Abstract
We describe a new approach to multiple 13C-15N distance measurements in uniformly labeled solids, frequency selective (FS) TEDOR. The method shares features with FS-REDOR and ZF- and BASE-TEDOR, which also provide quantitative 15N-13C spectral assignments and distance measurements in U-[13C,15N] samples. To demonstrate the validity of the FS-TEDOR sequence, we measured distances in [U-13C,15N]-asparagine which are in good agreement with other methods. In addition, we integrate high frequency dynamic nuclear polarization (DNP) into the experimental protocol and use FS-TEDOR to record a resolved correlation spectrum of the arg-13Cγ-15Nε region in [U-13C,15N]-bacteriorhodopsin. We observe six of the seven cross peaks expected from this membrane protein.
Keywords: heteronuclear distance measurement, chemical shift assignments, TEDOR, REDOR, DNP
1. Introduction
Heteronuclear dipole Hamiltonians are simple in comparison with their homonuclear counterparts in that they do not contain flip-flop terms; therefore, heteronuclear recoupling experiments are not subject to the phenomenon of dipolar truncation [2]. Accordingly, successful approaches to heteronuclear distance measurements exploit this simplicity and are usually based on REDOR [3] and its variants such as TEDOR [4; 5; 6]. However, even though dipolar truncation is absent, there remain complications when distance measurements in uniformly 13C-15N labeled systems are involved. In particular, evolution under the homonuclear 13C-13C J couplings imposes an overall modulation on the buildup or dephasing during the recoupling period. J-couplings can generate anti-phase coherences, which in turn give rise to phase-twisted line shapes. In order to circumvent these limitations, Jaroniec, et. al. introduced J-decoupled [7]and frequency-selective REDOR[8] experiments in which the detrimental effects of J-coupling are removed through the application of frequency-selective pulses. These effects were also consider by Mehta and Schaefer [9] in the development of the RDX version of REDOR. Another logical extension of these techniques has been the 3D ZF- and BASE TEDOR experiments [1; 10], in which multiple 13C-15N distances are measured simultaneously in a broadband or a band selective manner in the context of a 13C-15N correlation experiment. In these cases, the effects of J-coupling are removed through the application of a coherence filter or frequency selective 13C pulse, respectively. This family of sequences has been successfully employed in the de novo structural determination of a peptide [11], an amyloid fibril [12], and quantitative distance measurements in a membrane protein via MAS NMR [13], and is therefore potentially an important part of the repertoire of experiments for determining structures of proteins with MAS NMR.
However, even where the recoupled dipolar Hamiltonian for individual spin pair interactions commutes, strong dipolar couplings can compromise the sensitivity of the experiment. For example, in the TEDOR experiment, the intensity corresponding to a single 15N site is distributed over all 13C-15N cross peaks and is a function of the dipolar couplings, their mutual orientations, homonuclear J couplings, and the mixing time. Thus, in the practically important case of distance measurements between backbone 15N’s and 13C’s of an amino acid side chain, the presence of strong 13Cβ-15Nα couplings (2.5Å) in the TEDOR dynamics reduces the maximum intensity of polarization transfer due to weaker couplings (~3.3Å) to 1–3% of the initial intensity [12; 14]. This sometimes compromises the application of broadband techniques to membrane proteins and other systems in which sensitivity is limited. This effect is also particularly acute in amino acids whose side chains contain nitrogen (e.g. asparagine, glutamine lysine, arginine), as a single 13C site may be subject to several strong couplings that dominate the spin dynamics [15]. However, when the backbone or side chain 15N’’s are resolved, then a 15N-frequency-selective analogue of the 3D TEDOR experiment can simplify the spin dynamics and allow high sensitivity measurement of weak couplings.
2. Results and discussion
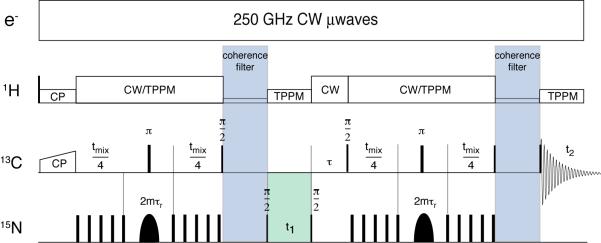
These deficiencies and opportunities have stimulated us to develop the frequency-selective (FS) TEDOR experiment illustrated in Figure 1 that shares many features with the FS-REDOR and BASE and ZF-TEDOR methods discussed above. Following ramped, 1H-13C cross polarization, we apply a REDOR train to generate 13C-15N antiphase coherence which evolves under the heteronuclear dipolar couplings. During the excitation period, a frequency selective pulse is applied on the 15N channel, such that only those 15N nuclei within the bandwidth contribute appreciably to the spin dynamics during the REDOR mixing period. Following the excitation period, optional frequency labeling with the 15N chemical shift occurs during t1. A second, frequency selective REDOR period converts the resulting coherence into observable magnetization on the 13C channel, which is detected during t2. A delay, τ, is necessary to ensure that the time between REDOR periods is an integer number of rotor cycles during which the signal is transferred to the 13C’s. Finally, 13C-13C J-couplings give rise to undesired zero-quantum and double-quantum coherences that, after the reconversion period, result in severe anti-phase distortions to the line shape and spurious cross-peaks in 2D spectra. Two variable-length coherence filters (z-filters) are applied to dephase these undesired coherences, resulting in undistorted and purely absorptive spectra. TPPM and CW decoupling is used where appropriate [16]. In addition, for the experiments on the membrane protein bacteriorhodopsin (bR), we enhance the overall signal sensitivity of the experiments by integrating dynamic nuclear polarization (DNP) into the protocol, as illustrated in Figure 1. In particular, we suspend the bR sample in 60% glycerol/H2O that forms a stable glass and serves to disperse the biradical polarizing agent TOTAPOL[17]. Irradiation with 250 GHz microwaves [18] leads to a polarization enhancement of ~43 in the signal intensities.
Figure 1.
Pulse sequence for 15N frequency selective TEDOR transfer. Only 15N spins within the bandwidth of the selective pulses contribute to the spin dynamics under REDOR. The sequence is described in the text. The REDOR pulses are phase cycled according to the XY-8 scheme and the overall phase cycle is documented elsewhere[1]. CW microwave irradiation was employed in the version of the experiment used to acquire the DNP enhanced spectrum.
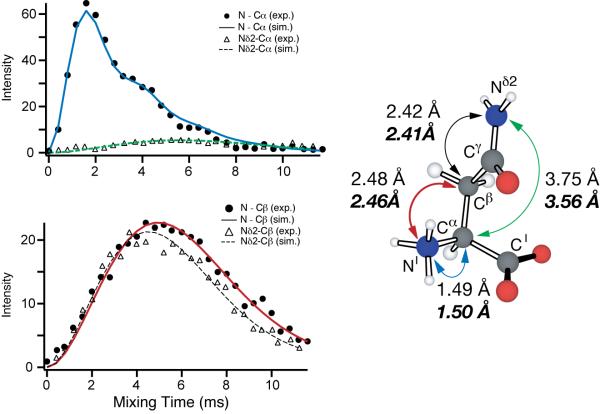
As an initial test of FS-TEDOR, all heteronuclear distances in [U-13C, 15N]-asparagine were measured in two experiments, with a selective pulse applied at the frequency of the backbone (-15NH3+) or side chain 15N (-15NH2). The results are summarized in Figure 2 and Table 1. All data were recorded on a 500 MHz (1H) home-built spectrometer (Cambridge Instruments) using a Varian triple-resonance MAS probe and in a sample of asparagine diluted to 10% in its natural abundance analogue to avoid the confounding effects of intermolecular couplings. The data were fit using a multispin simulation performed with the program SPINEVOLUTION [19]. Our FS-TEDOR distance is in good agreement with the distance measured by frequency-selective REDOR [8] which is a true two-spin experiment and with the neutron diffraction structure. We note that the FS-REDOR and FS-TEDOR will likely be more accurate that SPECIFIC-CP [20] experiments for distance measurements. A careful comparison with results from PAIN-CP [21] is in progress.
Figure 2.
FS-TEDOR experiment in 10% U-[13C,15N]-Asparagine. We employed 100 kHz TPPM decoupling during REDOR and other periods, a 1 ms Gaussian refocusing pulse (switched between the NH3 and NH2 sites), 50 kHz REDOR pulses. ωr/2π = 10 kHz
Table 1.
Heteronuclear distances measured with FS-TEDOR experiment as applied to [U-13C, 15N]-Asparagine
| FS-TEDOR (Å) | FS-REDOR (Å) | Neutron diffraction (Å) | |
|---|---|---|---|
| N-Cα | 1.50 | 1.50 | 1.49 |
| Nδ2-Cα | 3.56 | 3.58 | 3.75 |
| N-Cβ | 2.46 | 2.49 | 2.48 |
| Nδ2-Cβ | 2.41 | 2.44 | 2.42 |
To demonstrate the utility of FS-TEDOR as an approach for spectral editing in a more challenging and interesting case, we used the FS-TEDOR sequence, together with DNP, to record 2D 13C-15N spectra of the seven arginine side chains in the membrane protein bacteriorhodopsin (bR), a light-driven ion pump that has been studied extensively by magnetic resonance[22; 23; 24; 25; 26; 27; 28];[29]. Owing to their hydrophilicity, many of the lysine and arginine side chains in proteins are located at or near the surface. In contrast, those buried in the hydrophobic interior are frequently functionally important. bR contains a number of such examples, including Lys-216, to which the retinal chromophore is attached via a Schiff base linkage, and Arg-82, which is part of the complex counterion and is perturbed following deprotonation of the Schiff Base, a key step in the L->Mo photostate transition of bR [30]. In this and other systems, approaches to spectral editing based on selective cross polarization [20; 31; 32], magnetization preparation and/ or selective dephasing [13] , or both in combination[33], have been used to achieve similar ends. The optimal sequence to use in each case depends on the spin topology (e.g. 13C coupled to multiple 15Ns or the reverse) and desired selectivity.
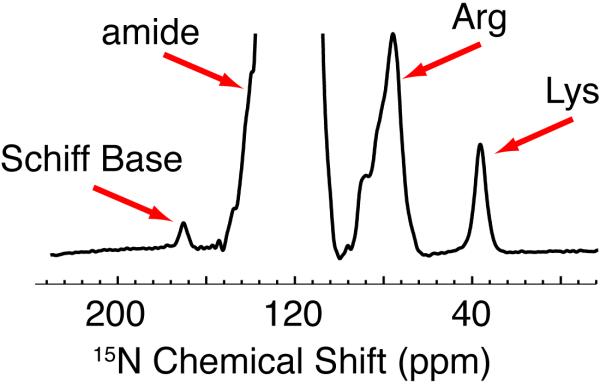
In this case, addition of DNP, which consists of CW irradiation of the EPR spectrum of TOTAPOL with 250 GHz microwaves to the FS-TEDOR experiment, enhances the signal sensitivity by a factor of ~43. Together with the lower temperatures, we obtain a factor of ~90 increase in signal intensity over our previous experiments on bR at 200 K. Figure 3 shows the 1D 15N spectrum of U-13C,15N-bR.
Figure 3.
1D DNP enhanced 15N spectra of bR illustrating the excellent S/N available with DNP that permits observation of the 15N signal of the single Schiff Base (K216). The Arg resonances irradiated with the selective pulse are labeled and lie at ~80 ppm.
We recorded correlation spectra, focusing on the 13Cγ-15Nε region of arginine. Details of instrumentation[18; 34; 35; 36], MAS probes[37], polarizing agents[38; 39], and sample preparation for applications of DNP to bacteriorhodopsin[33; 40; 41] are published elsewhere and not reviewed here. We empirically and separately optimized the TPPM decoupling [16] during the inter-pulse periods of the REDOR sequence, the selective pulse, and detection, to minimize the transverse relaxation (T2), avoid interference effects between the decoupling and multiple pulse sequence, and to maximize the sensitivity. We chose a Gaussian refocusing pulse whose length was optimized in the range 1.98 ms ± 0.265 ms [42](see Figure 4) and whose frequency was centered on the arginine side chain region (~90 ppm for 15Nε) (Figure 4). A pulse of this length sufficiently suppresses the amide backbone resonances, which otherwise would compromise the dynamic range of the experiment and obscure the arginine cross peaks, which are an order of magnitude less intense. Further, we selected a TEDOR mixing time of 4 ms, in which polarization transfer over two-bond distances (e.g. ~2.4–2.5 Å between arg-13Cγ-15Nε ) in U-13C,15N compounds is maximized.
Figure 4.
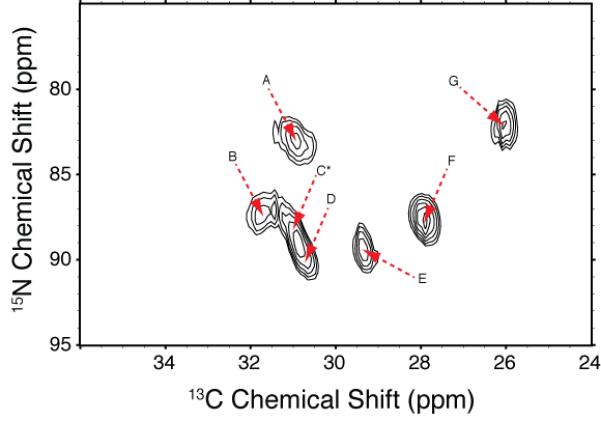
2D 15N-13C selective correlation experiment focused on the Arg-13Cγ-15Nε of bR. The sequence described in Figure 1 was employed with a 1.98 ms Gaussian refocusing pulse, and TPPM decoupling optimized to reduce transverse dephasing. The REDOR mixing time of 4 ms was chosen to favor short distances (~2.4–2.5 Å). The spinning frequency was 7.576 kHz, and all DNP experiments were conducted in dark-adapted bacteriorhodopsin (90 K), as described elsewhere.
The spectrum in Figure 4 reveals 15N-13C correlations which we assign to six of the seven expected Arg-13Cγ-15Nε cross peaks. Our assignment is based on our choice of parameters (a selective pulse centered at 90 ppm and a 4 ms TEDOR recoupling time) which favor Arg-13Cγ-15Nε magnetization transfer over Arg-13Cδ-15Nε,η1,η2, due to the mixing time and the presence of multiple 13Cδ-15N couplings. In addition, the chemical shifts observed agree with typical values for Arg-15Nε and Arg-13Cγ, indicated by amino acid chemical shift statistics for proteins [43; 44]which show Arg-15Nε chemical shifts as 90 ± 19 ppm vs. 75 ± 14 ppm and 75 ± 16 ppm for Arg-15Nη1 and Arg-15Nη2, respectively, and 27.2 ± 2.0 ppm for Arg-13Cγ vs. 43.2 ± 1.8 ppm for Arg-13Cδ. However, while both the 15N and 13C chemical shifts are only consistent with the Arg-13Cγ-15Nε assignments, their eventual validation will require site-specific assignment of all the Arg side chains, which we did not attempt here.
3. Conclusions
In summary, we have demonstrated that the FS-TEDOR experiment, a close analogue of the frequency-selective REDOR experiment, can be used quantitatively and qualitatively for 15N-13C correlation spectroscopy in crystalline solids and membrane proteins. Using DNP, we have recorded resolved arg-13Cγ-15Nε correlation spectra, tentatively identifying six of the seven expected resonances. Similar methods might be contemplated in lieu of specific isotopic labeling or suppression[47] to simplify the spin dynamics, or in contexts where quantitative distance information is required from a resolved 15N site, such as in distance measurements from the Schiff base of bR and other rhodopsins, or in peptides containing 15N-rich side chains[15; 48]. The decision to use FS-TEDOR over other methods depends on the topology of the spin system under consideration, the degree of selectivity desired, and the dephasing of transverse magnetization during the evolution times of the experiment.
Table 2.
Chemical shifts and intensities of crosspeaks in the Arg-13Cγ-15Nε region of 2D fs-TEDOR spectra bR (Figure 4). The chemical shifts and intensities were fit using SPARKY [45] following processing in NMRPipe [46]
| Peak |
15N Chemical Shift (ppm) |
13C Chemical Shift (ppm) |
Peak Volume (arbitrary units) |
|---|---|---|---|
| A | 82.9 | 30.9 | 2.9 |
| B | 87.3 | 31.7 | 2.7 |
| C* | 87.8 | 31.0 | 2.7 |
| D | 89.9 | 30.7 | 3.1 |
| E | 89.4 | 29.3 | 3.0 |
| F | 87.6 | 27.9 | 3.5 |
| G | 82.1 | 26.0 | 3.1 |
Cross peak C is not resolved from D.
Acknowledgements
The authors acknowledge invaluable technical assistance from Dr. D. Ruben, A. Thakkar, and J. Bryant, Dr. J. Sirigiri and Dr. R.J. Temkin, and discussions with Professor C.P. Jaroniec. VSB acknowledges the receipt of a PGS Fellowship from the Natural Sciences and Engineering Research Council of Canada. This research was supported by the National Institutes of Biomedical Imaging and Bioengineering through grants EB-001960, EB-002804, EB-001035, and EB-002026.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Jaroniec CP, Filip C, Griffin RG. 3D TEDOR NMR experiments for the simultaneous measurement of multiple carbon-nitrogen distances in uniformly C-13, N-15- labeled solids. Journal of the American Chemical Society. 2002;124:10728–10742. doi: 10.1021/ja026385y. [DOI] [PubMed] [Google Scholar]
- [2].Bayro MJ, Huber M, Ramachandran R, Davenport TC, Meier BH, Ernst M, Griffin RG. Dipolar truncation in magic-angle spinning NMR recoupling experiments. J. Chem. Phys. 2009;130:114506. doi: 10.1063/1.3089370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Gullion T, Schaefer J. Rotational-Echo Double-Resonance NMR. Journal of Magnetic Resonance. 1989;81:196–200. doi: 10.1016/j.jmr.2011.09.003. [DOI] [PubMed] [Google Scholar]
- [4].Hing AW, Vega S, Schaefer J. Transfer Echo Double Resonance NMR. J. Magn. Reson. 1992;96:205–209. [Google Scholar]
- [5].Li Y, Appleyard RJ, Shuttleworth WA, Evans JNS. Time-Resolved Solid-State REDOR NMR Measurements of 5-enolpyruvylshikimate 3-phosphate synthase. Journal of the American Chemical Society. 1994;116:10799–10800. [Google Scholar]
- [6].Michal CA, Jelinski LW. REDOR 3D: Heteronuclear distance measurements in uniformly labeled and natural abundance solids. Journal of the American Chemical Society. 1997;119:9059–9060. [Google Scholar]
- [7].Jaroniec CP, Tounge BA, Rienstra CM, Herzfeld J, Griffin RG. Measurement of C-13-N-15 distances in uniformly C-13 labeled biomolecules: J-decoupled REDOR. Journal of the American Chemical Society. 1999;121:10237–10238. [Google Scholar]
- [8].Jaroniec CP, Tounge BA, Herzfeld J, Griffin RG. Frequency selective heteronuclear dipolar recoupling in rotating solids: Accurate C-13-N-15 distance measurements in uniformly C-13,N-15-labeled peptides. Journal of the American Chemical Society. 2001;123:3507–3519. doi: 10.1021/ja003266e. [DOI] [PubMed] [Google Scholar]
- [9].Mehta AK, Schaefer J. Rotational-echo double resonance of uniformly labeled C-13 clusters. Journal of Magnetic Resonance. 2003;163:188–191. doi: 10.1016/s1090-7807(03)00077-6. [DOI] [PubMed] [Google Scholar]
- [10].Helmus JJ, Nadaud PS, Hofer N, Jaroniec CP. Determination of methyl C-13-N-15 dipolar couplings in peptides and proteins by three-dimensional and four-dimensional magic-angle spinning solid-state NMR spectroscopy. Journal of Chemical Physics. 2008;128 doi: 10.1063/1.2817638. [DOI] [PubMed] [Google Scholar]
- [11].Rienstra CM, Tucker-Kellogg L, Jaroniec CP, Hohwy M, Reif B, McMahon MT, Tidor B, Lozano-Perez T, Griffin RG. De novo determination of peptide structure with solid-state magic-angle spinning NMR spectroscopy. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:10260–10265. doi: 10.1073/pnas.152346599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Jaroniec CP, MacPhee CE, Bajaj VS, McMahon MT, Dobson CM, Griffin RG. High Resolution Molecular Structure of a Peptide in an Amyloid Fibril Determined by MAS NMR Spectroscopy. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:711–716. doi: 10.1073/pnas.0304849101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Jaroniec CP, Lansing JC, Tounge BA, Belenky M, Herzfeld J, Griffin RG. Measurement of dipolar couplings in a uniformly C-13,N-15- labeled membrane protein: Distances between the Schiff base and aspartic acids in the active site of bacteriorhodopsin. Journal of the American Chemical Society. 2001;123:12929–12930. doi: 10.1021/ja016923r. [DOI] [PubMed] [Google Scholar]
- [14].Jaroniec CP, MacPhee CE, Astrof NS, Dobson CM, Griffin RG. Molecular Conformation of a Peptide Fragment of Transthyretin in an Amyloid Fibril. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:16748–16753. doi: 10.1073/pnas.252625999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].van der Wel PCA, Lewandowski JR, Griffin RG. Solid-state NMR study of amyloid nanocrystals and fibrils formed by the peptide GNNQQNY from yeast prion protein Sup35p. Journal of the American Chemical Society. 2007;129:5117–5130. doi: 10.1021/ja068633m. [DOI] [PubMed] [Google Scholar]
- [16].Bennett AE, Rienstra CM, Auger M, Lakshmi KV, Griffin RG. Heteronuclear Decoupling in Rotating Solids. J. Chem. Phys. 1995;103:6951. [Google Scholar]
- [17].Song C, Hu K-N, Joo C-G, Swager TM, Griffin RG. TOTAPOL - A Biradical Polarizing Agent for Dynamic Nuclear Polarization Experiments in Aqueous Media. J. Am Chem. Soc. 2006;128:11385–90. doi: 10.1021/ja061284b. [DOI] [PubMed] [Google Scholar]
- [18].Bajaj VS, Hornstein MK, Kreischer KE, Sirigiri JR, Woskov PP, Mak-Jurkauskas ML, Herzfeld J, Temkin RJ, Griffin RG. 250 GHz CW gyrotron oscillator for dynamic nuclear polarization in biological solid state NMR. Journal of Magnetic Resonance. 2007;189:251–279. doi: 10.1016/j.jmr.2007.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Veshtort M, Griffin RG. SPINEVOLUTION: a powerful tool for simulations of solid and liquid state NMR experiments. J. Magn. Resonance. 2006;178:248–282. doi: 10.1016/j.jmr.2005.07.018. [DOI] [PubMed] [Google Scholar]
- [20].Baldus MA, Petkova AT, Herzfeld J, Griffin RG. Cross Polarization in the Tilted Frame: Assignment and Spectral Simplification in Heteronuclear Spin Systems. Molecular Physics. 1998;95:1197–1207. [Google Scholar]
- [21].Lewandowski JR, Paepe G.d., Griffin RG. Proton Assisted Insensitive Nuclei Cross Polarization. J. Am Chem. Soc. 2007;129:728–729. doi: 10.1021/ja0650394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Lanyi JK, Schobert B. Mechanism of proton transport in bacteriorhodopsin from crystallographic structures of the K, L, M1, M2, and M2′ intermediates of the photocycle. Journal of Molecular Biology. 2003;328:439–50. doi: 10.1016/s0022-2836(03)00263-8. [DOI] [PubMed] [Google Scholar]
- [23].Luecke H, Schobert B, Richter HT, Cartailler JP, Lanyi JK. Structure of bacteriorhodopsin at 1.55 angstrom resolution. Journal of Molecular Biology. 1999;291:899–911. doi: 10.1006/jmbi.1999.3027. [DOI] [PubMed] [Google Scholar]
- [24].Lugtenburg J, Muradinszweykowska M, Heeremans C, Pardoen JA, Harbison GS, Herzfeld J, Griffin RG, Smith SO, Mathies RA. Mechanism for the Opsin Shift of Retinals Absorption in Bacteriorhodopsin. Journal of the American Chemical Society. 1986;108:3104–3105. [Google Scholar]
- [25].Herzfeld J, Hu JG, Hatcher ME, Belenky M, Verdegem P, Lugtenburg J, Griffin RG. NMR evidence for an electrostatically controlled, torsion-based switch in light-driven proton transport by bacteriorhodopsin. Biophysical Journal. 2001;80:85. [Google Scholar]
- [26].Herzfeld J, Hu JG, Sun BQ, Petkova A, Hatcher ME, Bizounok M, Griffin RG. Solid-state NMR studies of energy transduction in bacteriorhodopsin. Biophysical Journal. 1998;74:A137–A137. [Google Scholar]
- [27].Herzfeld J, Lansing JC. Magnetic resonance studies of the bacteriorhodopsin pump cycle. Annual Review of Biophysics and Biomolecular Structure. 2002;31:73–95. doi: 10.1146/annurev.biophys.31.082901.134233. [DOI] [PubMed] [Google Scholar]
- [28].Herzfeld J, Tounge B. NMR probes of vectoriality in the proton-motive photocycle of bacteriorhodopsin: evidence for an ‘electrostatic steering’ mechanism. Biochimica Et Biophysica Acta-Bioenergetics. 2000;1460:95–105. doi: 10.1016/s0005-2728(00)00132-8. [DOI] [PubMed] [Google Scholar]
- [29].Stoeckenius W, Lozier RH, Niederberger W. Photoreactions of bacteriorhodopsin. Biophysics of Structure and Mechanism. 1977;3:65–68. doi: 10.1007/BF00536456. [DOI] [PubMed] [Google Scholar]
- [30].Petkova AT, Hu JGG, Bizounok M, Simpson M, Griffin RG, Herzfeld J. Arginine activity in the proton-motive photocycle of bacteriorhodopsin: Solid-state NMR studies of the wild-type and D85N proteins. Biochemistry. 1999;38:1562–1572. doi: 10.1021/bi981968z. [DOI] [PubMed] [Google Scholar]
- [31].Jehle S, Rehbein K, Diehl A, van Rossum BJ. Amino-acid selective experiments on uniformly C-13 and N-15 labeled proteins by MAS NMR: Filtering of lysines and arginines. Journal of Magnetic Resonance. 2006;183:324–328. doi: 10.1016/j.jmr.2006.08.015. [DOI] [PubMed] [Google Scholar]
- [32].Petkova AT, Baldus M, Belenky M, Hong M, Griffin RG, Herzfeld J. Backbone and Sidechain Assignment Strategies for Multiply Labeled Membrane Peptides and Proteins. J. Magn. Resonance. 2003;160:1–12. doi: 10.1016/s1090-7807(02)00137-4. [DOI] [PubMed] [Google Scholar]
- [33].Bajaj VS, Mak-Jurkauskas ML, Belenky ML, Herzfeld J, Griffin RG. Functional and shunt states of bacteriorhodopsin resolved by 250 GHz dynamic nuclear polarization-enhanced solid-state NMR. Proceedings of the National Academy of Sciences of the United States of America. 2009 doi: 10.1073/pnas.0900908106. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Bajaj VS, Farrar CT, Hornstein MK, Mastovsky I, Vieregg J, Bryant J, Elena B, Kreischer KE, Temkin RJ, Griffin RG. Dynamic nuclear polarization at 9T using a novel 250 GHz gyrotron microwave source. J. Mag. Res. 2003;160:85–90. doi: 10.1016/j.jmr.2011.09.010. [DOI] [PubMed] [Google Scholar]
- [35].Hornstein MK, Bajaj VS, Griffin RG, Temkin RJ. Continuous-wave operation of a 460-GHz second harmonic gyrotron oscillator. IEEE Transactions on Plasma Science. 2006;34:524–533. doi: 10.1109/TPS.2006.875769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Woskov PW, Bajaj VS, Hornstein MK, Temkin RJ, Griffin RG. Corrugated Waveguide and Directional Coupler for CW 250 GHz Gyrotron DNP Experiments. IEEE Transactions on Microwave Theory and Techniques. 2005;53:1863–69. doi: 10.1109/TMTT.2005.848097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Barnes AB, Mak-Jurkauskas ML, Matsuki Y, Bajaj VS, Wel P.C.A.v.d., DeRocher R, Bryant J, Sirigiri JR, Temkin RJ, Lugtenburg J, Herzfeld J, Griffin RG. Cryogenic sample exchange NMR probe for magic angle spinning dynamic nuclear polarization. Journal of Magnetic Resonance. 2009;198:261–270. doi: 10.1016/j.jmr.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Hu K-N, Bajaj VS, Rosay MM, Griffin RG. High Frequency Dynamic Nuclear Polarization Using Mixtures of TEMPO and Trityl Radicals. Journal of Chemical Physics. 2007;126:044512. doi: 10.1063/1.2429658. [DOI] [PubMed] [Google Scholar]
- [39].Hu KN, Yu HH, Swager TM, Griffin RG. Dynamic nuclear polarization with biradicals. Journal of the American Chemical Society. 2004;126:10844–10845. doi: 10.1021/ja039749a. [DOI] [PubMed] [Google Scholar]
- [40].Mak-Jurkauskas ML, Bajaj VS, Hornstein MK, Belenky M, Griffin RG, Herzfeld J. Gradual Winding of the Bacteriorhodopsin Chromophore in the First Half of Its Ion-Motive Photocycle: a Dynamic Nuclear Polarization-Enhanced Solid State NMR Study. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:883–888. doi: 10.1073/pnas.0706156105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Rosay M, Zeri AC, Astrof NS, Opella SJ, Herzfeld J, Griffin RG. Sensitivity-enhanced NMR of biological solids: Dynamic nuclear polarization of Y21M fd bacteriophage and purple membrane. Journal of the American Chemical Society. 2001;123:1010–1011. doi: 10.1021/ja005659j. [DOI] [PubMed] [Google Scholar]
- [42].Li Y, Wylie BJ, Rienstra CM. Selective refocusing pulses in magic-angle spinning NMR: Characterization and applications to multi-dimensional protein spectroscopy. Journal of Magnetic Resonance. 2006;179:206–216. doi: 10.1016/j.jmr.2005.12.003. [DOI] [PubMed] [Google Scholar]
- [43].Markley JL, Ulrich EL, Berman HM, Henrick K, Nakamura H, Akutsu H. BioMagResBank (BMRB) as a partner in the Worldwide Protein Data Bank (wwPDB): new policies affecting biomolecular NMR depositions. Journal of Biomolecular NMR. 2008;40:153–155. doi: 10.1007/s10858-008-9221-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, Livny M, Mading S, Maziuk D, Miller Z, Nakatani E, Schulte CF, Tolmie DE, Wenger RK, Yao HY, Markley JL. BioMagResBank. Nucleic Acids Research. 2008;36:D402–D408. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Goddard TD, Kneller DG. SPARKY Version 3.115. Univ. California; San Francisco: 2008. [Google Scholar]
- [46].Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- [47].Vuister GW, Kim SJ, Wu C, Bax A. 2D and 3D NMR study of phenylalanine residues in proteins by reserve isotopic labeling. Journal of the American Chemical Society. 1994;116:9206–9210. [Google Scholar]
- [48].van der Wel PCA, Hu KN, Lewandowski J, Griffin RG. Dynamic nuclear polarization of amyloidogenic peptide nanocrystals: GNNQQNY, a core segment of the yeast prion protein Sup35p. Journal of the American Chemical Society. 2006;128:10840–10846. doi: 10.1021/ja0626685. [DOI] [PubMed] [Google Scholar]