Abstract
A significant gap exists between genetics-based investigations of polyketide synthases (PKS) and nonribosomal peptide synthetase (NRPS) biosynthetic pathways and our understanding of their regulation, interaction, and activity in living systems. To help bridge this gap, here we present an Orthogonal Active Site Identification System (OASIS) for the proteomic identification and analysis of PKS/NRPS biosynthetic enzymes. OASIS probes target conserved features of PKS/NRPS active-sites to provide activity-based enrichment of modular synthases, followed by analysis through multidimensional protein identification technology (MudPIT) LC-MS/MS analysis. When applied to the model bacterium Bacillus subtilis, this functional proteomics method detects and quantifies all four modular synthases in the organism. Furthermore, tandem application of multiple OASIS probes enhances identification of specific PKS/NRPS modules from complex proteomic mixtures. By expanding the dynamic range of proteomic analysis for PKS/NRPS enzymes, OASIS offers a valuable tool for strain comparison, culture condition optimization, and enzyme discovery.
INTRODUCTION
Polyketides and nonribosomal peptides constitute two important classes of bioactive natural products. Due to the significant role these compounds play in promoting human health, the catalysts responsible for their biosynthesis, the polyketide synthase (PKS) and nonribosomal peptide synthetase (NRPS) classes, have been the subject of intense biochemical, genetic, and structural characterization over the past three decades (1,2). These studies have led to revolutionary advances in natural products research, including the production of novel bioactive small molecules through metabolic engineering (3). However, one area in which our understanding of these systems remains limited is at the proteomic level. Methods for the functional proteomic analyses of PKS and NRPS enzymes would advance our knowledge of natural product biosynthesis by providing insight into the dynamics and regulation of PKS and NRPS activity in the context of wild-type producers. In unsequenced organisms, such studies could facilitate discovery of natural product gene clusters on the LC-MS/MS time scale. In sequenced producers, such technologies would have applications in monitoring the expression of orphan gene clusters, improving fermentative microorganisms, and analyzing pathogenic bacteria for the production of small molecule virulence factors (4,5). PKS and NRPS enzymes present a technical challenge to traditional methods of proteomic analysis due to their unique biochemical properties, which cover the extreme ranges of molecular weight, pI, and protein abundance and also are functionally regulated by posttranslational modification (1,6). Here we introduce an orthogonal active site identification system (OASIS) for the proteomic identification and analysis of PKS and NRPS biosynthetic enzymes as a first step toward accessing the potential of natural product proteomics.
OASIS utilizes functional proteomic probes that directly target PKS and NRPS active-sites in combination with the gel-free LC-MS/MS platform multidimensional protein identification technology (MudPIT) (7) to identify and report on the functional state of PKS and NRPS enzymes in proteomic samples (Figure 1). When applied to the model bacterium Bacillus subtilis, this method allows for activity-based isolation and detection of enzymes encoded by all four known PKS and NRPS gene clusters found in this organism, greatly expanding the dynamic range of analysis for these enzymes compared to traditional activity assays. The active-site probes applied during OASIS are complementary, resulting in isolation of both common and distinct PKS and NRPS proteins, and can be used for the relative quantification of natural product synthase activity between strains. Finally, we demonstrate the ability of tandemly applied OASIS probes to enhance identification of peptides originating from specific PKS and NRPS modules in complex mixtures, providing a potentially powerful tool for natural product enzyme discovery in unsequenced organisms. By providing a thorough validation of these methods in this well-characterized natural product producer, this study will guide future efforts to identify and characterize these important biosynthetic assembly lines at the proteomic level.
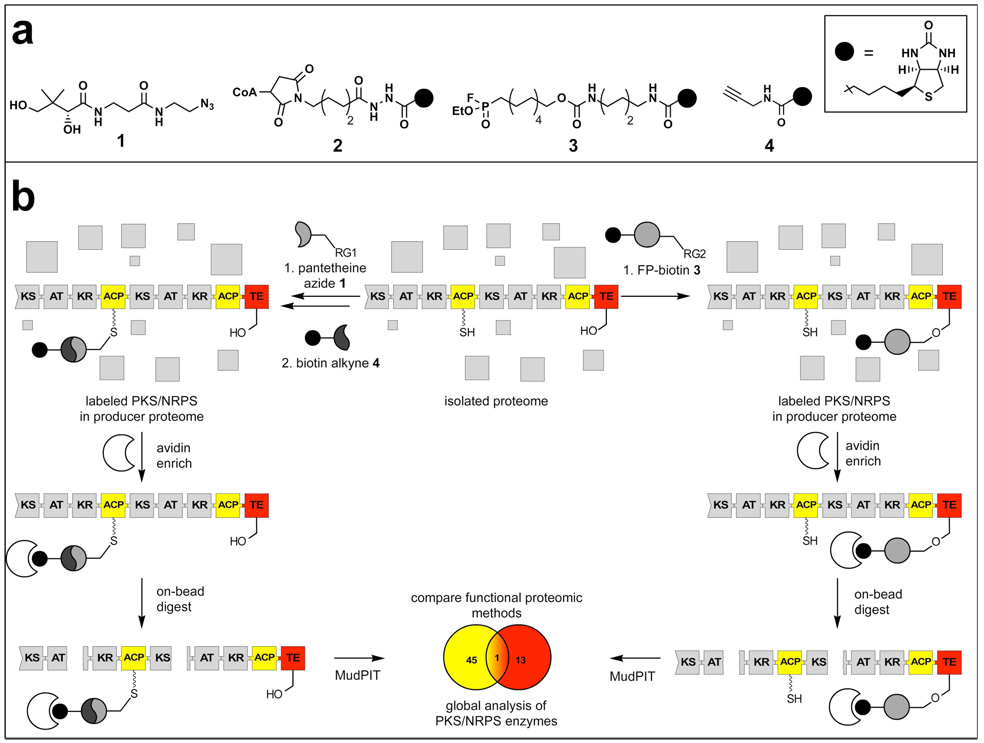
Figure 1.
Orthogonal Active Site Identification System for proteomic identification of PKS and NRPS enzymes. a) OASIS probes 1–4. Bioorthogonal CoA precursor 1 is added as a component of the bacterial growth media and is metabolically incorporated into CP domains in vivo. The azide-moiety allows for enrichment of labeled proteins via Cu(I)-catalyzed cycloaddition with biotin alkyne 4, followed by affinity chromatography. Biotin-CoA 2 provides quantitative labeling of PKS and NRPS CP domains in PPTase-deficient organisms (B. subtilis strain 168). Fluorophosphonate-biotin 3 labels TE domains through irreversible inhibition of the catalytic serine residue, and can be used directly for enrichment without further derivatization. b) Following biotinylation by OASIS CP (1–2) or TE (3) probes, enrichment by avidin affinity chromatography, followed by on-bead tryptic digest, results in tryptic peptides of PKS and NRPS enriched samples. Proteins are analyzed by multidimensional LC-MS (MudPIT), and the levels of enzyme activity are estimated by spectral counting analysis. Comparison of peptides enriched by orthogonal active site probes results in enhanced identification of multifunctional PKS/NRPS enzymes.
RESULTS AND DISCUSSION
Design of OASIS: A high-content proteomic platform for PKS/NRPS analysis
PKS/NRPS enzymes produce their seemingly disparate chemical structures by a similar modular biosynthetic logic, where multidomain enzymes activate, condense, and tailor a series of monomer units to produce the final natural product (Figure 1) (1,2). In order to establish a universal strategy for proteomic analysis of PKS/NRPS enzymes, we focused on two enzymatic domains commonly utilized by both enzyme types: carrier protein (CP) domains that serve to covalently tether biosynthetic intermediates to the PKS or NRPS during biosynthesis, and the thioesterase (TE) domains that catalyze the hydrolysis or macrocyclization of CP-bound polyketides and nonribosomal peptides at the end of the biosynthetic cycle (6,8). OASIS utilizes orthogonal active-site probes targeting each of these domains followed by enrichment by each probe to enhance analysis of peptides originating from CP/TE containing type I PKS and NRPS megasynthetases from complex proteomic mixtures. These samples are then analysed by MudPIT, and cross-references between the peptide identifications made from each probe allow for the identification of PKS and NRPS. Each tier of analysis contributes to the rich information content of OASIS, forming a versatile, high-content functional proteomics platform.
At the heart of this platform lie the OASIS CP and TE probes. CP domains are subject to posttranslational modification by a coenzyme A (CoA)-derived 4’-phosphopantetheine prosthetic group, whose introduction by a phosphopantetheinyltransferase (PPTase) converts apo-PKS/NRPS to the holo- form (9). Metabolic incorporation of bioorthogonal CoA precursor 1 augments this process (10), through uptake and transfer to CP domains by the action of the organism’s endogenous CoA biosynthetic pathway and PPTase enzymes (Supporting Information, Figures S1–S2) (11,12). Modified proteins may be visualized or enriched through chemoselective ligation of the azide group to alkyne-bearing reporters using the Cu(I)-catalyzed [3+2] cycloaddition reaction (“click chemistry,” Figure 1) (10,12). Alternately, biotinylated CoA analogue 2 can be used to label CP domains in proteomic samples in combination with exogenously added promiscuous PPTase enzyme Sfp (13). Notably, this chemoenzymatic approach is most effective in proteomes isolated from organisms deficient in PPTase activity such as B. subtilis strain 168, but has limited utility in most proteomic environments and thus is applied in this study only for comparative purposes.
TE domains can be functionally enriched using fluorophosphonate 3, which modifies the catalytic serine residue through irreversible inhibition of the conserved α,β-hydrolase motif (Figure 1) (14–16). This activity based protein profiling (ABPP) probe is well known for its class-wide reactivity with serine hydrolases, enriching not only terminal PKS/NRPS modules, but also the larger pool of cellular serine hydrolases. Importantly, each of these probes utilizes the endogenous activity of their cognate domains to label PKS/NRPS enzymes, providing an indirect report of each domain’s functional activity in natural product producer proteomes (17).
In order to expand the dynamic range of analysis and circumvent the limitations of gel-based strategies in analysis of high molecular weight PKS and NRPS proteins, both of these probes were applied in tandem with the gel-free protein identification platform known as MudPIT (Figure 1) (7). The increased resolution 2D-LC-MS/MS detection facilitates both protein detection and relative quantification between samples using spectral counting (18,19). As a subject for the evaluation of each of the OASIS methods we chose two strains of the gram-positive bacterium B. subtilis, 6051 and 168. While this organism is well known for its production of the nonribosomal peptides surfactin, bacillibactin, and plipastatin, as well as the hybrid polyketide-nonribosomal peptide natural product bacillaene (20,21), the secondary biosynthetic capabilities of these two strains are known to differ, as 168 contains an in-frame deletion in the gene encoding its secondary PPTase Sfp. This mutation decreases antibiotic production by curtailing efficient posttranslational modification of apo-PKS and NRPS enzymes, which in turn facilitates labeling of CP domains by probe 2. Notably, the terminal domains of the PKS and NRPS enzymes responsible for production of these compounds contain both CP and TE domains, allowing a direct comparison of protein labeling by probes 1 and 3, as well as facile analysis of the enhanced identification of CP-TE containing PKS/NRPS enzymes provided by OASIS (Figure 1).
Active site labeling, enrichment, and MudPIT of B. subtilis proteomes
Proteomes isolated from stationary phase cultures of B. subtilis were examined, as previous studies had observed polyketide and nonribosomal peptide production at this stage of growth (22,23). The wild-type strain 6051 was cultured in the presence of CoA precursor 1 for metabolic labeling of CP domains, while strain 168 was grown under identical conditions without 1. After initial verification of probe labeling by gel-based fluorescence profiling (Supplementary Figures S1 and S3), isolated proteomes were biotinylated by probes 2, 3, and 4 for enrichment (15). For CP-domain enrichment, metabolically labeled proteomes of strain 6051 were subjected to Cu(I)-catalyzed [3+2] cycloaddition reaction with biotin alkyne 4 (10,16). Proteomes of strain 168 were chemoenzymatically labeled with biotin CoA 2 using the PPTase enzyme Sfp, as described previously (13). For global enrichment of serine hydrolases, including TE domains, both proteomes were separately incubated with fluorophosphonate 3 (16). Each proteomic sample was then enriched on avidin-agarose, followed by tryptic digest and MudPIT analysis (Figure 1) (18). Multiple replicate analyses were performed for each enrichment method (n ≥ 4), as well as for probe-free controls in which 2, 3, and 4 were not added to proteomes (n ≥ 2). The number of tandem mass spectra assigned to each identified protein (spectral counts) were used as a measure of relative abundance (18,19). Spectral counts for enzymes identified from enriched samples were compared to probe-free controls to distinguish on-target activity of CP and TE-probes from endogenously biotinylated proteins and those non-specifically bound to avidin beads. In addition, an ad hoc cutoff value of ≥ 15 spectral counts was employed to restrict our semi-quantitative analyses to labeled proteins with good signal intensities. This value is similar to those used in previous ABPP studies (17), and was chosen based on the observation that with these criteria, >85% of the protein targets enriched from B. subtilis 6051 proteomes with fluorophosphonate 3 were annotated or showed strong homology to serine hydrolases. Replicate MudPIT data sets were averaged, and each method was assessed for number of PKS/NRPS proteins identified, total number of proteins observed, and reproducibility. The criteria and results of these analyses are summarized in Table 1 (for individual protein identification data see Supporting Information, Tables S1–S4).
Table 1.
Summary of proteins specifically enriched by probes 1, 2, and 3 in B. subtilis. Specific enrichment is based on the abundance of each protein in enriched compared to non-enriched samples, as indicated by spectral count analysis (total spectrum counts ≥ 15, ratio of enriched: probe-free control ≥ 10). S.E.M. = standard error of mean.
| Enrichment Probe |
B. subtilis Strain |
PKS/NRPS Proteins Identified | total proteins identified |
number SEM >30% |
||||
|---|---|---|---|---|---|---|---|---|
| srf (4)a | dhb (2) | pps (5) | pks (6) | total (16) |
||||
| 1 | 6051 | 2 | 2 | 3 | 5 | 12 | 58 | 1 |
| 2 | 168 | 3 | 2 | 4 | 2 | 11 | 45 | 15 |
| 3 | 6051 | 1 | 1 | 1 | 1 | 4 | 29 | 0 |
| 3 | 168 | 3 | 1 | 1 | 0 | 5 | 36 | 0 |
Abbreviations denote PKS/NRPS gene cluster of origin for identified proteins. Parentheses indicate total number of CP and TE containing open reading frames for each PKS and NRPS locus.
Comparative analysis of individual OASIS probes for identification of PKS/NRPS enzymes
Using the above methods, we first examined OASIS probes 1–3 for global identification of PKS and NRPS enzymes in the wild-type B. subtilis strain 6051. CP-probe 1 proved most effective in this regard, identifying 12 of the possible 16 PKS and NRPS CP-containing enzymes found in the B. subtilis genome, including hits from all four PKS and NRPS gene clusters (Table 1). This is a slight improvement compared to the activity of model CP-probe 2 in the mutant B. subtilis strain 168, a remarkable finding given that the mode of action of 1 requires it to cross the cell membrane and compete with endogenous CoA for labeling of CP-domains in vivo. Individual inspection of the proteins identified by OASIS probes 1–2 (Tables S1–S2) reveal a common underrepresentation of the discrete, low molecular weight (~9 kDa) CP domains of bacterial FAS and type II PKS biosynthesis (6), presumably due the low number of peptides these proteins produce upon tryptic digest. However, despite the bias of the analysis method (spectral counting) against this class of CPs, OASIS probe 1 identified DltC, a type II CP involved in cell wall biosynthesis using these criteria.
Hydrolase probe 3 also proved well-suited to the analysis of modular biosynthetic enzymes, identifying the TE-containing termination modules of 3 out of 4 PKS/NRPS enzymes in both strains of B. subtilis, as well as SrfAD, a type II TE enzyme involved in editing of misprimed CP domains (Table 1) (23). In strain 168 a non-TE containing NRPS (24), SrfAA, also showed specific enrichment by fluorophosphonate 3, but at greatly decreased spectral counts compared to its enrichment by CP-probe 2 (Tables S2 and S4). This disparity suggests SrfAA is not a covalent target of 3, but rather is enriched due to non-covalent interaction with a partner enzyme biotinylated by 3, possibly SrfAC. Previous studies have used similar co-immunoprecipitation of modular biosynthetic enzymes in B. subtilis as evidence for the macromolecular organization of these systems (25).
The targets of OASIS probes 1–3 were also analyzed to provide insight into the proteomic selectivity of each. ABPP probe 3 consistently enriched fewer proteins than CP-probes 1–2 (Table 1). The majority of enzymes labeled by 3 in both samples are annotated or homologous to serine hydrolases, consistent with its mode of action (14). Inspection of the individual peptide identifications made for PKS and NRPS enzymes following treatment with probes 1–3 also support their active-site directed mechanisms: peptides incorporating CP and TE catalytic serine residues are not observed. Surprisingly, only ~20–25 % of the total proteins identified by CP-probes 1–2 are annotated as CP-containing enzymes, indicating that these probe has substantial off-target reactivity (Tables S1-S2). This is not unexpected, as the Cu(I)-catalyzed chemoselective ligation reaction used for detection of enzymes labeled by metabolic probe 1 is known to show some nonselective labeling of proteomic samples, in particular when reporter-labeled alkynes such as 4 are applied (17).
CoA precursor 1 competes with endogenous intracellular CoA for labeling of PKS/NRPS enzymes (Supplementary Figure S2), and therefore labels only a percentage of total cellular CP-domains (11,12). In order to better gauge the relative labeling effiencies of these two agents, we compared the spectral count data from MudPIT analyses of 1 and 3-enriched enzymes from B. subtilis 6051 (Figure 2). Analysis of raw spectral counts observed for PksR, the highest abundance PKS/NRPS protein enriched by both probes 1 and 3 in this strain, reveals approximately equivalent enrichment using each probe. However, this raw spectral count must be corrected for the number of enrichable active-sites found on PksR (three CPs and only one TE – Figure 2.a), as well as the disparity in average sequence coverage observed following enrichment. The average sequence coverage of PksR generated by each probe was estimated by compiling a list of PksR peptides which were identified by database searching in at least two out of four MudPIT datasets following enrichment by probes 1 or 3 (Table S5). Using this analysis, one observes much greater average sequence coverage of PksR after enrichment by CP-probe 1 than with TE-probe 3 (33% compared to 17% - Supplementary Figure S4). Coverage arising from enrichment with TE-probe 3 is concentrated around the TE active-site (Figure 2b, red and yellow sequences), with 55% of the spectral counts comprised of four abundant peptides (Table S5). This observation suggests some PksR enzymes in the sample have undergone proteolysis, a finding consistent with previous gel-based fluorescence analysis of probe-labeling (Supplementary Figure S3) (15) as well as the lack of protease inhibitors added to the samples (omitted due to interference with probe 3 labeling). Correcting the spectral count data for sequence coverage (corrected = [average spectral counts]/[average sequence coverage * number of enrichable active-sites]) leads to the estimate that in vivo CP-labeling is 17% as effective as TE-labeling for quantitative labeling of PksR active-sites (Figure 2c). However, this reduced efficiency is compensated for by the presence of three CP active-sites on PksR, allowing probe 1 to pull down a greater number of proteolytic fragments than TE-probe 3.
Figure 2.
a) Domain organization for terminal PKS module of bacillaene synthase (PksR – hybrid PKS/NRPS). Abbreviations: ACP, acyl carrier protein; TE, thioesterase; MT, methyltransferase; KS, ketosynthase; AT; acyltransferase. b) Structure of bacillaene. c) Spectral count analysis of relative efficiencies of functional proteomics probes 1 and 3 for labeling PksR in B. subtilis 6051. CP-probe 1 labels PksR active sites less efficiently than TE-probe 3 (spectral counts per active site) but is equally competent in PKS identification (raw spectral counts) due to the greater number of CP-active sites per PKS/NRPS enzyme and ability to pull down many proteolytic fragments. d) Sequence coverage for PksR (terminal module of bacillaene synthase) generated by enrichment of B. subtilis 6051 using in vivo probe 1 (yellow) and activity-based probe 3 (red). Yellow = sequences enriched by CP-probe, Red = sequences enriched by TE-probe, Green = sequences enriched by both probes.
Finally, one caveat to the use of metabolic CP-probe 1 in functional proteomics applications is the possibility that its addition to the growth medium may perturb secondary metabolism, thereby providing an inaccurate reflection of the physiology of the organism being studied. To investigate this we performed additional MudPIT analyses utilizing unfractionated proteomic samples from B. subtilis 6051 grown with and without probe 1. The proteomes of B. subtilis grown with 1 demonstrated a slight, but consistent, increase in levels of CP-containing PKS/NRPS enzymes, as judged by spectral counting analysis (Table S6). This observation may reflect an upregulation of PKS/NRPS transcription upon inhibition of biosynthesis by 1. The whole proteome data also highlights the increase in PKS/NRPS dynamic range provided by CP-probe 1, as comparison of the two datasets shows greater global coverage of PKS/NRPS enzymes at much greater (two to tenfold) spectral counts following click chemistry enrichment of 1-labeled samples (Tables S1, S6).
Spectral count-based quantification of PKS/NRPS enzymes in Bacillus subtilis
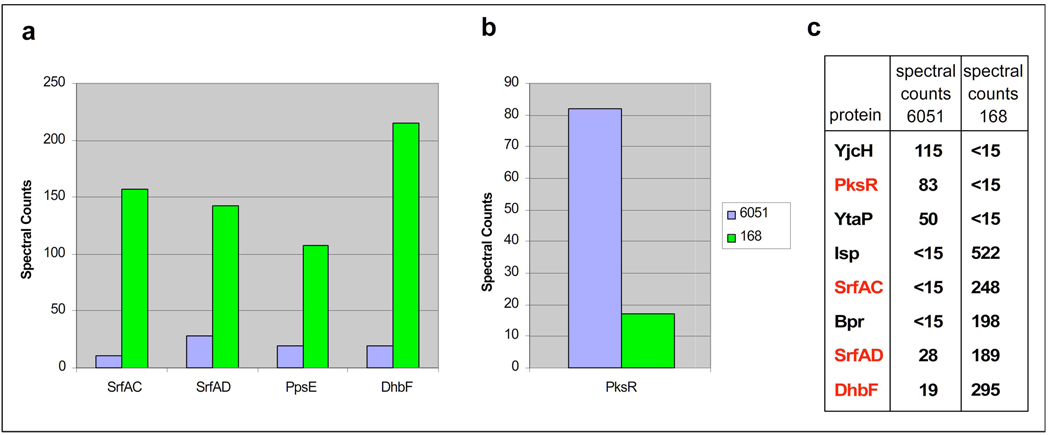
Next we applied these methods to analyze the relative abundance of PKS/NRPS enzymes in B. subtilis strains 6051 and 168. Because these two strains differ in PPTase capabilities required for CP-enrichment by OASIS probes 1–2, we focused on comparison of data generated by ABPP-MudPIT analysis using fluorophosphonate 3. As can be seen in Figure 3, the terminal NRPS enzymes SrfAC and DhbF show higher activity in strain 168, demonstrating enrichment at levels 5–10 times observed in strain 6051 as estimated by spectral count analysis. This finding is consistent with the higher levels of SrfAC observed in strain 168 during previous gel-based proteomic studies of this organism (15). The overall upregulation of the Srf operon is also evidenced by the greater spectral counts observed for the type II TE domain SrfAD in strain 168 (26). PksR, the PKS module responsible for chain termination in the biosynthesis of bacillaene, demonstrated the opposite relationship, with higher levels found in strain 6051 (Figure 3).
Figure 3.
Comparison of enzyme identifications and functional proteomics relationships revealed by interstrain activity-based protein profiling using OASIS probe 3. a) Comparison of NRPS levels in B. subtilis 168 and B. subtilis 6051 using TE-probe 3. All three NRPS termination modules of B. subtilis as well as the type II thioesterase encoded in the srf gene cluster (SrfAD) are highly upregulated in strain 168. b) Comparison of PKS levels (PksR) in B. subtilis 168 and B. subtilis 6051 using TE-probe 3. Wild-type organism 6051 shows higher levels of PksR than does 168 (<15 spectral counts). c) Truncated list of PKS/NRPS and non-PKS/NRPS proteases showing differential enrichment between strains 6051 and 168 using probe 3. For complete tabulated data see Supporting Tables S3 and S4.
In addition to providing a readout of PKS/NRPS enzymatic activity in proteomic samples, enrichment by fluorophosphonate 3 also provides insight into the functional activity of ancillary hydrolases in this organism. In particular, several serine hydrolases showed greater activity in strain 168, including Isp and Bpr, which were not functionally enriched in strain 6051 despite growth under equivalent conditions (Figure 3c). Bpr is known to be ribosomally produced as a zymogen, and its enrichment by fluorophosphonate 3 provides strong evidence that this enzyme has been proteolytically processed to its holo-form in late stationary phase cultures of strain 168 (27). In contrast, strain 6051 showed identification of an overlapping but distinct pool of hydrolases after enrichment by 3, including several unannotated hydrolases (YjcH, YtaP) that were among the most abundantly enriched enzymes in the sample (28).
Enhanced detection of peptides from PKS/NRPS termination modules by OASIS
Type I PKS and NRPS biosynthetic enzymes share the unique characteristic of possessing multiple active sites housed upon a single polypeptide (1,2). Since the modular architecture of PKS/NRPS enzymes can often be predicted from the structure of the small molecule, probes capable of utilizing this property for enzyme identification could be a powerful tool for discovery of unknown biosynthetic catalysts coding for specific polyketide and nonribosomal peptides in unsequenced organisms. For example, 4’-phosphopantetheinylation and hydrolase activities are ordinarily found on the same enzyme only in PKS/NRPS termination modules. Thus, cross-referencing peptide identifications made following enrichment by CP-probe 1 and TE-probe 3 should result in enhanced identification of type I PKS/NRPS modules coding for macrocyclic metabolites, while decreasing the number of non-specific background peptides observed (Figure 1).
As no protein database is available for unsequenced organisms, this identification strategy was analyzed on a peptide level by inspecting individual rather than average MudPIT datasets and discarding the ad hoc spectral counting criteria used in this functional proteomics work. Here we examined the number of unique peptides which were identified in every dataset generated by MudPIT of the B. subtilis 6051 proteome following enrichment by probes 1 and 3, but were identified in none of the datasets generated by MudPIT of the B. subtilis proteome following enrichment using an identical protocol but with no probe applied. The impact of the enrichment method used (i.e. CP or TE targeting) and the number of data sets collected/contrasted on the total number of unique peptides identified was analyzed. As can be seen in Table 2, collection of an increasing number of data sets reduces the number of unique peptides identified by both 1 and 3 (total peptides, Table 2, bottom table entry depicted in Figure 4a). As this peptide pool shrinks, the overall percentage represented by PKS and NRPS (% PKS/NRPS) increases, indicating reproducible isolation of a subset of PKS and NRPS peptides by each probe.
Table 2.
Analysis of OASIS enrichment on a peptide level. Collection and comparison of a greater number of OASIS MudPIT data sets using each enrichment method (1 or 3) decreases the total number of peptides (total peptides) uniquely identified in probe-labeled samples. The use of orthogonal active site probes biases the identified peptides towards PKS/NRPS enzymes (% PKS/NRPS increasing), particularly those containing CP-TE domains (% CP-TE). A list of peptides identified from the bottom row (consisting of 40 unique peptide identifications observed in all nine probe 1/3 enriched samples but not in no probe datasets) is provided in Table S8 (Supporting Information).
| # MudPIT Data Sets Collected | # peptides enriched | ||||
|---|---|---|---|---|---|
| Probe 1 (CP) |
Probe 3 (TE) |
No probe | Total peptides |
% PKS/NRPS (total) |
% CP-TE (total) |
| 1 | 1 | 1 | 599 | 7.5% (45) | 51% (23) |
| 2 | 2 | 2 | 216 | 8.3% (18) | 72% (13) |
| 3 | 3 | 3 | 66 | 13.6% (9) | 88% (8) |
| 4 | 4 | 4 | 47 | 17% (8) | 100% (8) |
| 5 | 4 | 5 | 39 | 20% (8) | 100% (8) |
Figure 4.
Enhanced detection of peptides from CP-TE containing PKS/NRPS enzymes by OASIS. (a) Venn diagrams comparing total number of unique and overlapping peptide identifications made by CP-probe 1 and TE-probe 3 in B. subtilis 6051. Unique peptides are classified as those found in each replicate MudPIT dataset after enrichment by CP-probe 1 (yellow, n=5 datasets) or TE-probe 3 (red, n=4 datasets) but not in any proteomic samples treated with no probe and similarly subjected to streptavidin enrichment (n=5 datasets). Overlapping peptides (green, n=9 datasets) were found in every sample enriched by either 1 or 3 but not in any no probe control samples. Applying multiple probes decreases the total number of peptides observed. (b) Analyzing unique PKS/NRPS peptides identified by OASIS. Of PKS/NRPS peptides identified by each method, OASIS hits (peptides identified by both analyses) contain exclusively peptides originating from CP-TE containing PKS/NRPS modules.
Examining the proteins to which these peptides are assigned indicates their enrichment accurately reflect the mechanism of the functional proteomic probes (1 and 3) utilized, as every PKS/NRPS peptide identified in the final two table entries originates from a CP- and TE-containing module (Table 2, Figure 4b). Unique peptides are observed from the terminal modules SrfAC, DhbF, PpsE, and PksR, which together make up every PKS/NRPS coded for in the B. subtilis genome (Table S8) (28). This is especially remarkable given the previously observed low abundance of the SrfAC and PpsE enzymes in B. subtilis 6051 samples, (vide infra). Because several non-PKS/NRPS background peptides are also observed, each of the 39 unique peptides observed in the final OASIS dataset (Table 2, bottom row; full list provided in Table S8) were also analyzed using the error tolerant Basic Local Alignment Search Tool MS-BLAST (29). PKS/NRPS homology was observed among the top hits only for peptides assigned by SEQUEST PKS/NRPS gene clusters (Table S9). This suggests that homology-based approaches may be used to distinguish PKS and NRPS peptides enriched and identified by OASIS from non-specific background, thus guiding oligonucleotide probe design for identification of PKS/NRPS clusters from unsequenced organisms.
Targeted Proteomic Profiling of PKS/NRPS Enzymes: Conclusions and Future Directions
In the current study, we used the model natural product producer B. subtilis as a subject to develop a high-content proteomics platform, OASIS, for the study of PKS and NRPS enzymes. By exploiting endogenous mechanisms of PKS/NRPS activity for enzyme labeling, OASIS probes provide enrichment as well as a functional readout of CP and TE activity, offering a significant improvement over whole proteome methods of analysis. Furthermore, spectral counting analysis can be used to compare relative levels of PKS/NRPS activity, providing a new strain comparison. To demonstrate this capacity we applied OASIS probe 3 to comparatively profile PKS/NRPS hydrolase activity in B. subtilis strains 6051 and 168. These strains showed substantial differences in activity-based enrichment of terminal modules from the surfactin, bacillibactin, plipastatin, and bacillaene PKS/NRPS pathways (Figure 3). Focusing on surfactin, which has been recently proposed to play a role in B. subtilis intracellular communication (30), we observed heightened levels of the biosynthetic hydrolases SrfAC and SrfAD in strain 168. This is broadly consistent with previous gel-based analyses, as well as the known properties of this lab strain, which include high competence (31). A major regulator of competence is the transcription factor ComA, whose upregulation has been found to also stimulate expression of the srf operon (20). While these observations support the accuracy of OASIS probes, further studies are needed to evaluate the metabolic relevance of these strain variations. However, these studies indicate that functional proteomics could serve as an effective tool to compliment contemporary biochemical and molecular genetics methods.
This study also demonstrates the ability of CP-probe 1 to label PKS and NRPS biosynthetic enzymes in vivo. 1 shows excellent overall detection of type I PKS/NRPS biosynthetic enzymes in B. subtilis 6051, even surpassing the number of PKS/NRPS enzymes identified following enrichment by model CP-probe 2 with B. subtilis 168 (Table 1). Additionally, the enrichment of multiple CP-attachment sites by OASIS probe 1 results in broad PKS/NRPS sequence coverage (Figure 2b) as well as the largest number of distinct PKS/NRPS peptides of any probe (Figure 4a). All three OASIS probes enriched PKS/NRPS enzymes consistent with their proposed mechanisms of action. However, metabolic CP-probe 1 showed a lower proteome-wide level of fidelity than activity-based TE-probe 3, enriching many enzymes which were not annotated targets of phosphopantetheinyation. While Yin et al. previously identified non-canonical targets of phosphopantetheinylation in B. subtilis through a phage display approach (32), no overlap was seen with the non-PKS/NRPS enzymes enriched by 1, leading us to attribute their enrichment to nonselective labeling by reactive alkyne 4 (16). Furthermore, whole proteome analysis suggests that alone probe 1 functions as a low-level agonist of type I PKS/NRPS biosynthetic enzyme production (Table S6). Due to these factors metabolic CP-probe 1 may be best suited for PKS/NRPS discovery applications, while activity-based probe 3 is likely to be more useful in the comparative profiling of PKS/NRPS expression. The utility of CP-probe 1 in proteomic discovery approaches is aided by the ability of a single CP active-site labeling event to result in enrichment of an entire PKS/NRPS megasynthase protein (producing a large number of tryptic peptides suitable for LC-MS/MS sampling and analysis). Additionally, 1 has the capacity to enrich spatially separated active-sites on multimodular PKS/NRPS enzymes even in proteolytic or chemically damaged samples.
After investigating the activity of each probe individually, we demonstrated the power of the OASIS method by cross-referencing peptide identifications made by 1 and 3 to enhance detection of PKS/NRPS peptides originating from termination modules (Table 2). This approach provides a potentially new avenue for discovery of natural product gene clusters, indicating that PKS/NRPS enzymes coding for specific products may be rationally isolated and identified from complex biological samples based on proposed aspects of their biosynthetic pathways. Alternately, the global detection of hydrolytic PKS/NRPS termination modules provided by OASIS may be used to identify and clone orphan PKS/NRPS clusters, whose genetic organization could then be used to guide the search for new natural products. Each of these applications will require overcoming a key technical challenge, namely that while this initial study made mass spectral assignments based on database searching (33), in unsequenced organisms no such database exists. The incorporation of alternate spectral assignment strategies such as error tolerant database searching (34) and de novo sequencing (35) into the OASIS workflow will therefore be necessary in order to perform similar analyses on unsequenced or environmental samples. This is a non-trivial obstacle, and here our B. subtilis datasets provide a valuable control for the further training and assessment these bioinformatic methods for identification of PKS/NRPS peptides from unsequenced organisms.
Finally, we note some potential limitations of OASIS as presently constituted. While the utility of CP-probe 1 has been previously been demonstrated in a number of bacterial organisms (10), its effectiveness may be impacted in organisms lacking a permissive CoA biosynthetic pathway or appropriate uptake mechanisms. Therefore, we are currently exploring the development of additional OASIS probes designed against other domains in these pathways to complement metabolic labeling by 1 to develop PKS and NRPS-specific OASIS methods. Additionally, the high information content of OASIS proteomic studies are partially offset by the low throughput of MudPIT LC-MS/MS methods. Analysis of the number of replicate MudPIT datasets necessary for enhanced identification of peptides originating from PKS/NRPS termination modules (Table 2) should help guide future efforts. This limitation may also be addressed by the continued development of bifunctional enrichment protocols, allowing the labeling profiles of orthogonal OASIS probes to be initially analyzed using SDS-PAGE or blotting techniques, such that candidate PKS/NRPS-containing proteomic samples may be preselected for OASIS LC-MS/MS analysis (36). Bridging the gap between our detailed understanding of PKS/NRPS enzymes in vitro and the less developed knowledge of their activity in living systems, OASIS represents a promising new tool for the discovery and engineering natural product biosynthetic pathways.
Supplementary Material
Acknowledgments
This work was supported by NIH GM075797 and NSF CAREER MCB-0347681.
Footnotes
Supporting Information Available
Supplementary Figures and Tables as well as full experimental methods for materials, cell culture conditions, proteome enrichments, and LC-MS/MS analysis are provided in the Supporting Information. This material is available free of charge via the internet at http://pubs.acs.org.
References
- 1.Fischbach MA, Walsh CT. Assembly-line enzymology for polyketide and nonribosomal Peptide antibiotics: logic, machinery, and mechanisms. Chem.Rev. 2006;106:3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- 2.Sieber SA, Marahiel MA. Molecular mechanisms underlying nonribosomal peptide synthesis: approaches to new antibiotics. Chem.Rev. 2005;105:715–738. doi: 10.1021/cr0301191. [DOI] [PubMed] [Google Scholar]
- 3.Tang L, Shah S, Chung L, Carney J, Katz L, Khosla C, Julien B. Cloning and heterologous expression of the epothilone gene cluster. Science. 2000;287:640–642. doi: 10.1126/science.287.5453.640. [DOI] [PubMed] [Google Scholar]
- 4.Gross H. Strategies to unravel the function of orphan biosynthesis pathways: recent examples and future prospects. Appl. Microbiol. Biotechnol. 2007;75:267–277. doi: 10.1007/s00253-007-0900-5. [DOI] [PubMed] [Google Scholar]
- 5.Cisar JS, Tan DS. Small molecule inhibition of microbial natural product biosynthesis-an emerging antibiotic strategy. Chem. Soc.Rev. 2008;37:1320–1329. doi: 10.1039/b702780j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mercer AC, Burkart MD. The ubiquitous carrier protein--a window to metabolite biosynthesis. Nat. Prod.Rep. 2007;24:750–773. doi: 10.1039/b603921a. [DOI] [PubMed] [Google Scholar]
- 7.Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 8.Kopp F, Marahiel MA. Macrocyclization strategies in polyketide and nonribosomal peptide biosynthesis. Nat. Prod.Rep. 2007;24:735–749. doi: 10.1039/b613652b. [DOI] [PubMed] [Google Scholar]
- 9.Lambalot RH, Gehring AM, Flugel RS, Zuber P, LaCelle M, Marahiel MA, Reid R, Khosla C, Walsh CT. A new enzyme superfamily - the phosphopantetheinyl transferases. Chem. Biol. 1996;3:923–936. doi: 10.1016/s1074-5521(96)90181-7. [DOI] [PubMed] [Google Scholar]
- 10.Mercer AC, Meier JL, Torpey JW, Burkart MD. In vivo modification of native carrier protein domains. Chembiochem. 2009;10:1091–1100. doi: 10.1002/cbic.200800838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clarke KM, Mercer AC, La Clair JJ, Burkart MD. In vivo reporter labeling of proteins via metabolic delivery of coenzyme A analogues. J. Am. Chem. Soc. 2005;127:11234–11235. doi: 10.1021/ja052911k. [DOI] [PubMed] [Google Scholar]
- 12.Meier JL, Mercer AC, Rivera H, Jr, Burkart MD. Synthesis and evaluation of bioorthogonal pantetheine analogues for in vivo protein modification. J. Am. Chem. Soc. 2006;128:12174–12184. doi: 10.1021/ja063217n. [DOI] [PubMed] [Google Scholar]
- 13.La Clair JJ, Foley TL, Schegg TR, Regan CM, Burkart MD. Manipulation of carrier proteins in antibiotic biosynthesis. Chem. Biol. 2004;11:195–201. doi: 10.1016/j.chembiol.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 14.Liu Y, Patricelli MP, Cravatt BF. Activity-based protein profiling: the serine hydrolases. Proc. Natl. Acad. Sci.USA. 1999;96:14694–14699. doi: 10.1073/pnas.96.26.14694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meier JL, Mercer AC, Burkart MD. Fluorescent profiling of modular biosynthetic enzymes by complementary metabolic and activity based probes. J. Am. Chem. Soc. 2008;130:5443–5445. doi: 10.1021/ja711263w. [DOI] [PubMed] [Google Scholar]
- 16.Speers AE, Cravatt BF. Profiling enzyme activities in vivo using click chemistry methods. Chem. Biol. 2004;11:535–546. doi: 10.1016/j.chembiol.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 17.Evans MJ, Cravatt BF. Mechanism-based profiling of enzyme families. Chem.Rev. 2006;106:3279–3301. doi: 10.1021/cr050288g. [DOI] [PubMed] [Google Scholar]
- 18.Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 19.Jessani N, Niessen S, Wei BQ, Nicolau M, Humphrey M, Ji Y, Han W, Noh DY, Yates JR, 3rd, Jeffrey SS, Cravatt BF. A streamlined platform for high-content functional proteomics of primary human specimens. Nat. Methods. 2005;2:691–697. doi: 10.1038/nmeth778. [DOI] [PubMed] [Google Scholar]
- 20.Stein T. Bacillus subtilis antibiotics: structures, syntheses and specific functions. Mol. Microbiol. 2005;56:845–857. doi: 10.1111/j.1365-2958.2005.04587.x. [DOI] [PubMed] [Google Scholar]
- 21.Butcher RA, Schroeder FC, Fischbach MA, Straight PD, Kolter R, Walsh CT, Clardy J. The identification of bacillaene, the product of the PksX megacomplex in Bacillus subtilis. Proc. Natl. Acad. Sci. USA. 2007;104:1506–1509. doi: 10.1073/pnas.0610503104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barr JG. Changes in the extracellular accumulation of antibiotics during growth and sporulation of Bacillus subtilis in liquid culture. J. Appl. Bacteriol. 1975;39:1–13. doi: 10.1111/j.1365-2672.1975.tb00539.x. [DOI] [PubMed] [Google Scholar]
- 23.Schwarzer D, Mootz HD, Linne U, Marahiel MA. Regeneration of misprimed nonribosomal peptide synthetases by type II thioesterases. Proc. Natl. Acad. Sci.USA. 2002;99:14083–14088. doi: 10.1073/pnas.212382199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeigler DR, Prágai Z, Rodriguez S, Chevreux B, Muffler A, Albert T, Bai R, Wyss M, Perkins JB. The origins of 168, W23, and other Bacillus subtilis legacy strains. J. Bacteriol. 2008;190:6983–6995. doi: 10.1128/JB.00722-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Straight PD, Fischbach MA, Walsh CT, Rudner DZ, Kolter R. A singular enzymatic megacomplex from Bacillus subtilis. Proc. Natl. Acad. Sci.USA. 2007;104:305–310. doi: 10.1073/pnas.0609073103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakano MM, Marahiel MA, Zuber P. Identification of a genetic locus required for biosynthesis of the lipopeptide antibiotic surfactin in Bacillus subtilis. J. Bacteriol. 1988;170:5662–5668. doi: 10.1128/jb.170.12.5662-5668.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roitsch CA, Hageman JH. Bacillopeptidase F: two forms of a glycoprotein serine protease from Bacillus subtilis 168. J. Bacteriol. 1983;155:145–152. doi: 10.1128/jb.155.1.145-152.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, Azevedo V, Bertero MG, Bessieres P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell SC, Bron S, Brouillet S, Bruschi CV, Caldwell B, Capuano V, Carter NM, Choi SK, Codani JJ, Connerton IF, Danchin A, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 29.Shevchenko A, Sunyaev S, Loboda A, Shevchenko A, Bork P, Ens W, Standing KG. Charting the proteomes of organisms with unsequenced genomes by MALDI-quadrupole time-of-flight mass spectrometry and BLAST homology searching. Anal. Chem. 2001;73:1917–1926. doi: 10.1021/ac0013709. [DOI] [PubMed] [Google Scholar]
- 30.Buttner K, Bernhardt J, Scharf C, Schmid R, Mader U, Eymann C, Antelmann H, Volker A, Volker U, Hecker M. A comprehensive two-dimensional map of cytosolic proteins of Bacillus subtilis. Electrophoresis. 2001;22:2908–2935. doi: 10.1002/1522-2683(200108)22:14<2908::AID-ELPS2908>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 31.Hamoen LW, Venema G, Kuipers OP. Controlling competence in Bacillus subtilis: shared use of regulators. Microbiology. 2003;149:9–17. doi: 10.1099/mic.0.26003-0. [DOI] [PubMed] [Google Scholar]
- 32.Yin J, Straight PD, Hrvatin S, Dorrestein PC, Bumpus SB, Jao C, Kelleher NL, Kolter R, Walsh CT. Genome-Wide High-Throughput Mining of Natural-Product Biosynthetic Gene Clusters by Phage Display. Chem. Biol. 2007;14:303–312. doi: 10.1016/j.chembiol.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 33.Ducret A, Van Oostveen I, Eng JK, Yates JR, 3rd, Aebersold R. High throughput protein characterization by automated reverse-phase chromatography/electrospray tandem mass spectrometry. Protein Sci. 1998;7:706–719. doi: 10.1002/pro.5560070320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsur D, Tanner S, Zandi E, Bafna V, Pevzner PA. Identification of post-translational modifications by blind search of mass spectra. Nat. Biotechnol. 2005;23:1562–1567. doi: 10.1038/nbt1168. [DOI] [PubMed] [Google Scholar]
- 35.Frank A, Pevzner P. PepNovo: de novo peptide sequencing via probabilistic network modeling. Anal. Chem. 2005;77:964–973. doi: 10.1021/ac048788h. [DOI] [PubMed] [Google Scholar]
- 36.Shute TS, Matsushita M, Dickerson TJ, La Clair JJ, Janda KD, Burkart MD. A site-specific bifunctional protein labeling system for affinity and fluorescent analysis. Bioconjug. Chem. 2005;16:1352–1355. doi: 10.1021/bc050213j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.