system. One function of RGS proteins is to accelerate the activation–deactivation kinetics of G-protein-coupled inwardly rectifying potassium (GIRK) channels. The opening of GIRK channels reduces the firing rates of neurons. Recent studies suggest that RGS proteins also modulate the coupling efficiency between GABAB (gamma-amino butyric acid-type B) receptors and GIRK channels in dopamine neurons of the ventral tegmental area (VTA), the initial target for addictive drugs in the brain reward pathway. Chronic drug exposure can dynamically regulate the expression levels of RGS. Functional and behavioral studies now reveal that levels of RGS2 protein, through selective association with GIRK3, critically determine whether GABAB agonists are excitatory or inhibitory in the VTA. The regulation of RGS protein in the reward pathway may underlie adaptation to different types of addictive drugs.
Introduction
The brain contains an intrinsic reward system that originates in the VTA and is activated by unexpected natural rewards, such as food and sex, and addictive drugs (Box 1). Within the VTA, DA neuron activity is controlled by GABA interneurons. Recently, we have shown that γ-hydroxybutyrate (GHB), an addictive club-drug, can activate DA neurons through its action on GABAB receptors [1,2]. GABAB receptors are members of a large family of G-protein-coupled receptors (GPCRs), which contain seven transmembrane domains and signal via heterotrimeric G-proteins (Gαβγ). Activation of GPCRs promotes the exchange of GTP for GDP on the Gα subunit. Activated G-proteins then dissociate into Gα-GTP and Gβγ dimers, which interact with a wide-range of effectors, including cyclases, lipases and ion channels. The Gα subunit terminates the activation by hydrolyzing GTP into GDP and reassembling the inactive heterotrimer (Gα-GDP*Gβγ).
Box 1 The mesocorticolimbic dopamine system in health and disease.
The mesocorticolimbic dopamine system originates in the ventral tegmental area (VTA), and projects to the nucleus accumbens (NAc), the prefrontal cortex, the septum, the amygdala and the hippocampus. The majority of projection neurons release the neuromodulator dopamine (DA) when the basal firing activity converts to burst firing. Under physiological conditions, this burst-firing of DA neurons occurs when a reward is received by surprise. If a conditioned stimulus (CS) predicts the reward, burst-firing activation occurs with the CS instead of the now expected reward. If, on the other hand, a CS is presented and no reward delivered, DA neurons reduce firing. Taken together, one function of DA neurons is to code for the prediction error of reward rather than reward itself [38].
All addictive drugs increase DA concentrations in target nuclei of the mesocorticolimbic system. A leading hypothesis to explain addiction is that the release of DA, even when reward is expected, generates a pathological learning signal that represents the first step towards compulsion [39]. At the cellular level, inappropriate release of DA may trigger adaptive phenomena, such as drug-evoked synaptic plasticity in the VTA and its target nuclei.
The molecular determinants underlying the increase of DA levels are specific for each class of drug, but occur through three main cellular mechanisms [31]. Opioids, cannabinoids, GHB, and probably benzodiazepines primarily decrease the activity of GABA interneurons, leading to disinhibition of DA neurons. Nicotine directly depolarizes DA neurons. Psychostimulants, like amphetamines, cocaine and ecstasy, interfere with DA uptake, leading to elevated levels of synaptic DA.
Addictive drugs also induce dependence, which in contrast to addiction, is defined by the occurrence of a withdrawal syndrome upon abrupt termination of drug exposure. Dependence is typically associated with tolerance, which requires that subjects increase the dose to obtain the same drug effect. The observation that chronic GHB exposure makes acute administration of GHB inhibit rather than excite DA neurons, may reflect a special form of tolerance that cannot be overcome by increasing the GHB dose.
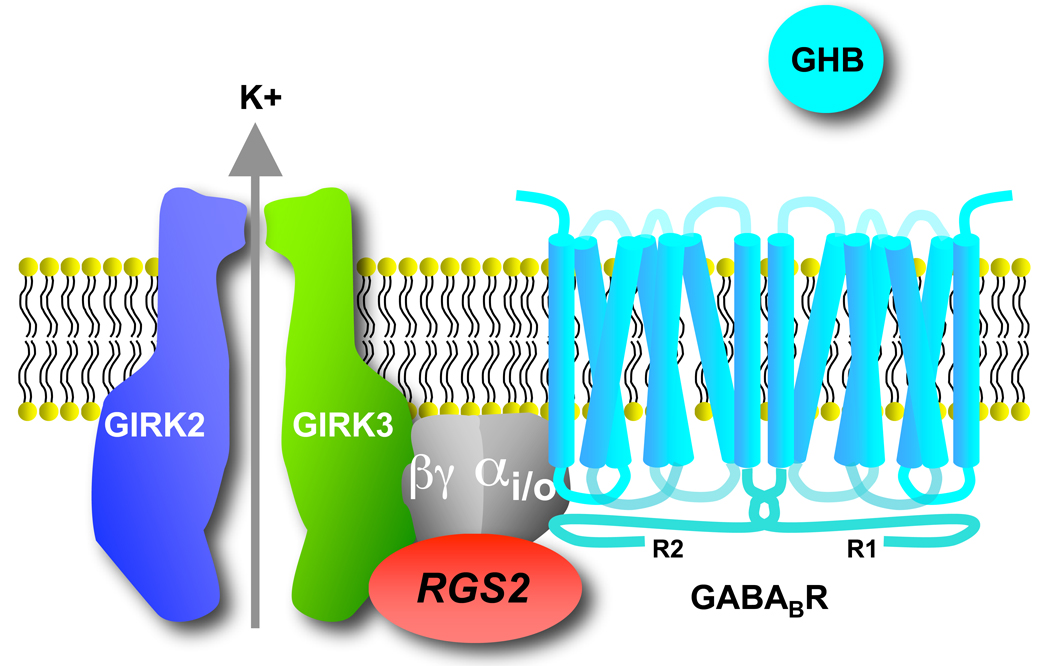
Activation of GABAB receptors leads to opening of GIRK channels. Primarily three different GIRK channels exist in the mammalian brain (GIRK1-3) that assemble into homo-tetramers (GIRK2) or hetero-tetramers (GIRK1/2, GIRK1/3, and GIRK2/3) [3]. GIRK channels preferentially allow K+ ions to enter the neuron (referred to as inward rectification). However, the small outward flow of K+ ions is of physiological relevance because it reduces the excitability of neurons. Stimulation of GPCRs that communicate through pertussis toxin (PTX)-sensitive G-proteins (the Gi/o family), such as the GABAB receptor, activates GIRK channels through the direct binding of G-protein Gβγ dimers to the channel [3]. More recent evidence suggests the PTX-sensitive Gα subunits also associate directly with the channel, suggesting the inactive heterotrimer is situated near the channel forming a signaling complex [4,5] (Figure 1). In summary, GIRK channels contribute to the resting membrane potential of neurons and, upon receptor stimulation, generate a hyperpolarizing postsynaptic potential [3].
Figure 1. Macromolecular signaling complex.
A preassembled complex exists between the dimeric GABAB R1 and R2 subunits, the heterotrimeric G-proteins, GIRK2/3 channels and RGS2. Note that RGS2 interacts selectively with GIRK3 subunits, and heterotrimeric G-proteins associate directly with GIRK and GABAB receptors, thus enabling RGS to modulate the coupling efficiency (i.e. EC50) between GABAB receptors and GIRK channels.
Changing the strength of GPCR signaling in the VTA may affect the response to addictive drugs. In principle, there are three distinct mechanisms for modifying GPCR signaling. One way is through GPCR desensitization, which involves both clathrin-mediated endocytosis [6] and uncoupling of G-proteins [7]. Another mechanism is through changes in effector activity, such as down-regulation of GIRK and calcium channels [5,8]. Lastly, changes in G-protein availability can alter GPCR signaling. G-protein availability can be influenced by guanine nucleotide dissociation inhibitor proteins (GDI) [9] and by RGS proteins. The family of RGS proteins contains a GTPase-activating protein (GAP) domain that promotes the formation of the inactive G protein heterotrimer [10,11]. In addition to the GAP domain, different RGS proteins contain a wide-range of other signaling domains [12]. RGS proteins have received much attention as key proteins in the response to addictive drugs [13]. The functional consequences of changes in RGS proteins are not well understood.
We have recently discovered that regulation of a RGS protein subtype in the brain reward pathway (see Box 1) dramatically alters GIRK channel signaling in dopamine neurons [2]. In this opinion essay, we discuss the evidence for the involvement of RGS proteins in addiction, the role of RGS2 proteins in directly modifying signaling through GIRK channels, and postulate the role of RGS proteins in regulating GPCR signaling in other pathways.
An extended family of RGS proteins
RGS proteins comprise a large family of proteins that contain over 37 different members [12]. Each RGS protein has a conserved 120 amino acid core domain, commonly called the RGS domain, which is responsible for the GAP activity [14,15]. RGS proteins are widely expressed throughout the brain, where they can potentially modulate GPCR-mediated signaling [16]. Individual RGS proteins interact with particular Gα subunits, which may be determined by specific sequences in the RGS domain, their selective expression, and the corresponding Gα subunit. For example, two RGS9 splice variants, RGS9-1 and RGS9-2, differ only in the C terminal tail, which confers selectivity with interaction proteins [17]. In addition, cell-type specific expression distinguishes the splice variants. RGS9-1 is observed almost exclusively in the retina and is responsible for acceleration of hydrolysis of GTP by Gαt. RGS9-2, in turn, is strongly expressed in the striatum and regulates D2 dopamine and µ-opioid receptors coupled to Gαi/o [18]. Thus, the combination of cell-specific expression and selective protein-protein association for different RGS proteins suggest key functional roles for specific RGS proteins in the brain.
Using GIRK channels as ultra-sensitive detectors (nM sensitivity) for Gβγ subunits, the effect of RGS proteins on GPCR signaling can be easily observed. In heterologous cells expressing GIRK channels and a GPCR, RGS proteins accelerate both activation and deactivation rates up to 100 fold [19]. For example, RGS1 through RGS5 and RGS8 proteins accelerate both activation and deactivation kinetics of GIRK currents after stimulation of M2 muscarinic or serotonin A1 receptors [20,21]. The acceleration of deactivation kinetics agrees well with the GAP activity in the RGS. The faster rate of activation with RGS, on the other hand, is not well understood. The GAP activity of the RGS would be expected to slow, not accelerate, the rate of activation. RGS proteins will increase the pool of available G proteins, which could accelerate activation [22]. Alternatively, for GIRK channels, the formation of a receptor–G-protein–GIRK complex has been proposed to promote faster activation [20,23]. Interestingly, both RGS7 and RGS8 accelerate the activation of GIRK current but RGS8 more prominently accelerates deactivation [24]. These findings suggest that functional domains other than GAP could be involved in modulation.
RGS proteins: a member of the macromolecular signaling club
An emerging theme with GIRK signaling is that GPCRs, G-proteins and GIRK channels exist in a macromolecular signaling complex [25] (Figure 1). Now, recent studies suggest that RGS proteins and GIRK channels may also interact directly within this complex. For example the degradation-resistant RGS4 co-precipitates with several GPCRs and GIRKs forming stable macromolecular complexes, while RGS3s does not interact with the GPCR-GIRK channel complex [26]. These observations suggest a “precoupling” model in the case of RGS4 versus a “collision coupling” model for RGS3. “Precoupling” can accelerate GIRK channel gating with a 100-fold higher potency [26]. Further support for a macromolecular complex comes from fluorescence resonance energy transfer (FRET) studies that demonstrate the interaction between RGS4 and GABAB R1 or R2 subunits [27], or between RGS2 and GIRK3 but not GIRK2 [2]. Thus, a subset of RGS proteins appears to interact selectively with GIRK channels.
Targeting RGS proteins to GIRK channels in a macromolecular complex may facilitate the RGS-mediated changes in the coupling efficiency between a GPCR and GIRK channel. Here, the coupling efficiency is determined by the EC50 concentration; that is, the agonist concentration required to activate 50% of the maximal GIRK current. Low coupling efficiency and therefore high EC50 reflect poor G-protein availability and low GIRK channel affinity for Gβγ dimers. The value of the EC50 depends on several additional parameters, including receptor number, receptor-ligand affinity, and agonist efficacy, which however remain constant under the conditions studied here.
Direct evidence that RGS proteins can specifically influence receptor-GIRK coupling comes from several studies [28][29][2]. First, co-transfection of RGS3s reduces the coupling efficiency (i.e. increases the EC50) between muscarinic M2 or 5HT1A receptors and GIRK1/2 heteromeric channels in vitro [28,30]. Interestingly, the increase in EC50 was greater for RGS4 even though both RGS3s and RGS4 strongly accelerate the deactivation kinetics. Second, in acute slices of the VTA, selective expression of RGS2 in DA neurons modulates the coupling between GABAB receptors and GIRK channels [2]. Pharmacological inhibition of RGS proteins or genetic ablation of RGS2 in DA neurons decreases the EC50, from ~15 µM in control neurons to ~7 µM. Thus, the presence of RGS2 opposes G-protein activation of GIRK channels, leading to a higher EC50. The lower Gβγ affinity for GIRK2/GIRK3 heteromeric channels also contributes to the shift in EC50 for baclofen activation [1][29] and DA neurons uniquely express GIRK2/GIRK3 channels. We observed a similar decrease in EC50 in GIRK3 knockout mice as well as in RGS2/GIRK3 double-knockout mice [2]. These findings suggest that RGS2 specifically modulates the coupling to channels containing GIRK3. Indeed, a close association between GIRK3 and RGS2 is detected using FRET spectroscopic measurements [2]. Thus, specific protein-protein interactions among RGS proteins, G-proteins, GIRK channels and GABAB receptors may be involved in establishing macromolecular signaling complexes that fine-tune the coupling efficiency of GPCR signaling.
Ups and downs of RGS expression
Due to their role in modifying GPCR signaling, the expression of RGS proteins could be altered by chronic exposure to addictive drugs. Indeed, numerous studies have demonstrated that the expression levels of RGS genes and proteins in the brain are dynamically regulated with drugs of abuse [13]. Both elevations and reductions in RGS transcripts and protein have been described with exposure to addictive drugs, however, suggesting an important but poorly understood adaptive function with chronic drug exposure. We will consider two general classes of addictive drugs (see Box 1, [31]): psychostimulants, such as cocaine and methamphetamine, which lead to elevated dopamine (DA) levels and prolonged DA signaling; and morphine and the club drug GHB, which activate opioid and GABAB receptors, respectively. Acute administration of cocaine and amphetamines reduces RGS in some regions of the brain while increasing RGS in other regions (Table 1). Chronic exposure to psychostimulants generally increases levels of RGS transcripts. Acute administration of morphine typically increases RGS expression, while chronic exposure reduces RGS expression – the opposite response of psychostimulants. Exceptions to these general observations, however, may reflect species differences or subtle variations in the treatment protocols. We found that chronic GHB treatment (injections twice-daily for one week) leads to a reduction of RGS2 mRNA in DA neurons of the VTA (Figure 2, [2]). In summary, addictive drugs lead to bidirectional changes in the levels of RGS transcripts and protein, suggesting complex changes in G-protein signaling, depending on the cell type and region of the brain.
Table 1. Effects of addictive drugs on RGS expression.
Compilation of studies reporting changes in RGS-mRNA expression levels with acute or chronic exposure to addictive drugs. For discussion see text. CPu=caudate putamen; DCG=dorsal central gray; NAc=nucleus accumbens; PAG=periaqueductal gray; RtTg=reticulo-tegmental nucleus
| Drug | Treatment | RGS mRNA | Brain region | Animal model | Ref |
|---|---|---|---|---|---|
| Morphine | Acute | ↑ RGS9-2 protein | NAc, PAG and dorsal striatum |
C57BL/6J mice |
[40] |
| Striatum and thalamus | Albino mice CD-1 |
[41] | |||
| ↓ RGS9-2 | Cortex | ||||
| ↑ RGS4 | NAc and DCG | Sprague– Dawley rats |
[42] | ||
| ↓ RGS4 | RtTg and LC | ||||
| Chronic | ↓RGS9-2 | NAc, PAG and dorsal striatum |
Sprague– Dawley rats |
[40] | |
| ↑ RGS9-2 | Striatum, Thalamus, PAG, Cortex |
Albino mice CD-1 |
[41] | ||
| ↑ RGS4 | LC | Sprague– Dawley rats |
[42] | ||
| ↑ RGS4 protein | LC | Sprague– Dawley rats |
[43] | ||
| ↓ RGS2 | VTA (DA neurons) | C57BL/6J mice |
[2] | ||
| GHB | Chronic | ↓ RGS2 | VTA (DA neurons) | C57BL/6J mice |
[2] |
| Cocaine | Acute | ↑ RGS4 | NAc and DCg | Sprague– Dawley rats |
[42] |
| ↓ RGS4 | RtTG and LC | ||||
| ↓RGS2 | Hippocampus, cortex, and striatum |
Sprague Dawley or Fischer-344 rats |
[44] | ||
| Chronic | ↑RGS4 | NAc and CPu | C57BL/6J mice |
[45] | |
| ↑RGS4 | LC | Sprague– Dawley rats |
[42] | ||
|
Amphetamine/ Methamphetamine |
Acute | ↑ RGS2, RGS3 and RGS5 |
Striatum | Fischer rats | [46] |
| ↓ RGS4 | Forebrain | Sprague– Dawley rats |
[47] | ||
| ↑ RGS4 | NAc and DCG | Sprague– Dawley rats |
[42] | ||
| Chronic | ↑ RGS4 | LC | Sprague– Dawley rats |
[42] |
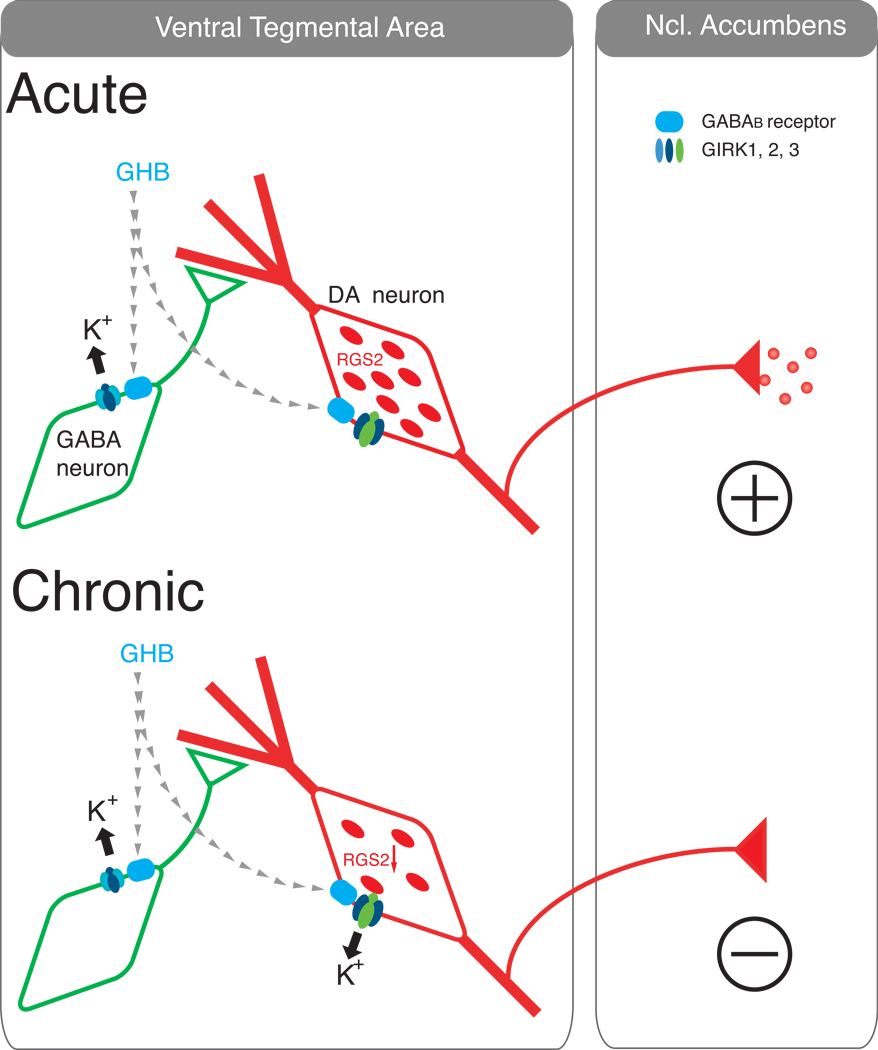
Figure 2. Chronic GHB-dependent decrease in RGS2 protein in VTA-DA neurons changes the neuronal excitability and behavioural response to GHB.
Acute administration of GHB stimulates GABAB receptors (upper panel) coupled to GIRK channels in GABA neurons green). DA neurons (red) are unresponsive to GHB because high levels of RGS2 reduce the coupling efficiency (high EC50) between GABAB receptors and GIRK channels. Opening of GIRK channels in GABA neurons disinhibits DA neurons and promotes DA release in the NAc. Chronic exposure to GHB (lower panel) reduces RGS2 expression in DA neurons, strengthening the GABAB receptor-GIRK coupling efficiency. In this situation, acute administration of GHB also stimulates GABAB receptors coupled to GIRK channels in DA neurons, thereby directly inhibiting DA activity and reducing DA release in the NAc. The unique expression of GIRK2 and GIRK3 channels in DA neurons, the lower Gβγ affinity of GIRK2/3 heteromeric channels, and the selective association of RGS2 with GIRK3 enable the up- or down-regulation of DA activity.
While it is clear that RGS levels can be up- or down-regulated in specific regions of the brain in response to drugs of abuse, the functional consequence of this change in expression has remained more elusive. Few studies have examined the behavioral consequences of changes in RGS protein levels. In one study, overexpression of RGS9-2 in the NAc reduced the locomotor responses to cocaine while RGS9-deficient mice showed augmented locomotor and rewarding responses to cocaine [32]. Conversely, we found that chronic exposure to GHB decreases RGS2 protein (Figure 2, [2]), which significantly enhances the coupling efficiency between GABAB receptors and GIRK channels in the DA neurons of the VTA. This shift in the EC50 to lower concentrations (increase in coupling efficiency) is sufficient such that low concentrations of GHB, which normally do not activate GIRK currents in DA neurons, can now hyperpolarize DA neurons and decrease DA firing rates. Remarkably, this decrease in DA neuron excitability is associated with a behavioral loss of drinking preference for GHB.
The mechanism by which addictive drugs change RGS expression is not well understood but likely involves DA receptors (D1 and D2), activation of cAMP via adenylyl cyclase, PKA-dependent phosphorylation and CREB [33][34][35][36]. RGS regulation via DA receptors may be unique for RGS2, which is the only member of the RGS family found thus far to contain a cAMP-responsive promoter region. During withdrawal, when cAMP levels are elevated due to super-sensitization of adenylyl cyclase, RGS2 levels could quickly recover and exceed baseline values due to the cAMP-responsive promoter region. More studies are needed to delineate the mechanism of drug-dependent changes in RGS expression.
Conclusions
Numerous studies have shown that sustained exposure to addictive drugs can regulate mRNA expression of several members of the RGS family. We have recently discovered that drug-dependent down-regulation of RGS2 protein significantly enhances coupling efficiency of GABAB receptors with GIRK channels. These changes were associated with a polarity switch in the output of the VTA, in which the behavioral response to GHB is converted from reinforcing to aversive in animals chronically treated with GHB. The unique expression of GIRK2/3 channels and RGS2 proteins in the VTA-DA neurons, combined with selective molecular interactions between GIRK3 and RGS2 in a signaling complex, enable the electrophysiological and behavioral polarity switch in DA neurons from the VTA. These studies highlight RGS proteins as powerful regulators of GPCR-GIRK coupling efficiency and suggest a mechanism for a special form of tolerance (Box 1).
Demonstrating that RGS proteins participate in the adaptive response to addictive drugs suggest RGS proteins could be novel targets for treating addiction. Harnessing the change in coupling efficiency in DA neurons could provide a new avenue of research for treating addiction. For example, selective inhibition of the GAP domain in RGS2 would be expected to enhance GPCR signaling in the VTA, reducing DA neuron excitability. Alternatively, selective inhibition of RGS proteins could modify tolerance and dependence to certain drugs. More selective pharmacological tools will be needed, however, to avoid negative side-effects. Future studies developing inhibitors targeted to some of the unique domains in RGS proteins could be one solution [37]. Lastly, if RGS-dependent changes in coupling efficiency between GPCRs and other effectors are found, then RGS proteins may become interesting targets for a wide range of pathologies from drug addiction to hypertension and heart disease.
References
- 1.Cruz HG, et al. Bi-directional effects of GABA(B) receptor agonists on the mesolimbic dopamine system. Nat. Neurosci. 2004;7:153–159. doi: 10.1038/nn1181. [DOI] [PubMed] [Google Scholar]
- 2.Labouebe G, et al. RGS2 modulates coupling between GABA(B) receptors and GIRK channels in dopamine neurons of the ventral tegmental area. Nat. Neurosci. 2007 doi: 10.1038/nn2006. [DOI] [PubMed] [Google Scholar]
- 3.Stanfield PR, et al. Constitutively active and G-protein coupled inward rectifier K+ channels: Kir2.0 and Kir3.0. Rev. Physiol. Biochem. Pharmacol. 2002;145:47–179. doi: 10.1007/BFb0116431. [DOI] [PubMed] [Google Scholar]
- 4.Peleg S, et al. G(alpha)(i) controls the gating of the G protein-activated K(+) channel, GIRK. Neuron. 2002;33:87–99. doi: 10.1016/s0896-6273(01)00567-0. [DOI] [PubMed] [Google Scholar]
- 5.Clancy SM, et al. Coregulation of natively expressed pertussis toxin-sensitive muscarinic receptors with G-protein-activated potassium channels. J. Neurosci. 2007;27:6388–6399. doi: 10.1523/JNEUROSCI.1190-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanyaloglu AC, von Zastrow M. Regulation of GPCRs by endocytic membrane trafficking and its potential implications. Annu. Rev. Pharmacol. Toxicol. 2008;48:537–568. doi: 10.1146/annurev.pharmtox.48.113006.094830. [DOI] [PubMed] [Google Scholar]
- 7.Kelly E, et al. Agonist-selective mechanisms of GPCR desensitization. Br. J. Pharmacol. 2008;153 Suppl 1:S379–S388. doi: 10.1038/sj.bjp.0707604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altier C, et al. ORL1 receptor-mediated internalization of N-type calcium channels. Nat. Neurosci. 2006;9:31–40. doi: 10.1038/nn1605. [DOI] [PubMed] [Google Scholar]
- 9.Willard FS, et al. Return of the GDI: the GoLoco motif in cell division. Annu. Rev. Biochem. 2004;73:925–951. doi: 10.1146/annurev.biochem.73.011303.073756. [DOI] [PubMed] [Google Scholar]
- 10.Ross EM, Wilkie TM. GTPase-activating proteins for heterotrimeric G proteins: regulators of G protein signaling (RGS) and RGS-like proteins. Annu. Rev. Biochem. 2000;69:795–827. doi: 10.1146/annurev.biochem.69.1.795. [DOI] [PubMed] [Google Scholar]
- 11.Pierce KL, et al. Seven-transmembrane receptors. Nat. Rev. Mol. Cell. Biol. 2002;3:639–650. doi: 10.1038/nrm908. [DOI] [PubMed] [Google Scholar]
- 12.Willars GB. Mammalian RGS proteins: multifunctional regulators of cellular signalling. Semin. Cell. Dev. Biol. 2006;17:363–376. doi: 10.1016/j.semcdb.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 13.Traynor JR, Neubig RR. Regulator of G protein signaling and drugs of abuse. Mol. Interv. 2005;5:30–41. doi: 10.1124/mi.5.1.7. [DOI] [PubMed] [Google Scholar]
- 14.Dohlman HG, et al. Sst2, a negative regulator of pheromone signaling in the yeast Saccharomyces cerevisiae: expression, localization, and genetic interaction and physical association with Gpa1 (the G-protein alpha subunit) Mol. Cell. Biol. 1996;16:5194–5209. doi: 10.1128/mcb.16.9.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koelle MR, Horvitz HR. EGL-10 regulates G protein signaling in the C. elegans nervous system and shares a conserved domain with many mammalian proteins. Cell. 1996;84:115–125. doi: 10.1016/s0092-8674(00)80998-8. [DOI] [PubMed] [Google Scholar]
- 16.Hollinger S, Hepler JR. Cellular regulation of RGS proteins: modulators and integrators of G protein signaling. Pharmacol. Rev. 2002;54:527–559. doi: 10.1124/pr.54.3.527. [DOI] [PubMed] [Google Scholar]
- 17.Bouhamdan M, et al. Brain-specific regulator of G-protein signaling 9-2 selectively interacts with alpha-actinin-2 to regulate calcium-dependent inactivation of NMDA receptors. J. Neurosci. 2006;26:2522–2530. doi: 10.1523/JNEUROSCI.4083-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie GX, Palmer PP. How regulators of G protein signaling achieve selective regulation. J. Mol. Biol. 2007;366:349–365. doi: 10.1016/j.jmb.2006.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mark MD, Herlitze S. G-protein mediated gating of inward-rectifier K+ channels. Eur. J. Biochem. 2000;267:5830–5836. doi: 10.1046/j.1432-1327.2000.01670.x. [DOI] [PubMed] [Google Scholar]
- 20.Doupnik CA, et al. RGS proteins reconstitute the rapid gating kinetics of gbetagamma- activated inwardly rectifying K+ channels. Proc. Natl. Acad. Sci. U S A. 1997;94:10461–10466. doi: 10.1073/pnas.94.19.10461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herlitze S, et al. New roles for RGS2, 5 and 8 on the ratio-dependent modulation of recombinant GIRK channels expressed in Xenopus oocytes. J. Physiol. 1999;517:341–352. doi: 10.1111/j.1469-7793.1999.0341t.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chuang HH, et al. Evidence that the nucleotide exchange and hydrolysis cycle of G proteins causes acute desensitization of G-protein gated inward rectifier K+ channels. Proc. Natl. Acad. Sci. U S A. 1998;95:11727–11132. doi: 10.1073/pnas.95.20.11727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Biddlecome GH, et al. Regulation of phospholipase C-beta1 by Gq and m1 muscarinic cholinergic receptor. Steady-state balance of receptor-mediated activation and GTPase-activating protein-promoted deactivation. J. Biol. Chem. 1996;271:7999–8007. doi: 10.1074/jbc.271.14.7999. [DOI] [PubMed] [Google Scholar]
- 24.Saitoh O, et al. RGS7 and RGS8 differentially accelerate G protein-mediated modulation of K+ currents. J. Biol. Chem. 1999;274:9899–9904. doi: 10.1074/jbc.274.14.9899. [DOI] [PubMed] [Google Scholar]
- 25.Doupnik CA. GPCR-Kir channel signaling complexes: defining rules of engagement. J Recept. Signal Transduct. Res. 2008;28:83–91. doi: 10.1080/10799890801941970. [DOI] [PubMed] [Google Scholar]
- 26.Jaen C, Doupnik CA. RGS3 and RGS4 differentially associate with G protein-coupled receptor-Kir3 channel signaling complexes revealing two modes of RGS modulation. Precoupling and collision coupling. J. Biol. Chem. 2006;281:34549–34560. doi: 10.1074/jbc.M603177200. [DOI] [PubMed] [Google Scholar]
- 27.Fowler CE, et al. Evidence for association of GABA(B) receptors with Kir3 channels and regulators of G protein signalling (RGS4) proteins. J. Physiol. 2007;580:51–65. doi: 10.1113/jphysiol.2006.123216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doupnik CA, et al. Measuring the modulatory effects of RGS proteins on GIRK channels. Methods Enzymol. 2004;389:131–154. doi: 10.1016/S0076-6879(04)89009-8. [DOI] [PubMed] [Google Scholar]
- 29.Jelacic TM, et al. Functional and biochemical evidence for G-protein-gated inwardly rectifying K+ (GIRK) channels composed of GIRK2 and GIRK3. J. Biol. Chem. 2000;275:36211–3626. doi: 10.1074/jbc.M007087200. [DOI] [PubMed] [Google Scholar]
- 30.Jaen C, Doupnik CA. Neuronal Kir3.1/Kir3.2a channels coupled to serotonin 1A and muscarinic m2 receptors are differentially modulated by the "short" RGS3 isoform. Neuropharmacology. 2005;49:465–476. doi: 10.1016/j.neuropharm.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 31.Lüscher C, Ungless MA. The Mechanistic Classification of Addictive Drugs. PLoS Med. 2006;3:e437. doi: 10.1371/journal.pmed.0030437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rahman Z, et al. RGS9 modulates dopamine signaling in the basal ganglia. Neuron. 2003;38:941–952. doi: 10.1016/s0896-6273(03)00321-0. [DOI] [PubMed] [Google Scholar]
- 33.Pepperl DJ, et al. Regulation of RGS mRNAs by cAMP in PC12 cells. Biochem. Biophys. Res. Commun. 1998;243:52–55. doi: 10.1006/bbrc.1997.8056. [DOI] [PubMed] [Google Scholar]
- 34.Taymans JM, et al. Striatal gene expression of RGS2 and RGS4 is specifically mediated by dopamine D1 and D2 receptors: clues for RGS2 and RGS4 functions. J. Neurochem. 2003;84:1118–1127. doi: 10.1046/j.1471-4159.2003.01610.x. [DOI] [PubMed] [Google Scholar]
- 35.Thirunavukkarasu K, et al. Analysis of regulator of G-protein signaling-2 (RGS-2) expression and function in osteoblastic cells. J. Cell. Biochem. 2002;85:837–850. doi: 10.1002/jcb.10176. [DOI] [PubMed] [Google Scholar]
- 36.Carlezon WAJ, et al. The many faces of CREB. Trends Neurosci. 2005;28:436–445. doi: 10.1016/j.tins.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 37.Zhong H, Neubig RR. Regulator of G protein signaling proteins: novel multifunctional drug targets. J. Pharmacol. Exp. Ther. 2001;297:837–845. [PubMed] [Google Scholar]
- 38.Schultz W. Behavioral theories and the neurophysiology of reward. Annu. Rev. Psychol. 2006;57:87–115. doi: 10.1146/annurev.psych.56.091103.070229. [DOI] [PubMed] [Google Scholar]
- 39.Redish AD. Addiction as a computational process gone awry. Science. 2004;306:1944–1947. doi: 10.1126/science.1102384. [DOI] [PubMed] [Google Scholar]
- 40.Zachariou V, et al. Essential role for RGS9 in opiate action. Proc. Natl. Acad. Sci. U S A. 2003;100:13656–13661. doi: 10.1073/pnas.2232594100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lopez-Fando A, et al. Expression of neural RGS-R7 and Gbeta5 Proteins in Response to Acute and Chronic Morphine. Neuropsychopharmacology. 2005;30:99–110. doi: 10.1038/sj.npp.1300515. [DOI] [PubMed] [Google Scholar]
- 42.Bishop GB, et al. Abused drugs modulate RGS4 mRNA levels in rat brain: comparison between acute drug treatment and a drug challenge after chronic treatment. Neurobiol Dis. 2002;10:334–343. doi: 10.1006/nbdi.2002.0518. [DOI] [PubMed] [Google Scholar]
- 43.Gold SJ, et al. Regulation of RGS proteins by chronic morphine in rat locus coeruleus. Eur. J. Neurosci. 2003;17:971–980. doi: 10.1046/j.1460-9568.2003.02529.x. [DOI] [PubMed] [Google Scholar]
- 44.Ingi T, et al. Dynamic regulation of RGS2 suggests a novel mechanism in G-protein signaling and neuronal plasticity. J. Neurosci. 1998;18:7178–7188. doi: 10.1523/JNEUROSCI.18-18-07178.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang D, et al. Repeated cocaine administration induces gene expression changes through the dopamine D1 receptors. Neuropsychopharmacology. 2005;30:1443–1454. doi: 10.1038/sj.npp.1300680. [DOI] [PubMed] [Google Scholar]
- 46.Burchett SA, et al. RGS mRNA expression in rat striatum: modulation by dopamine receptors and effects of repeated amphetamine administration. J. Neurochem. 1999;72:1529–1533. doi: 10.1046/j.1471-4159.1999.721529.x. [DOI] [PubMed] [Google Scholar]
- 47.Schwendt M, et al. Acute amphetamine down-regulates RGS4 mRNA and protein expression in rat forebrain: distinct roles of D1 and D2 dopamine receptors. J. Neurochem. 2006;96:1606–1615. doi: 10.1111/j.1471-4159.2006.03669.x. [DOI] [PubMed] [Google Scholar]