Summary
Activity-regulated gene expression is believed to play a key role in the development and refinement of neuronal circuitry. Nevertheless, the transcriptional networks that regulate synaptic plasticity remain largely uncharacterized. We show here that the CREB- and activity-regulated microRNA, miR132, is induced during periods of active synaptogenesis. Moreover, miR132 is necessary and sufficient for hippocampal spine formation. Expression of the miR132 target, p250GAP, is inversely correlated with miR132 levels and spinogenesis. Furthermore, knockdown of p250GAP increases spine formation while introduction of a p250GAP mutant unresponsive to miR132 attenuates this activity. Inhibition of miR132 decreases both mEPSC frequency and the number of GluR1-positive spines, while knockdown of p250GAP has the opposite effect. Additionally, we show that the miR132/p250GAP circuit regulates Rac1 activity and spine formation by modulating synapse-specific Kalirin7-Rac1 signaling. These data suggest that neuronal activity regulates spine formation, in part, by increasing miR132 transcription, which in turn activates a Rac1-Pak actin remodeling pathway.
Introduction
Activity-regulated gene expression has been implicated in synapse formation and long-lasting synaptic plasticity. The majority of excitatory presynaptic terminals in the CNS impinge on small actin-enriched spines (Nimchinsky et al., 2002; Yuste and Bonhoeffer, 2001, 2004). Regulation of spine number and size is associated with use-dependent plasticity, neurological disorders, and mental retardation (Fiala et al., 2002; Maletic-Savatic et al., 1999; Toni et al., 1999). Although synapse development is partly controlled by intrinsic factors, neuronal activity also plays a critical role (Katz and Shatz, 1996; Yuste and Bonhoeffer, 2001). Indeed, the timing of afferent innervation and synapse formation coincides with the period of maximum dendritic remodeling (Pokorny and Yamamoto, 1981; Wong and Ghosh, 2002).
The transcription factor CREB regulates dendritic growth (Redmond et al., 2002; Wayman et al., 2006) and spine formation in hippocampal neurons (Marie et al., 2005). How this occurs remains poorly defined, however. Recently, microRNAs (miRNAs) have been implicated in regulating dendritic morphology (Fiore et al., 2009; Schratt et al., 2006; Vo et al., 2005; Wayman et al., 2008). microRNAs are 19–25 bp molecules that play an important role in development and differentiation (Du and Zamore, 2005). In this study, we examine whether the CREB-miR132 pathway regulates activity-dependent formation of excitatory synapses.
We report that inhibition of miR132 attenuates activity-induced spine formation and that introduction of miR132 increases spine formation in organotypic hippocampal neurons. Likewise, depletion of the Rho-family GTPase Activating Protein, p250GAP, a miR132 target, markedly increases mEPSC frequency and the prevalence of GluR1 positive spines. Expression of a miR132 insensitive version of the p250GAP mRNA decreases spine formation. These data suggest that the miR132-p250GAP pathway regulates both morphological and functional synapse formation. Although p250GAP has previously been proposed to regulate spine morphology, how this occurs was not clear (Nakazawa et al., 2008). We show that transcription of miR132 or downregulation of its target, p250GAP, increases Rac1 activity. We further show that the Rac1-PAK pathway is downstream of miR132 and that the RacGEF, Kalirin-7, is essential for miR132-regulated spine formation.
RESULTS
Expression of miR132 is associated with activity-dependent synaptogenesis
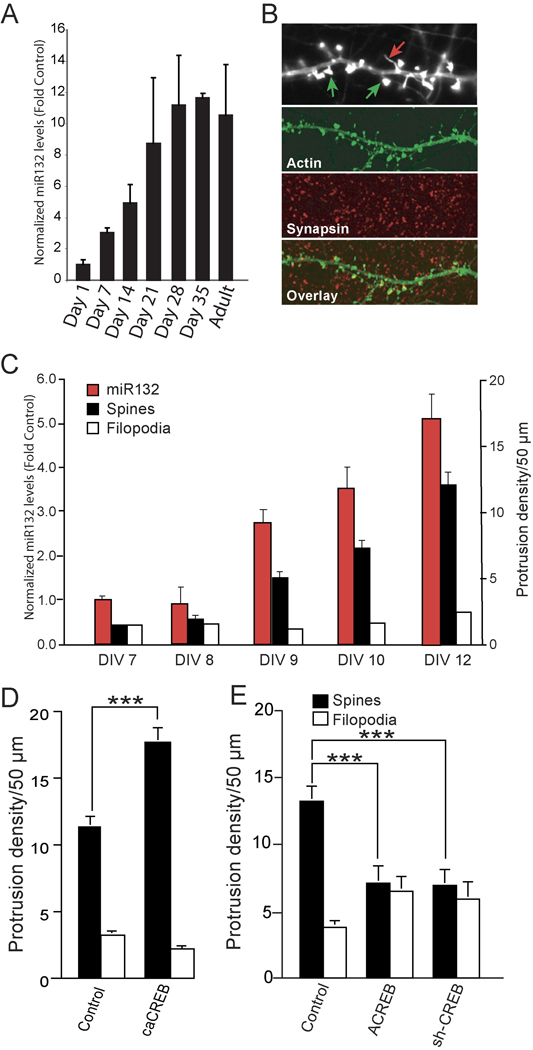
The induction of miR132 by synaptic activity led us to investigate whether miR132 expression is increased during periods of active synaptogenesis. Spine formation in the developing rat hippocampus is minimal during the first postnatal week but accelerates dramatically between weeks 2 through 4 (Pokorny and Yamamoto, 1981; Steward and Falk, 1991). Basal levels of the mature-miR132 were low in the hippocampus before post-natal day 7 (Fig. 1A). At day 7, there was a statistically significant increase in miR132 levels. miR132 levels continued to increase and, at postnatal day 21, reached levels equivalent to those seen in the adult. Thus, the kinetics of miR132 expression correlated with periods of active synapse formation (Pokorny and Yamamoto, 1981; Steward and Falk, 1991).
Fig. 1. miR132 expression correlates with spine formation in vivo and in vitro.
(A) Mature miR132 levels are enhanced within the hippocampus during the period of afferent innervation. Hippocampi were isolated from rats (2 animals/developmental time point beginning at post natal day 1). RNA was isolated, reverse–transcribed, and analyzed by Taqman real-time PCR with miR132 cDNA primers. The data is normalized to GAPDH cDNA levels. (B) Cultured hippocampal neurons were transfected with YFP-γ actin to visualize dendritic protrusions, fixed on DIV 12, and immunostained for the presynaptic marker synapsin 1. (Upper panel) Spines (green arrows, protrusion head size 2 fold > shaft) were identified as mushroom-shaped projections with intense expression of EYFP-γ actin at their tips, whereas filopodia (red arrows, protrusion head size 2 fold ≤ shaft) were thin with low expression of EYFP-γ actin throughout. (C) Hippocampal neurons were transfected with EYFP-γ actin on DIV 6 and then cultured 7–12 days. At each time point, the cultures were either fixed and analyzed for spine and filopodia (black and white bars), or RNA was isolated and reverse transcribed and analyzed by Taqman-real time-PCR with mature-miR132 cDNA primers (red bars). For RT-PCR experiments, the data were normalized to GAPDH cDNA levels (± SEM n=5–6). (D, E) Hippocampal neurons were transfected with EYFP-γ actin ± empty vector (Control) ± ACREB ± siCREB or ± caCREB on DIV 7 and then fixed on DIV12. Dendrites were imaged, and at least three different 50 µm sections per neuron (25–30 neurons per condition) were analyzed. Quantitation of dendritic spines and filopodia are shown (± SEM, *** P < 0.001).
The increase in miR132 levels may reflect an increase in endogenous synaptic activity. To test this possibility, we examined miR132 expression in cultured neurons where the kinetics of synaptogenesis can be measured precisely. To assess dendritic spine development, we transfected hippocampal neurons with low levels of a plasmid expressing YFP-γ actin. On day 12 in vitro (DIV 12), these neurons exhibit extensive dendritic arborization with numerous dendritic protrusions. We identified spines by their mushroom-shaped morphology and the presence of intense puncta of YFP-γ actin at their tip heads (Fig. 1B, green arrow), whereas filopodia (Fig. 1B, red arrow) were long, thin protrusions with low and homogenous YFP-γ actin expression. When the cultures were also stained for the presynaptic marker synapsin 1 (Fig. 1B) 97% of spines, but only 14% of filopodia, showed opposing staining. Between DIV 7–8, pre-miR132 and mature-miR132 expression was low (Fig. 1C and data not shown). After 9 days, both pre-miR132 and mature miR132 levels increased and remained elevated. Thus, the increase in miR132 levels coincided with the onset of spine formation.
Inhibition of CREB suppresses spinogenesis
In agreement with the findings of others (Cardinaux et al., 2000; Marie et al., 2005), expression of caCREB (constitutively active CREB, CREBDIEDML) increased spine density (Fig. 1D). We also found that expression of a dominant negative CREB mutant (ACREB) or a CREB-targeted shRNA inhibited spine formation (Fig. 1E). ACREB or shCREB did not significantly regulate nuclear pyknosis, cleaved caspase 3 levels or dramatically alter gross dendritic morphology in hippocampal neurons grown in the presence of B27 (Lee et al., 2005 and data not shown). These data suggest that CREB-dependent transcription is necessary for the formation of dendritic spines. Taken together with our previously published finding that CREB regulates dendrite growth by increasing transcription of miR132 (Vo et al., 2005; Wayman et al., 2008), these results suggested that miR132 might also be involved in spine formation.
miR132 regulates spine formation by down-regulating p250GAP
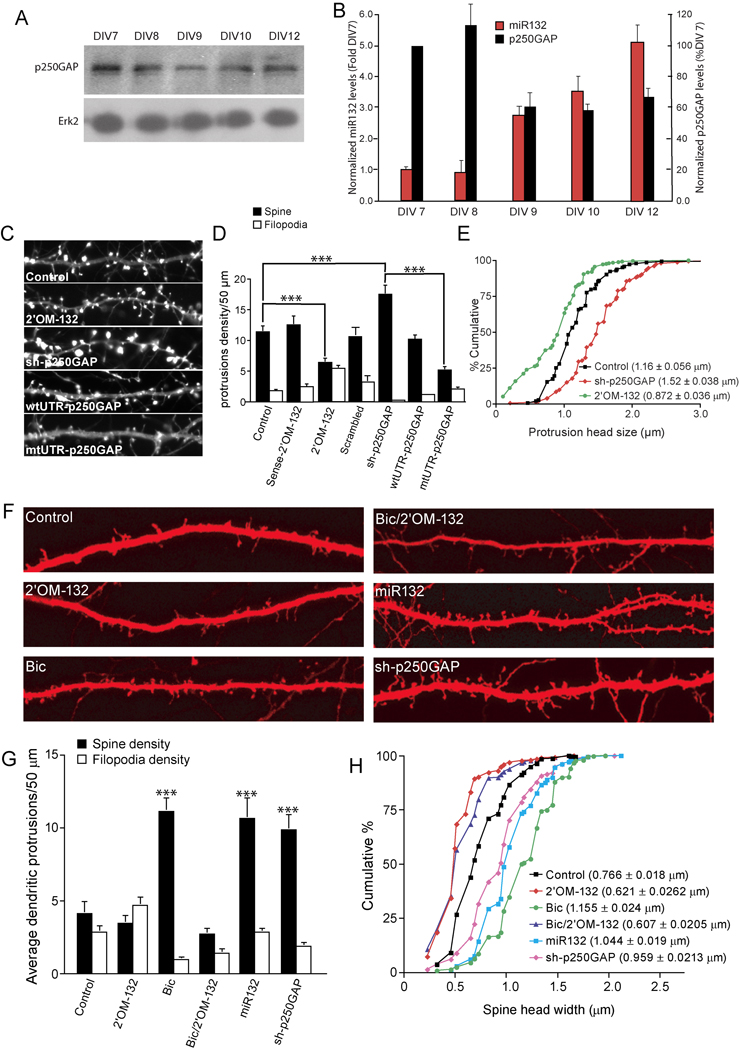
We recently demonstrated that miR132 regulates activity-dependent dendritic growth by inhibiting translation of its target, p250GAP (Wayman et al., 2008). Because miR132 levels increase during periods of spine formation, it might be expected that a reduction in p250GAP protein would also occur. As shown in Fig. 2A and B, p250GAP levels are highest at DIV7–8, decrease by 40% at DIV 9, and remain reduced through DIV 12. These experiments led us to examine whether regulation of p250GAP by miR132 also affected spine formation.
Fig. 2. miR132 regulates spine formation by repressing p250GAP.
(A) Developmentally timed hippocampal cultures were analyzed by Western blot for expression of p250GAP. (B) p250GAP expression was normalized to Erk2 levels and plotted with normalized mature-miR132 levels. (C–E) Hippocampal neurons DIV 7 were transfected with EYFP-γ actin and either empty vector (Control), Sense-2’OM-132 (sense control), 2’OM-132 (Antisense 2’OM-132), sh-p250GAP wtUTR-p250GAP, or mtUTR-p250GAP. On DIV12, the neurons were fixed and imaged. D,E, Effects of indicated transfections on (D) protrusion density and (E) spine head width. F–H, Hippocampal slices were cultured for 3 days and subjected to biolistic transfection with TFP ± empty vector (Control) or other plasmids as indicated. Slices were allowed to recover for 1 day and then stimulated (where indicated) with 20 µM bicuculline for 2 days. On DIV 7, dendritic protrusions were analyzed for spines and filopodia. (F) Representative examples of apical CA1 dendrites from pyramidal neurons in hippocampal organotypic sliced cultures are shown. Summary of the effects of transfection on protrusion density (G) and spine head width (H) are shown (*** denotes significance between test and control conditions). Statistical analyses utilized ANOVA and Tukey’s post-test. (± SEM, *** P < 0.001).
Because phenotypic effects of miR132 expression can easily be saturated (Wayman et al. 2008 and data not shown), we utilized organotypic slice cultures, which have both low levels of spontaneous activity and low basal miR132 expression (Supplemental Fig. 1), to examine whether miR132 can trigger de novo spine formation. Organotypic hippocampal slices retain the cellular and morphological organization of the intact hippocampus and have been used extensively to study use-dependent synaptic plasticity (Bahr, 1995; Caeser and Aertsen, 1991). Hippocampal slices from post natal day 4 rat pups were cultured for 4 days and then treated with 20 µM bicuculline to mimic afferent input (see Fig. 2F–H). Bicuculline treatment dramatically increased synaptic activity (Supplemental Fig. 2) and miR132 levels (Wayman et al., 2008). To analyze spinogenesis in slice culture, we segregated dendritic protrusions on apical dendrites of CA1 pyramidal neurons into spines (head size ≥ 2-fold larger than shaft) or filopodia (head size 2-fold ≤ than shaft). Under basal conditions, approximately 50% of dendritic protrusions are mushroom-shaped spines (Fig. 2F and G). Bicuculline promoted an approximately 50 % increase in spine density and protrusion size (0.766 ±0.018 vs. 1.155 ± 0.024 µm respectively, Fig. 2H). This increase was generated by synaptic activity because pretreatment with APV or TTX strongly suppressed spine formation (data not shown). Importantly, transfection of miR132 mimicked the effects of bicuculline on spine density and size (Fig. 2F–H). These data show that introduction of miR132 is sufficient to trigger de novo spine formation in hippocampal neurons.
Consistent with the idea that miR132 regulated spine formation, transfection of 2’O-methyl anti-sense-miR132 oligo inhibitors (2’OM-132) but not 2’O-methyl- sense-miR132 oligo (Sense-2’OM-132) attenuated spine formation, decreased spine head size, and increased the number of filopodia in cultured hippocampal neurons (Fig. 2C–E). Taqman real time PCR analyses showed that the 2'OM-132 inhibitor down-regulated mature miR132 levels, but not unrelated miRNAs (Wayman et al., 2008) and data not shown). In cultured hippocampal slices, where basal miR132 levels are low, transfection of the 2’OM-132 inhibitor had little effect on basal spine density (Fig. 2F–H) or spine head size (Fig. 2H) but transfection of this oligo completely blocked bicuculline-stimulated increases in spine density and head size (Fig. 2F–H).
We next tested the role of p250GAP in miR132-mediated spine formation. shRNA-mediated knockdown of p250GAP (Supplemental Fig. 3) markedly increased spine density and spine size while reducing filopodia density in cultured hippocampal neurons (Fig. 2C–E) and organotypic hippocampal pyramidal cells (Fig. 2F–H). We had previously shown that point mutations in the miR132 miRNA response element of p250GAP (mUTR-p250GAP) attenuated its downregulation in response to both miR132 and synaptic activity (Wayman et al., 2008). Given the reciprocal expression of miR132 and p250GAP (Fig. 2B), introduction of this miR132-insensitive mutant might be expected to increase steady state p250GAP levels. Accordingly, expression of mtUTR-p250GAP, but not wild-type p250GAP (wtUTR-p250GAP), inhibited spine formation (Fig. 2C, D). These experiments support the idea that suppression of p250GAP is essential for the effects of miR132 on spine formation.
miR132 regulates the formation of functional synapses
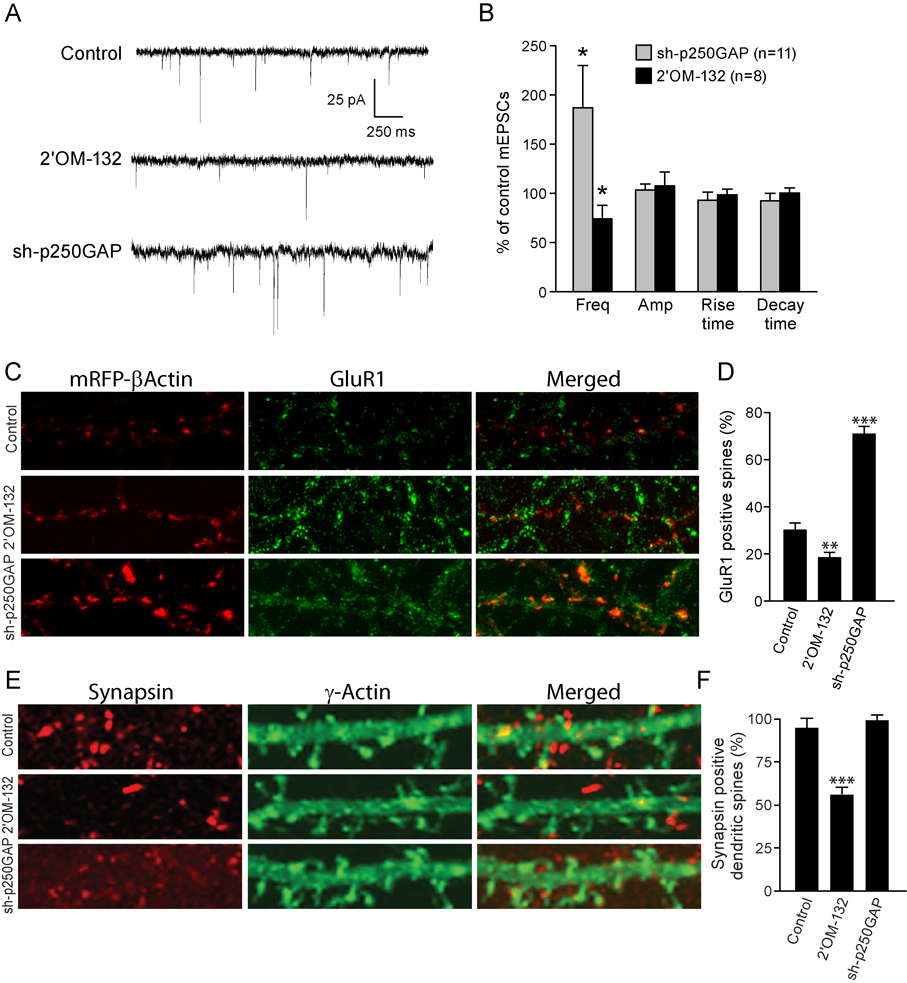
To address whether miR132 and p250GAP also regulates synaptic function, we recorded miniature excitatory post-synaptic currents (mEPSCs) from synaptically mature hippocampal neurons. Targeted knockdown of p250GAP increased mEPSC frequency by approximately 2-fold (Fig. 3A, B). In contrast, transfection of 2’OM-132 caused a ~30% reduction in mEPSC frequency. Neither inhibiting the expression of p250GAP nor blocking miR132 function significantly affected mEPSC amplitude, rise time, or decay time (Fig. 3B). We also examined the number of dendritic spines containing surface-expressed GluR1 receptors. In agreement with our electrophysiological findings, introduction of 2’OM-132 decreased, and shRNA-mediated knockdown of p250GAP increased, the number of GluR1-positive spines (Fig. 3C, D). To characterize the effects of miR132 and p250GAP on synapse formation further, we immunostained neurons for the presynaptic protein, synapsin I, and correlated this staining with mRFP actin-staining of dendritic spines. Under basal conditions ~95% of mRFP-labeled dendritic spines had juxtaposed synapsin I positive puncta. Thus, it was not surprising that p250GAP knockdown did not have a discernable effect (Fig. 3E, F). Transfection with 2’OM-132 reduced the juxtaposition of synapsin I and mRFP-labeled spines to 55%.
Fig. 3. miR132 regulates synaptic function and morphology.
(A, B) Effect of 2’OM-132 and sh-p250GAP on mEPSCs. Cultured neurons (DIV 7) were transfected with EYFP-γ actin and either empty vector (Control), 2’OM-132 or sh-p250GAP and mEPSCs were recorded on DIV12. (A) Representative traces of mEPSCs recorded from control, 2’OM-132, and sh-p250GAP transfected neurons. (B) Frequencies, amplitudes, rise times, and decay times of mEPSCs normalized to control. Statistical analyses utilized ANOVA and Tukey’s post-test. (± SEM, * P < 0.05). (C) Hippocampal neurons DIV 7 were transfected with mRFP-β actin and either empty vector (Control), 2’OM-132 or sh-p250GAP. The neurons were fixed on DIV 12 and stained for GluR1 using an N-terminal anti-GluR1 antibody. (D) Co-localization of GluR1 and actin-stained spines. (E) Hippocampal neurons DIV 7 were transfected with mRFP-β actin and either empty vector (Control) 2’OM-132 or sh-p250GAP. Neurons were fixed on DIV 12 and stained for the presynaptic protein, synapsin I. F, Effects of 2’OM-132 and sh-p250GAP on dendritic spines that show associated synapsin I staining. (± SEM, ** P < 0.01 , *** P < 0.001).
miR132 regulates spine formation by activating the Rac-PAK pathway
p250GAP has been shown to regulate the activity of Rac, RhoA and Cdc42 by increasing their endogenous GTPase activity (Moon et al., 2003; Nakayama et al., 2000; Nakazawa et al., 2003; Okabe et al., 2003; Zhao et al., 2003). Therefore, we asked which GTPases are regulated by p250GAP in our hippocampal neuron cultures. We first tested whether expression of miR132 or sh-p250GAP regulated spinogenesis by inhibiting RhoA. In our culture system, transfection of miR132 or sh-p250GAP did not significantly affect RhoA activity (data not shown). Moreover, consistent with previous studies (Elia et al., 2006; Fu et al., 2007; Kang et al., 2009; Sfakianos et al., 2007; Zhang and Macara, 2008; Nakayama et al., 2000; Pilpel and Segal, 2004; Tashiro et al., 2000), we found that constitutively active RhoA (caRhoA, RhoAG14V,) inhibited spinogenesis, whereas inhibition of RhoA activity by expression of either dominant negative RhoA (dnRhoA, RhoAT19N) or sh-RhoA increased spine density and spine head size (Supplemental Fig. 4A–C). Moreover, caRhoA inhibited spinogenesis stimulated by sh-p250GAP (Supplemental Fig. 4D). Therefore, we conclude that miR132 does not regulate hippocampal neuron spinogenesis via RhoA activity.
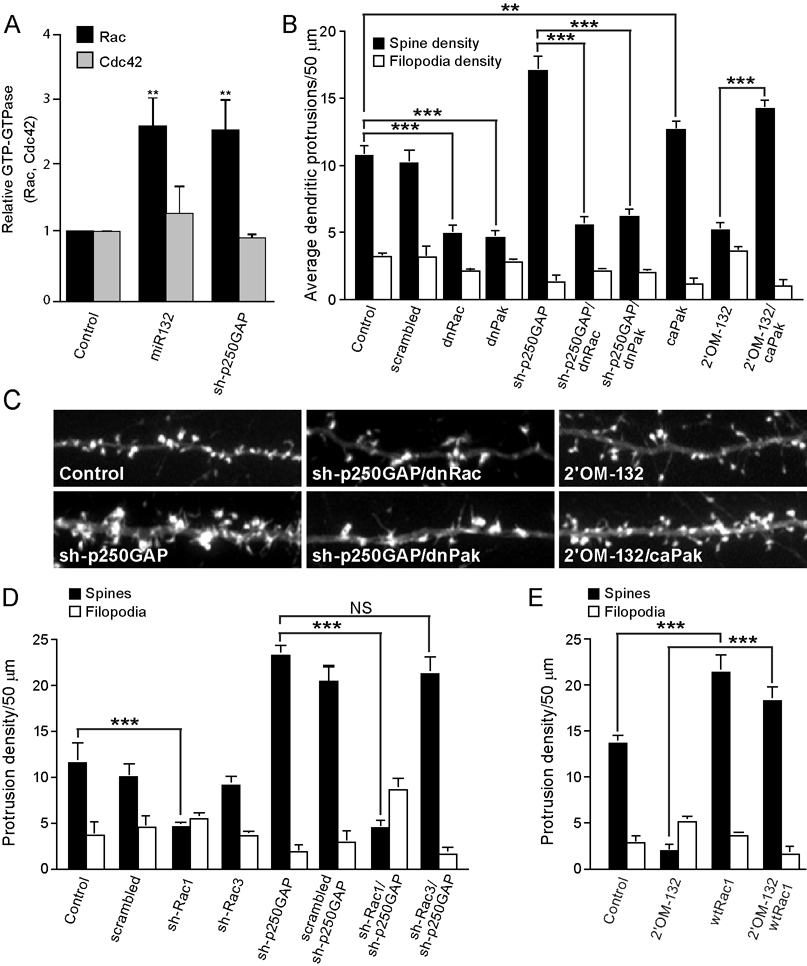
Because p250GAP has been reported to be a GTPase activating protein for both Rac and Cdc42 (Moon et al., 2003; Nakayama et al., 2000; Nakazawa et al., 2003; Okabe et al., 2003; Zhao et al., 2003), we examined whether these small G-proteins contributed to miR132 regulation of spine formation. GST-PAK pull down experiments showed that miR132 transfection significantly increased Rac activity but did not affect Cdc42 activity (Fig. 4A and Supplemental Fig. 5). Interestingly, shRNA-mediated knockdown of p250GAP similarly increased Rac activity. These data suggest that miR132 regulates Rac activity, at least in part, via its ability to repress p250GAP translation and implies that p250GAP functions as a Rac-GAP in hippocampal neurons. We next utilized dominant interfering mutants to evaluate whether Rac or Pak mediate miR132-stimulated spine formation. As expected, expression of inactive Rac (dnRac, RacT17N) or Pak (dnPakK299R) strongly suppressed the number of spines under basal conditions (Fig. 4B and C). Likewise, the increase in spine formation triggered by p250GAP knockdown was completely blocked by dnRac and dnPak. Conversely, expression of a constitutively active Pak (caPak, PakT423E) rescued the deficit in spine formation caused by miR132 inhibition.
Fig. 4. miR132 and p250GAP regulate the Rac-PAK pathway.
Hippocampal cultures were transfected with myc-Rac or myc-Cdc42 ± empty vector (Control), miR132 or ± sh-p250GAP on DIV 5. On DIV7, Rac and Cdc42 activity were assayed using Rac/Cdc42 pull down assays. (A) Quantitation of Rac and Cdc42 activity from three independent experiments. (B, C) Rac and Pak activity is required for miR132 stimulated dendritic growth. Hippocampal neurons were transfected DIV 7 with plasmids encoding mRFP-β actin ± scrambled shRNA, miR132, sh-p250GAP, 2’OM-132, dnRac, dnPak, or caPak. Dendritic spine density was quantified on DIV 12. (D) Rac1 (and not Rac3) is downstream of p250GAP. Hippocampal neurons were transfected DIV 7 with plasmids encoding mRFP-β actin ± sh-p250GAP and, where indicated, shRNA constructs targeting Rac1 or Rac3 or a scrambled control. Dendritic spine density was quantified on DIV 12. Quantitation of dendritic spine and filopodia density is shown (± SEM). (E) Activation of Rac1 is sufficient to stimulate spine formation in the absence of miR132. Hippocampal neurons were transfected DIV 7 with plasmids encoding mRFP- actin ± 2’OM-132 and, where indicated, wild-type Rac1. Dendritic spine and filopodia density on DIV 12 is shown (± SEM, ** P < 0.01 , *** P < 0.001).
There are three isoforms of Rac, but only Rac1 and Rac3 are expressed in neurons (Hajdo-Milasinovic et al., 2007; Schwamborn and Puschel, 2004). To determine which Rac isoform contributes to miR132-stimulated spinogenesis we utilized specific sh-RNAs (Supplemental Fig. 6). shRNA-mediated knockdown of Rac1, but not Rac3, inhibited basal and si-p250GAP-stimulated spine formation (Fig. 4D). These data place Rac1 downstream of miR132 and imply that Rac1 activity should occlude the effect of inhibiting miR132 function. Consistent with this model, spine formation triggered by wtRac1 was unaffected by 2’OM-132 transfection (Fig. 4E). Taken together, these data suggest that Rac1 is downstream of miR132 and p250GAP.
Kalirin-7 is required for activity- and miR132-stimulated spine formation
The Rac GEF, Kalirin-7, is expressed selectively in spines and has been proposed to play a critical role in hippocampal spine formation (Ma et al., 2003; Penzes et al., 2001; Xie et al., 2007a; Xie et al., 2007b). Kalirin-7 is an alternatively spliced variant of the Kalirin gene that is expressed at high levels in the hippocampus (Ma et al., 2003). We utilized shRNA constructs that selectively targeted the unique 3’ untranslated region of Kalirin-7 to determine whether it contributes to activity- and miR132-stimulated spine formation. Kalirin-7 knockdown attenuated basal spine formation as well as that triggered by bicuculline, miR132 transfection, and p250GAP knockdown (Fig. 5A–C). Conversely, overexpression of Kalirin-7 not only increased spine formation, but also occluded the effect of miR132 inhibition. These data indicate that activity-induced spine formation requires both miR132-mediated disinhibition, and Kalirin-7-mediated activation, of Rac1.
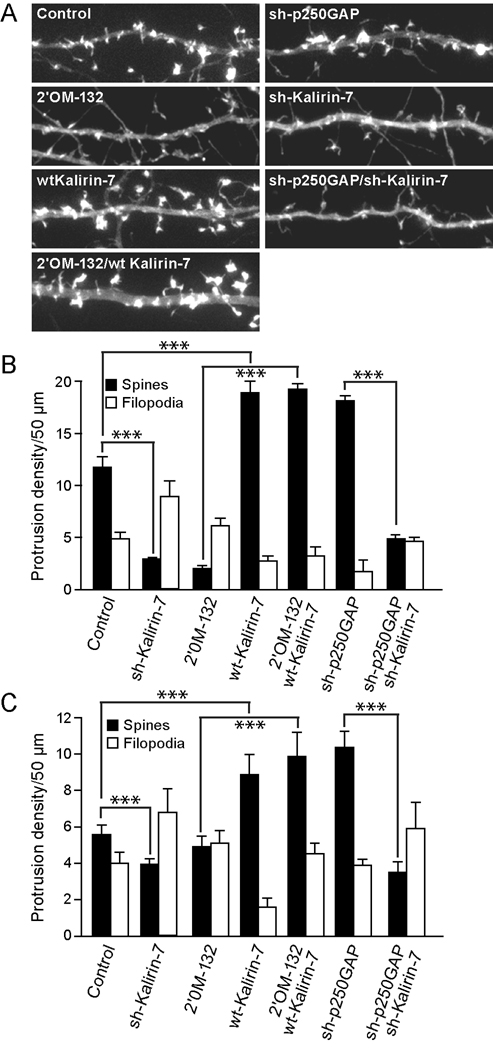
Fig. 5. miR132-regulated spine formation requires the RacGEF, Kalirin-7.
(A, B) Hippocampal neurons DIV 7 were transfected with mRFP-β actin and either empty vector (Control) 2’OM-132, sh-p250GAP, wtKalirin-7, sh-Kalirin-7, or combinations where indicated. On DIV12, the neurons were fixed and imaged. Representative dendrites are shown in A and quantified in B. (C) Hippocampal slices were cultured for 3 days and subjected to biolistic transfection with TFP ± other plasmids as indicated. Slices were allowed to recover for 1 day and then stimulated (where indicated) with 20 µM bicuculline for 2 days. On DIV 6, dendritic protrusions were analyzed for spines and filopodia (± SEM, *** P < 0.001).
Discussion
Activity-dependent regulation of miR132
Activity-regulated gene expression is believed to be a critical regulator of synapse clustering, synaptogenesis, and synaptic plasticity. Although several activity-regulated genes have been found to regulate synaptogenesis, for the most part, the products of these genes have encoded proteins. In this study, we show that transcription of miR132 is tightly associated with periods of synaptogenesis in hippocampal neurons. We also show that miR132 inhibitors inhibit synapse formation while introduction of miR132 into quiescent hippocampal neurons markedly enhances this process. We describe an activity-regulated miRNA pathway that regulates synapse structure and function by inhibiting translation of the GTPase activating protein, p250GAP (Fig. 6). We previously showed that p250GAP is a bona fide miR132 target (Vo et al., 2005; Wayman et al., 2008) and others have shown that this protein is enriched in the post-synaptic density (Nakazawa et al., 2003; Okabe et al., 2003). Moreover, the onset of spine formation in cultured hippocampal neurons closely correlates with an increase in miR132 and a decrease in p250GAP levels. p250GAP interacts with multiple synaptic proteins that are effectors of synaptic plasticity including the NMDA NR2B receptor subunit, the scaffold protein PSD-95 (Nakazawa et al., 2003; Okabe et al., 2003), the NR2B kinase Fyn (Taniguchi et al., 2003), and the signaling intermediate beta-catenin (Okabe et al., 2003). p250GAP activity is also regulated by CaM kinase II phosphorylation and its localization at the post-synaptic density may be regulated by NMDA receptor signaling (Nakazawa et al., 2003). We suggest that down-regulation of p250GAP function is a critical mechanism by which neuronal activity modulates structural plasticity at the synapse.
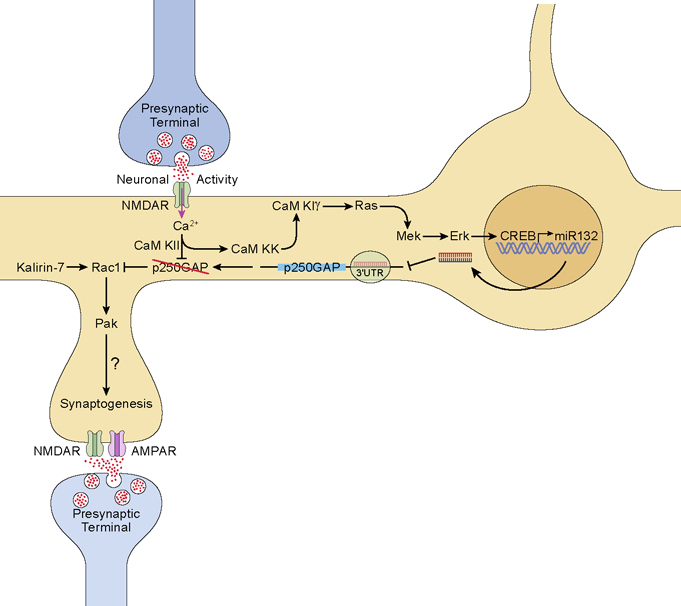
Fig. 6. Regulation of dendritic spine development by miR132.
Increased synaptic activity stimulates glutamate release which, in turn, activates NMDA receptors. Ca2+ influx through the NMDAR activates CaMKII, CaMKK and CaMKIγ. CaMKIγ activates the MEK/Erk pathway to stimulate CREB-dependent synthesis of miR132. miR132 suppresses translation of p250GAP and CaMKII directly phosphorylates and inhibits p250GAP thereby releasing inhibition of Rac1. In the presence of Kalirin-7, Rac1 is activated and subsequently activates Pak to drive local actin polymerization and spine formation.
miR132 regulates activity-dependent spine plasticity via the Rac-Pak pathway
p250GAP is an inhibitor of Rho family GTPases (Moon et al., 2003; Nakayama et al., 2000; Nakazawa et al., 2003; Okabe et al., 2003; Zhao et al., 2003) and cultured cerebellar granule neurons from p250GAP knockout mice show increased Cdc42 activity (Nasu-Nishimura et al., 2006). Rho family GTPases play a critical role in the regulation of spine structure via their ability to regulate actin dynamics (Van Aelst and Cline, 2004). Rho appears to have a negative effect on spine formation and maintenance whereas Rac1 has a positive effect (Nakayama et al., 2000; Pilpel and Segal, 2004; Tashiro et al., 2000; Elia et al., 2006; Fu et al., 2007; Kang et al., 2009; Sfakianos et al., 2007; Zhang and Macara, 2008). The effects of Cdc42 on spine morphogenesis are variable (Cheng et al., 2003; Irie and Yamaguchi, 2002; Scott et al., 2003; Tashiro et al., 2000). We utilized Pak pulldowns to show that, in cultured hippocampal neurons, miR132 expression or p250GAP depletion specifically activates Rac1.
We propose that miR132 causes localized increases in Rac activation by suppressing p250GAP. In accordance with this model, expression of Rac/Pak dominant negative mutants or shRNA-mediated knockdown of Rac1 reduces spine formation triggered by p250GAP knockdown. Likewise, constitutive expression of Rac1 and constitutively active Pak ameliorates the reduction in spine formation triggered by miR132 inhibition, suggesting that the Rac1-Pak pathway is downstream of miR132. Interestingly, other regulators of spine morphogenesis, such as, EphB and Tiam1, also show selectivity for Rac in hippocampal neurons (Tolias et al., 2007). We cannot exclude a role for other downstream effectors of Rac, including Lim-kinase, myosin heavy chain IIb, and Wave (Govek et al., 2005). Nevertheless, our data suggests that Pak, in particular, plays an essential role.
Although multiple studies have shown that neuronal activity stimulates Rac activation, how this occurs has remained obscure. In this study, we show that neuronal activity increases expression of a miRNA that controls Rac1 activity by downregulating translation of a Rac GAP. We propose that this is a major mechanism linking activity to the Rac actin-remodeling pathway. Recently, it has been reported that p250GAP may regulate spine morphology by inhibiting RhoA (Nakayama et al., 2008). Consistent with previous studies, we find that expression of constitutively active RhoA inhibits spine formation while shRNA-mediated RhoA knockdown or expression of dnRhoA increases spine density (Nakayama et al., 2000; Pilpel and Segal, 2004; Tashiro et al., 2000; Zhang and Macara, 2008). It is conceivable that the differential regulation of spine formation seen by Nakazawa et al. reflects their use of embryonic neurons. Furthermore, differences between our data and those of Nakazawa et al with respect to Rac and RhoA regulation by p250GAP may reflect the fact that our GTPase assays were performed in hippocampal neurons while Nakazawa et al. used cortical neurons. Thus, we propose that miR132 regulates spine formation in hippocampal neurons by down regulating p250GAP and activating the Rac1-PAK actin remodeling pathway.
Requirement for Kalirin-7 in miR132 dependent synaptogenesis
Removal of a Rac inhibitor, by itself, would not increase Rac activity unless an active RacGEF was localized at or near p250GAP. The RacGEF, Kalirin-7, is concentrated in dendritic spines of forebrain hippocampal neurons and has a subcellular localization reminiscent of p250GAP (Nakazawa et al., 2003; Penzes et al., 2001). Kalirin-7 is the most abundant kalirin isoform in the adult brain and its temporal expression mirrors miR132 expression within the hippocampus (Ma et al., 2003; Vo et al., 2005) and (Fig. 1A). Kalirin-7 interacts with multiple PDZ domain containing proteins, including PSD-95, SAP-102, AP-97, chapsyn-110 and S-SCAM, which might link Kalirin-7 with the NMDA receptor (Penzes et al., 2001). Inhibiting Kalirin-7 function reduces, and overexpressing Kalirin-7, increases dendritic spine density (Ma et al., 2003; Ma et al., 2008; Penzes et al., 2001; Xie et al., 2007b). Recently, Xie et al proposed that CaM kinase II (CaMKII) modulates dendritic spine morphology by phosphorylating Kalirin-7 but the mechanism is unclear (Xie et al., 2007). We show that Kalirin-7 is essential for spine formation triggered by miR132 expression or p250GAP knockdown. Selectively inhibiting the expression Kalirin-7 blocks both miR132 and sh-p250GAP stimulated spinogenesis. Because Kalirin-7-stimulated spine formation is unaffected by inhibition of miR132 function, we propose that Kalirin-7 is the Rac1GEF responsible for activating Rac1 following miR132-mediated downregulation of p250GAP.
Regulation of structural and functional synaptogenesis by miR132
The CREB transcriptional pathway regulates dendritic growth in cortical neurons (Redmond et al., 2002) and CREB activity is necessary and sufficient for activity-regulated dendritic plasticity (Wayman et al., 2008; Wayman et al., 2006). A positive role for CREB in spine and synapse formation has been suggested previously (Marie et al., 2005), and we show here that inhibition of CREB function using dominant negatives or shRNA blocks spine formation. In this study, we propose that CREB and neuronal activity regulate spine formation in part by activating miR132 expression. Our observation that expression of a miR132 insensitive p250GAP mutant attenuates spine formation supports an essential role for the CREB-miR132 pathway in morphological plasticity.
In cultured hippocampal neurons, the period of maximal synaptogenesis coincided with peak miR132 levels, and 2’O-methyl RNA miR132 inhibitors selectively decreased spine formation, decreased spine head size, and increased filopodia density. This suggests that miR132 may also regulate filopodial growth or the conversion of filopodia to spines. Likewise, depletion of the miR132 target, p250GAP, increased spine formation and spine head size, and introduction of the p250GAP MRE mutant, but not wild-type p250GAP, selectively impaired spine formation. Because a previous study showed that CREB activation potentiates NMDA receptor-driven mEPSCs (Marie et al., 2005), we examined whether miR132 regulates basal synaptic function. Depletion of p250GAP increased, and transfection of miR132 2’O-methyl inhibitors reduced, mEPSC frequency. These changes could result from alterations in the number or size of presynaptic contacts or changes in presynaptic activity.
Afferent tetanization and experience-dependent activity regulate spine formation in the hippocampus and other brain areas (Engert and Bonhoeffer, 1999; Kozorovitskiy et al., 2005; Leuner et al., 2003; Maletic-Savatic et al., 1999; Toni et al., 1999). Nevertheless, it is not entirely clear how activity and spine formation are linked. We show that bicuculline-mediated induction of afferent activity increases spine size and number in the hippocampal slice preparation. Transfection of miR132 markedly increased spine formation, and inhibition of miR132 attenuated spine formation induced by bicuculline. Knockdown of the miR132 target, p250GAP, mimicked activity-dependent spine formation. These experiments suggest that miR132 plays a critical role in spine formation and possibly functional synaptogenesis.
miRNA regulation of gene expression
Several components of the RISC complex are enriched in dendrites, suggesting that miRNAs might repress gene expression in specific cellular compartments. In particular, miR134 has been proposed to directly regulate dendritic translation of its target, LIMK and Pum2 (Fiore et al., 2009; Schratt et al., 2006). Recent studies suggest that formation of functional miRNA-RISC complexes might also be regulated by intracellular signaling (Bhattacharyya et al., 2006). It is conceivable that miRNA function could be regulated by signals limited to the dendritic or synaptic compartment (Ashraf et al., 2006).
Although our data indicate that miR132 regulates morphogenesis by selectively repressing translation of p250GAP, miR132 can also repress MeCP2 (Klein et al., 2007) Mutations in MeCP2 underlie Rett syndrome, an X-linked neurodevelopment disorder which is associated with defective dendritic arborization and spine formation (Armstrong et al., 1995; Belichenko et al., 1994). We have not examined whether miR132 regulation of MeCP2 contributes to the effects on spine development.
miR132 and synaptic plasticity
Our observation that miR132 is an activity-dependent regulator of synapse morphogenesis suggests that it could also contribute to synaptic plasticity in vivo. CREB-dependent transcription has been suggested to play a role in synapse morphogenesis in invertebrate systems and hippocampal neurons (Bartsch et al., 1998; Marie et al., 2005; Martin, 2002; Martin et al., 1997). We also recently showed that miR132 inhibitors negatively regulate light-induced phase shifting, suggesting that the miR132 pathway regulates experience-dependent synaptic responses in the SCN (Cheng et al., 2007). Whether the CREB-miR132 pathway directly regulates use- or experience-dependent synaptic function remains to be determined.
Experimental Methods
Reagents and Plasmids
The following reagents were purchased from the indicated sources: U0126, Calbiochem; STO-609, from Tocris Cookson. Map2B-GFP (Wayman et al., 2006) and caCREB (Cardinaux et al., 2000) plasmids have been described previously. The pCAG-ACREB (Arthur et al., 2004) and pCAG-miR132 (Vo et al., 2005) plasmids were also described previously. The pCAG-EYFP-γ actin plasmid was constructed by replacing GFP in the EGFP-γ actin described in (Fischer et al., 2000). The p250GAP miR132 MRE sequence was amplified from a human cDNA with primers GCCCCGGGAGCAATAGAGTT and TGGGAGGGGAAGGTGGTGAT and cloned into the XbaI site of pRL-TK (Promega). The p250GAP mutant was generated by PCR-based mutagenesis of GFP-tagged p250GAP (Nakazawa et al., 2003) with the following primers: GGTTATTGAAAAAAATAGAAGTCCACTGTCCAGCAGAGG and CTC TGCTGGACAGTGGACTTCTATTTTTTTCAATAACC. Wt-Kalirin-7 and sh-Kalirin7 were gifts from the labs of R. E. Mains and B. A. Eipper and were previously described (Ma et al., 2008). sh-Rac1, sh-Rac3 and sh-RhoA were described by Hajdo-Milasinovic et al. (2007) The sequences of the 2’-O-methyl oligoribonucletodies (IDT) are: antisense, GGGCGACCAUGGCUGUAGACUGUUACUGUGG; sense, UCCAUUGUCAGAUGUCGGUACCAGCGGGGCG.
Cell Culture
Hippocampal neurons (2×105 cells per square centimeter) were cultured from P1–2 Sprague Dawley rats on plates coated with poly-L-lysine (Sigma; molecular weight 300,000) as described previously (Wayman et al., 2006). Hippocampal neurons were maintained in Neurobasal A media (Invitrogen) supplemented with B27 (Invitrogen), 0.5 mM L-glutamine, and 5 µM cytosine- D-arabinofuranoside (Sigma; added at 2 DIV). Hippocampal neurons were then cultured a further 3–7 days, at which time they were either transfected or treated with various pharmacological reagents as specified in the text or figure legends.
Transfection
Primary hippocampal neurons were transfected with LipofectAMINE 2000 (Invitrogen) according to the manufacturer’s protocols. In each experiment, we optimized DNA amounts, transfection reagent amounts, and transfection duration to minimize toxicity and maximize transfection efficiency. None of the transfections or drug treatments caused apoptosis as assessed by Hoechst staining. miR132 and sh-p250GAP expression vectors were electroporated into P1 hippocampal neurons as described in the rat hippocampal neuron Amaxa nucleofection protocol. Lipofectamine 2000 transfection efficiency was 0.5–5% and Amaxa electroporation efficiency was 60–75%
Slice Culture and Transfection
Organotypic hippocampal slices from P4 Sprague-Dawley rats were cultured for 3 days as described previously (Wayman et al., 2006). To visualize spine morphology, we transfected slices with pCAG-TFP using a Helios Gene Gun (BioRad), according to the manufacturer’s protocol. Following transfection, slices were allowed to recover for 24 hr before stimulation with 20 µm bicuculline-methchloride (Tocris) for 2 days. Slices were fixed, mounted, and imaged using a confocal microscope. Dendritic spine density and morphology were measured as described below.
Reverse Transcription
Neurons were treated as described, and total RNA was isolated using Trizol (Invitrogen) according to manufacturer’s instructions. RNA (50 ng to 1 µg) was reverse transcribed with Moloney murine leukemia virus reverse transcriptase (Invitrogen) and 50–250 ng random primers (Invitrogen).
Quantitative real-time PCR
PCRs (10 µl) contained 1 µl 10× PCR buffer (Invitrogen), 2.5 mM MgCl2, 200 mM dNTP (Roche), 0.125–0.25 mM primer (IDT), 1× SYBR green I (Invitrogen), and 1 U platinum Taq (Invitrogen). All RT PCR was run on an Opticon OP346 (MJ Research) with an initial 35 s 95°C denaturation step followed by 30–50 cycles at 94°C for 15 s and 68°C for 40 s. Real-time PCR was analyzed using the linear standard curve method. All standard curves had an R2 of at least .995, were composed of a minimum of 5 points, and were linear for at least 3 orders of magnitude. To avoid plateau effects, the Ct was always positioned in the logarithmic component of the sigmoid fluorescence curve. The Ct was selected solely based on the maximal linearity of standard curve. Taqman reverse transcription and real time PCR for mature miR132 was conducted according to the manufacturer’s instructions (Applied Biosystems). RT-PCR data was normalized to GAPDH cDNA levels also detected by real-time PCR (normalization against 18s RNA showed similar results). All RT-PCR data showed at least 100-fold higher levels of product than no reverse transcriptase controls. The following primers were used: miR132-precursor-1 CCTCCGGTTCCCACAGTAACAA, miR132-precursor-2 CCGCGTCTCCAGGGCAAC, GAPDH-1 AGTGCCAGCCTCGTCCCGTAG, GAPDH-2 CCAAATCCGTTCACACCGACCTT.
Quantification of spine density and morphology
High density hippocampal neurons were transfected with either EYFP-γactin or mRFP-βactin ± test plasmids or oligos. Expression of fluorescently tagged actin allows visualization of dendritic spine density and size. Expression of low levels of either γactin or βactin has no significant effect on either spine density or size. Neurons were transfected on DIV 6–7 then fixed (4% paraformaldehyde, 3% sucrose, 60 mM PIPES, 25 mM HEPES, 5 mM EGTA, 1 mM MgCl2, pH7.4) on DIV12 for 20 min at room temperature. Fluorescent images were acquired using a cooled CCD camera (Hamamatsu Photonics) attached to a Zeiss Axioplan2 (Carl Zeiss) inverted microscope with a 63× oil immersion lens. Morphometric measurements were performed using Openlab software (Improvision). Dendritic spine density was measured on primary and secondary dendrites at a distance of at least 150 µm form the soma. At least 3×50 µm of dendritic length from each of at least 25–35 neurons were analyzed for each data point reported. Each experiment was repeated at least three times using independent preparations.
Immunocytochemistry
Hippocampal neurons were fixed in 4% paraformaldehyde, 4% sucrose, phosphate-buffered saline (PBS), and 50 mM HEPES pH 7.5 at 37 °C for 15 min. The cells were rinsed 3 times for 5 min in PBS, blocked for 1 hr in blocking buffer (PBS containing 10% BSA). The cells were then stained for surface-expressed GluR1 using a rabbit anti N-terminal anti-GluR1 antibody (Calbiochem) in blocking buffer overnight at room temperature and washed 3 times for 5 min in blocking buffer. For synapsin-I staining, cells were permeabilized with 1% BSA solution in PBS containing 0.2% Triton X-100. Cells were then stained with antisynapsin I (rabbit polyclonal; 1:1,000; Chemicon International). Coverslips were mounted on glass slides and analyzed by fluorescence microscopy as described above.
Pulldown assays
DIV 5 neurons were transfected with either miR132 or sh-p250GAP and either myc-Rac or myc-Cdc42. After two days in culture, the neurons were rapidly harvested with ice-cold lysis buffer containing 1% NP-40, 0.5% CHAPS, 25mM Tris-Cl pH 7.5, 125mM NaCl, 25mM MgCl2, 25mM NaF, 1mM β-glycerol phosphate, plus protease inhibitors (leupeptin, aprotinin, benzamidine, pepstatin-A and antipain). The lysates were centrifuged at 20,000g for 10 minutes to clear insoluble material. Cleared lysates were applied to pre-loaded glutathione sepharose beads containing 20ug of GST fusion protein of the CRIB domain of PAK1(GST-PAK-PBD) or RhoA binding domain of Rhotekin and were incubated with rocking for 1 hour at 4°C. Resin was washed 3× with lysis buffer and extracted with 2× SDS sample buffer. Rac1 and Cdc42 bound to PAK-PBD were detected by Western blotting with Anti-myc monoclonal antibody. RhoA bound to GST-Rhotekin were detected by Western blotting with anti-HA monoclonal antibody.
Electrophysiology
Whole-cell voltage clamp recordings were performed on cultured hippocampal neurons as described above using an Axopatch-200b amplifier (Molecular Devices, Sunnyvale, CA). Cells were continuously perfused (1 ml min−1) with an external buffer solution that contained (in mM): 140 NaCl, 2.5 KCl, 3 CaCl, 1 MgCl2, 25 Hepes and 33 glucose; pH was adjusted to 7.3 using NaOH. Osmolarity was adjusted to 310 mosmol l−1 by the addition of NaCl. For isolating miniature excitatory postsynaptic currents (mEPSCs), picrotoxin (50 µM), strychnine (1 µM), and tetrodotoxin (0.5 µM) were added to the external buffer to block GABAA receptor, glycine receptor, and Na channel activity, respectively.
Patch electrodes were pulled from thin-walled borosilicate glass capillaries (tip resistance ranged from 4 – 6 MΩ) and filled with internal buffer solution that contained (in mM): 140 potassium gluconate, 10 KCl, 0.5 EGTA , and 11 Hepes; pH was adjusted to 7.2 using NaOH. All experiments were carried out at room temperature (25 °C). Whole-cell recordings were only established after a high-resistance seal (>2 GΩ) was achieved. Only cells that had an input resistance of >200 MΩ and resting membrane potentials <−50mV were considered for experiments. Resting membrane potentials were measured immediately upon breaking into whole-cell mode by setting the current to 0 pA. Cells were then voltage clamped at a holding potential of −65 mV for the duration of the experiment. Access resistance (Ra) typically ranged between 10 and 15 MΩ and was compensated by ≥60% and monitored at the beginning and end of each experiment with small voltage pulses. Cells were rejected from analysis if Ra increased by more than 15%, or if the input resistance fell below 200 MΩ.
Statistical Analyses
Raw data approximated normality (Shapiro-Wilk’s test) and the assumption of equal variances (Levene’s test). Analysis of variance and Tukey’s post test were used to test the null hypothesis.
Supplementary Material
Basal miR132 levels are low in organotypic hippocampal slice cultures compared to DIV12 disassociated hippocampal cultures. RNA was isolated from either DIV 4 organotypic hippocampal slice cultures (SC) or DIV 12 disassociated hippocampal cultures (DC), reversed-transcribed, and analyzed by Taqman-real time PCR with miR132 cDNA primers. The data was normalized to GAPDH cDNA levels and presented (± SEM).
Bicuculline increase synaptic activity in organotypic hippocampal slice cultures. Representative voltage record of spontaneous postsynaptic potentials demonstrating the increase in cell excitability following the application of 20 µM bicuculline. Bottom, expanded voltage record illustrating that bicuculline (BIC) induced both single and short bursts of action potentials.
Transfection of si-p250GAP inhibits the expression of p250GAP in hippocampal neurons. Hippocampal neurons were transfected by electroporation on the day of plating. On DIV3 the expression of endogenous p250GAP was analyzed by Western blot. (A) Representative Western blots. (B) Quantitation of p250GAP expression from three independent experiments, normalized to empty vector transfection control and Erk2 (± SEM).
RhoA inhibits spinogenesis. (A–D) Hippocampal neurons were transfected on DIV 7 with mRFP-β actin, ±empty vector (Control) ± caRho(A) ± dnRhoA, ± shRhoA ± sh-p250GAP. On DIV12, the neurons were fixed and imaged. Analysis of the effects of the indicated transfections on protrusion density (B and D), and spine head width (C) indicate that RhoA inhibits spinogenesis. Statistical analyses utilized ANOVA and Tukey’s post-test. (± SEM, *** P < 0.001).
miR132 and sh-p250GAP increase endogenous Rac activity. Hippocampal neurons were transfected by electroporation on the day of plating with Myc-Rac1 ± empty vector (control) ± miR132 ± sh-p250GAP. On DIV 3 Rac activity was assayed by Rac pull down assay. (A) Representative Western blots. (B) Quantitation of Rac activity from duplicate samples in three independent experiments. (± SEM, * P < 0.05, ** P < 0.01).
Transfection of shRNA constructs targeting Rac1, Rac3 or RhoA inhibits the expression of their respective targets in hippocampal neurons. Hippocampal neurons were transfected on DIV7 with Myc-Rac1, Myc-Rac3 or HA-RhoA ± sh-Rac1, ± sh-Rac3 ± sh-RhoA or a scrambled control. On DIV8 the expression of Myc-Rac1 and Myc-Rac3 was analyzed by Western blot. Representative Western blots of the knockdown and Erk2 as a loading control are shown.
Acknowledgments
We thank Jami Dwyer, Amir Bashar and Kay Shi for technical assistance and Gail Mandel for comments and suggestions. This work was supported by grants from the NIH and the Hope for Depression Research Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Armstrong D, Dunn JK, Antalffy B, Trivedi R. Selective dendritic alterations in the cortex of Rett syndrome. J Neuropathol Exp Neurol. 1995;54:195–201. doi: 10.1097/00005072-199503000-00006. [DOI] [PubMed] [Google Scholar]
- Arthur JS, Fong AL, Dwyer JM, Davare M, Reese E, Obrietan K, Impey S. Mitogen- and stress-activated protein kinase 1 mediates cAMP response element-binding protein phosphorylation and activation by neurotrophins. J Neurosci. 2004;24:4324–4332. doi: 10.1523/JNEUROSCI.5227-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashraf SI, McLoon AL, Sclarsic SM, Kunes S. Synaptic protein synthesis associated with memory is regulated by the RISC pathway in Drosophila. Cell. 2006;124:191–205. doi: 10.1016/j.cell.2005.12.017. [DOI] [PubMed] [Google Scholar]
- Bahr BA. Long-term hippocampal slices: a model system for investigating synaptic mechanisms and pathologic processes. J Neurosci Res. 1995;42:294–305. doi: 10.1002/jnr.490420303. [DOI] [PubMed] [Google Scholar]
- Bartsch D, Casadio A, Karl KA, Serodio P, Kandel ER. CREB1 encodes a nuclear activator, a repressor, and a cytoplasmic modulator that form a regulatory unit critical for long-term facilitation. Cell. 1998;95:211–223. doi: 10.1016/s0092-8674(00)81752-3. [DOI] [PubMed] [Google Scholar]
- Belichenko PV, Oldfors A, Hagberg B, Dahlstrom A. Rett syndrome: 3-D confocal microscopy of cortical pyramidal dendrites and afferents. Neuroreport. 1994;5:1509–1513. [PubMed] [Google Scholar]
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- Caeser M, Aertsen A. Morphological organization of rat hippocampal slice cultures. J Comp Neurol. 1991;307:87–106. doi: 10.1002/cne.903070109. [DOI] [PubMed] [Google Scholar]
- Cardinaux JR, Notis JC, Zhang Q, Vo N, Craig JC, Fass DM, Brennan RG, Goodman RH. Recruitment of CREB binding protein is sufficient for CREB-mediated gene activation. Mol Cell Biol. 2000;20:1546–1552. doi: 10.1128/mcb.20.5.1546-1552.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CM, Mervis RF, Niu SL, Salem N, Jr, Witters LA, Tseng V, Reinhardt R, Bondy CA. Insulin-like growth factor 1 is essential for normal dendritic growth. J Neurosci Res. 2003;73:1–9. doi: 10.1002/jnr.10634. [DOI] [PubMed] [Google Scholar]
- Cheng HY, Papp JW, Varlamova O, Dziema H, Russell B, Curfman JP, Nakazawa T, Shimizu K, Okamura H, Impey S, Obrietan K. microRNA modulation of circadian-clock period and entrainment. Neuron. 2007;54:813–829. doi: 10.1016/j.neuron.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du T, Zamore PD. microPrimer: the biogenesis and function of microRNA. Development. 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- Elia LP, Yamamoto M, Zang K, Reichardt LF. p120 catenin regulates dendritic spine and synapse development through Rho-family GTPases and cadherins. Neuron. 2006;51:43–56. doi: 10.1016/j.neuron.2006.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engert F, Bonhoeffer T. Dendritic spine changes associated with hippocampal long-term synaptic plasticity. Nature. 1999;399:66–70. doi: 10.1038/19978. [DOI] [PubMed] [Google Scholar]
- Fiala JC, Spacek J, Harris KM. Dendritic spine pathology: cause or consequence of neurological disorders? Brain Res Brain Res Rev. 2002;39:29–54. doi: 10.1016/s0165-0173(02)00158-3. [DOI] [PubMed] [Google Scholar]
- Fiore R, Khudayberdiev S, Christensen M, Siegel G, Flavell SW, Kim TK, Greenberg ME, Schratt G. Mef2-mediated transcription of the miR379-410 cluster regulates activity-dependent dendritogenesis by fine-tuning Pumilio2 protein levels. EMBO J. 2009 doi: 10.1038/emboj.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M, Kaech S, Wagner U, Brinkhaus H, Matus A. Glutamate receptors regulate actin-based plasticity in dendritic spines. Nat Neurosci. 2000;3:887–894. doi: 10.1038/78791. [DOI] [PubMed] [Google Scholar]
- Fu WY, Chen Y, Sahin M, Zhao XS, Shi L, Bikoff JB, Lai KO, Yung WH, Fu AK, Greenberg ME, Ip NY. Cdk5 regulates EphA4-mediated dendritic spine retraction through an ephexin1-dependent mechanism. Nat Neurosci. 2007;10:67–76. doi: 10.1038/nn1811. [DOI] [PubMed] [Google Scholar]
- Govek EE, Newey SE, Van Aelst L. The role of the Rho GTPases in neuronal development. Genes Dev. 2005;19:1–49. doi: 10.1101/gad.1256405. [DOI] [PubMed] [Google Scholar]
- Hajdo-Milasinovic A, Ellenbroek SI, van Es S, van der Vaart B, Collard JG. Rac1 and Rac3 have opposing functions in cell adhesion and differentiation of neuronal cells. J Cell Sci. 2007;120:555–566. doi: 10.1242/jcs.03364. [DOI] [PubMed] [Google Scholar]
- Irie F, Yamaguchi Y. EphB receptors regulate dendritic spine development via intersectin, Cdc42 and N-WASP. Nat Neurosci. 2002;5:1117–1118. doi: 10.1038/nn964. [DOI] [PubMed] [Google Scholar]
- Kang MG, Guo Y, Huganir RL. AMPA receptor and GEF-H1/Lfc complex regulates dendritic spine development through RhoA signaling cascade. Proc Natl Acad Sci U S A. 2009;106:3549–3554. doi: 10.1073/pnas.0812861106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz LC, Shatz CJ. Synaptic activity and the construction of cortical circuits. Science. 1996;274:1133–1138. doi: 10.1126/science.274.5290.1133. [DOI] [PubMed] [Google Scholar]
- Klein ME, Lioy DT, Ma L, Impey S, Mandel G, Goodman RH. Homeostatic regulation of MeCP2 expression by a CREB-induced microRNA. Nat Neurosci. 2007;10:1513–1514. doi: 10.1038/nn2010. [DOI] [PubMed] [Google Scholar]
- Kozorovitskiy Y, Gross CG, Kopil C, Battaglia L, McBreen M, Stranahan AM, Gould E. Experience induces structural and biochemical changes in the adult primate brain. Proc Natl Acad Sci U S A. 2005;102:17478–17482. doi: 10.1073/pnas.0508817102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuner B, Falduto J, Shors TJ. Associative memory formation increases the observation of dendritic spines in the hippocampus. J Neurosci. 2003;23:659–665. doi: 10.1523/JNEUROSCI.23-02-00659.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XM, Huang J, Wang Y, Eipper BA, Mains RE. Kalirin, a multifunctional Rho guanine nucleotide exchange factor, is necessary for maintenance of hippocampal pyramidal neuron dendrites and dendritic spines. J Neurosci. 2003;23:10593–10603. doi: 10.1523/JNEUROSCI.23-33-10593.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XM, Wang Y, Ferraro F, Mains RE, Eipper BA. Kalirin-7 is an essential component of both shaft and spine excitatory synapses in hippocampal interneurons. J Neurosci. 2008;28:711–724. doi: 10.1523/JNEUROSCI.5283-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maletic-Savatic M, Malinow R, Svoboda K. Rapid dendritic morphogenesis in CA1 hippocampal dendrites induced by synaptic activity. Science. 1999;283:1923–1927. doi: 10.1126/science.283.5409.1923. [DOI] [PubMed] [Google Scholar]
- Marie H, Morishita W, Yu X, Calakos N, Malenka RC. Generation of silent synapses by acute in vivo expression of CaMKIV and CREB. Neuron. 2005;45:741–752. doi: 10.1016/j.neuron.2005.01.039. [DOI] [PubMed] [Google Scholar]
- Martin KC. Synaptic tagging during synapse-specific long-term facilitation of Aplysia sensory-motor neurons. Neurobiol Learn Mem. 2002;78:489–497. doi: 10.1006/nlme.2002.4088. [DOI] [PubMed] [Google Scholar]
- Martin KC, Casadio A, Zhu H, Yaping E, Rose JC, Chen M, Bailey CH, Kandel ER. Synapse-specific, long-term facilitation of aplysia sensory to motor synapses: a function for local protein synthesis in memory storage. Cell. 1997;91:927–938. doi: 10.1016/s0092-8674(00)80484-5. [DOI] [PubMed] [Google Scholar]
- Moon SY, Zang H, Zheng Y. Characterization of a brain-specific Rho GTPase-activating protein, p200RhoGAP. J Biol Chem. 2003;278:4151–4159. doi: 10.1074/jbc.M207789200. [DOI] [PubMed] [Google Scholar]
- Nakayama AY, Harms MB, Luo L. Small GTPases Rac and Rho in the maintenance of dendritic spines and branches in hippocampal pyramidal neurons. J Neurosci. 2000;20:5329–5338. doi: 10.1523/JNEUROSCI.20-14-05329.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama M, Goto TM, Sugimoto M, Nishimura T, Shinagawa T, Ohno S, Amano M, Kaibuchi K. Rho-kinase phosphorylates PAR-3 and disrupts PAR complex formation. Dev Cell. 2008;14:205–215. doi: 10.1016/j.devcel.2007.11.021. [DOI] [PubMed] [Google Scholar]
- Nakazawa T, Kuriu T, Tezuka T, Umemori H, Okabe S, Yamamoto T. Regulation of dendritic spine morphology by an NMDA receptor-associated Rho GTPase-activating protein, p250GAP. J Neurochem. 2008;105:1384–1393. doi: 10.1111/j.1471-4159.2008.05335.x. [DOI] [PubMed] [Google Scholar]
- Nakazawa T, Watabe AM, Tezuka T, Yoshida Y, Yokoyama K, Umemori H, Inoue A, Okabe S, Manabe T, Yamamoto T. p250GAP, a novel brain-enriched GTPase-activating protein for Rho family GTPases, is involved in the N-methyl-d-aspartate receptor signaling. Mol Biol Cell. 2003;14:2921–2934. doi: 10.1091/mbc.E02-09-0623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasu-Nishimura Y, Hayashi T, Ohishi T, Okabe T, Ohwada S, Hasegawa Y, Senda T, Toyoshima C, Nakamura T, Akiyama T. Role of the Rho GTPase-activating protein RICS in neurite outgrowth. Genes Cells. 2006;11:607–614. doi: 10.1111/j.1365-2443.2006.00966.x. [DOI] [PubMed] [Google Scholar]
- Nimchinsky EA, Sabatini BL, Svoboda K. Structure and function of dendritic spines. Annu Rev Physiol. 2002;64:313–353. doi: 10.1146/annurev.physiol.64.081501.160008. [DOI] [PubMed] [Google Scholar]
- Okabe T, Nakamura T, Nishimura YN, Kohu K, Ohwada S, Morishita Y, Akiyama T. RICS, a novel GTPase-activating protein for Cdc42 and Rac1, is involved in the beta-catenin-N-cadherin and N-methyl-D-aspartate receptor signaling. J Biol Chem. 2003;278:9920–9927. doi: 10.1074/jbc.M208872200. [DOI] [PubMed] [Google Scholar]
- Penzes P, Johnson RC, Sattler R, Zhang X, Huganir RL, Kambampati V, Mains RE, Eipper BA. The neuronal Rho-GEF Kalirin-7 interacts with PDZ domain-containing proteins and regulates dendritic morphogenesis. Neuron. 2001;29:229–242. doi: 10.1016/s0896-6273(01)00193-3. [DOI] [PubMed] [Google Scholar]
- Pilpel Y, Segal M. Activation of PKC induces rapid morphological plasticity in dendrites of hippocampal neurons via Rac and Rho-dependent mechanisms. Eur J Neurosci. 2004;19:3151–3164. doi: 10.1111/j.0953-816X.2004.03380.x. [DOI] [PubMed] [Google Scholar]
- Pokorny J, Yamamoto T. Postnatal ontogenesis of hippocampal CA1 area in rats. I. Development of dendritic arborisation in pyramidal neurons. Brain Res Bull. 1981;7:113–120. doi: 10.1016/0361-9230(81)90075-7. [DOI] [PubMed] [Google Scholar]
- Redmond L, Kashani AH, Ghosh A. Calcium regulation of dendritic growth via CaM kinase IV and CREB-mediated transcription. Neuron. 2002;34:999–1010. doi: 10.1016/s0896-6273(02)00737-7. [DOI] [PubMed] [Google Scholar]
- Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- Schwamborn JC, Puschel AW. The sequential activity of the GTPases Rap1B and Cdc42 determines neuronal polarity. Nat Neurosci. 2004;7:923–929. doi: 10.1038/nn1295. [DOI] [PubMed] [Google Scholar]
- Scott EK, Reuter JE, Luo L. Small GTPase Cdc42 is required for multiple aspects of dendritic morphogenesis. J Neurosci. 2003;23:3118–3123. doi: 10.1523/JNEUROSCI.23-08-03118.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfakianos MK, Eisman A, Gourley SL, Bradley WD, Scheetz AJ, Settleman J, Taylor JR, Greer CA, Williamson A, Koleske AJ. Inhibition of Rho via Arg and p190RhoGAP in the postnatal mouse hippocampus regulates dendritic spine maturation, synapse and dendrite stability, and behavior. J Neurosci. 2007;27:10982–10992. doi: 10.1523/JNEUROSCI.0793-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward O, Falk PM. Selective localization of polyribosomes beneath developing synapses: a quantitative analysis of the relationships between polyribosomes and developing synapses in the hippocampus and dentate gyrus. J Comp Neurol. 1991;314:545–557. doi: 10.1002/cne.903140311. [DOI] [PubMed] [Google Scholar]
- Taniguchi S, Liu H, Nakazawa T, Yokoyama K, Tezuka T, Yamamoto T. p250GAP, a neural RhoGAP protein, is associated with and phosphorylated by Fyn. Biochem Biophys Res Commun. 2003;306:151–155. doi: 10.1016/s0006-291x(03)00923-9. [DOI] [PubMed] [Google Scholar]
- Tashiro A, Minden A, Yuste R. Regulation of dendritic spine morphology by the rho family of small GTPases: antagonistic roles of Rac and Rho. Cereb Cortex. 2000;10:927–938. doi: 10.1093/cercor/10.10.927. [DOI] [PubMed] [Google Scholar]
- Tolias KF, Bikoff JB, Kane CG, Tolias CS, Hu L, Greenberg ME. The Rac1 guanine nucleotide exchange factor Tiam1 mediates EphB receptor-dependent dendritic spine development. Proc Natl Acad Sci U S A. 2007;104:7265–7270. doi: 10.1073/pnas.0702044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toni N, Buchs PA, Nikonenko I, Bron CR, Muller D. LTP promotes formation of multiple spine synapses between a single axon terminal and a dendrite. Nature. 1999;402:421–425. doi: 10.1038/46574. [DOI] [PubMed] [Google Scholar]
- Van Aelst L, Cline HT. Rho GTPases and activity-dependent dendrite development. Curr Opin Neurobiol. 2004;14:297–304. doi: 10.1016/j.conb.2004.05.012. [DOI] [PubMed] [Google Scholar]
- Vo N, Klein ME, Varlamova O, Keller DM, Yamamoto T, Goodman RH, Impey S. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc Natl Acad Sci U S A. 2005;102:16426–16431. doi: 10.1073/pnas.0508448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayman GA, Davare M, Ando H, Fortin D, Varlamova O, Cheng HY, Marks D, Obrietan K, Soderling TR, Goodman RH, Impey S. An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proc Natl Acad Sci U S A. 2008;105:9093–9098. doi: 10.1073/pnas.0803072105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayman GA, Impey S, Marks D, Saneyoshi T, Grant WF, Derkach V, Soderling TR. Activity-dependent dendritic arborization mediated by CaM-kinase I activation and enhanced CREB-dependent transcription of Wnt-2. Neuron. 2006;50:897–909. doi: 10.1016/j.neuron.2006.05.008. [DOI] [PubMed] [Google Scholar]
- Wong RO, Ghosh A. Activity-dependent regulation of dendritic growth and patterning. Nat Rev Neurosci. 2002;3:803–812. doi: 10.1038/nrn941. [DOI] [PubMed] [Google Scholar]
- Xie Y, Vessey JP, Konecna A, Dahm R, Macchi P, Kiebler MA. The GTP-binding protein Septin 7 is critical for dendrite branching and dendritic-spine morphology. Curr Biol. 2007a;17:1746–1751. doi: 10.1016/j.cub.2007.08.042. [DOI] [PubMed] [Google Scholar]
- Xie Z, Srivastava DP, Photowala H, Kai L, Cahill ME, Woolfrey KM, Shum CY, Surmeier DJ, Penzes P. Kalirin-7 controls activity-dependent structural and functional plasticity of dendritic spines. Neuron. 2007b;56:640–656. doi: 10.1016/j.neuron.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuste R, Bonhoeffer T. Morphological changes in dendritic spines associated with long-term synaptic plasticity. Annu Rev Neurosci. 2001;24:1071–1089. doi: 10.1146/annurev.neuro.24.1.1071. [DOI] [PubMed] [Google Scholar]
- Yuste R, Bonhoeffer T. Genesis of dendritic spines: insights from ultrastructural and imaging studies. Nat Rev Neurosci. 2004;5:24–34. doi: 10.1038/nrn1300. [DOI] [PubMed] [Google Scholar]
- Zhang H, Macara IG. The PAR-6 polarity protein regulates dendritic spine morphogenesis through p190 RhoGAP and the Rho GTPase. Dev Cell. 2008;14:216–226. doi: 10.1016/j.devcel.2007.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Ma H, Bossy-Wetzel E, Lipton SA, Zhang Z, Feng GS. GC-GAP, a Rho family GTPase-activating protein that interacts with signaling adapters Gab1 and Gab2. J Biol Chem. 2003;278:34641–34653. doi: 10.1074/jbc.M304594200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Basal miR132 levels are low in organotypic hippocampal slice cultures compared to DIV12 disassociated hippocampal cultures. RNA was isolated from either DIV 4 organotypic hippocampal slice cultures (SC) or DIV 12 disassociated hippocampal cultures (DC), reversed-transcribed, and analyzed by Taqman-real time PCR with miR132 cDNA primers. The data was normalized to GAPDH cDNA levels and presented (± SEM).
Bicuculline increase synaptic activity in organotypic hippocampal slice cultures. Representative voltage record of spontaneous postsynaptic potentials demonstrating the increase in cell excitability following the application of 20 µM bicuculline. Bottom, expanded voltage record illustrating that bicuculline (BIC) induced both single and short bursts of action potentials.
Transfection of si-p250GAP inhibits the expression of p250GAP in hippocampal neurons. Hippocampal neurons were transfected by electroporation on the day of plating. On DIV3 the expression of endogenous p250GAP was analyzed by Western blot. (A) Representative Western blots. (B) Quantitation of p250GAP expression from three independent experiments, normalized to empty vector transfection control and Erk2 (± SEM).
RhoA inhibits spinogenesis. (A–D) Hippocampal neurons were transfected on DIV 7 with mRFP-β actin, ±empty vector (Control) ± caRho(A) ± dnRhoA, ± shRhoA ± sh-p250GAP. On DIV12, the neurons were fixed and imaged. Analysis of the effects of the indicated transfections on protrusion density (B and D), and spine head width (C) indicate that RhoA inhibits spinogenesis. Statistical analyses utilized ANOVA and Tukey’s post-test. (± SEM, *** P < 0.001).
miR132 and sh-p250GAP increase endogenous Rac activity. Hippocampal neurons were transfected by electroporation on the day of plating with Myc-Rac1 ± empty vector (control) ± miR132 ± sh-p250GAP. On DIV 3 Rac activity was assayed by Rac pull down assay. (A) Representative Western blots. (B) Quantitation of Rac activity from duplicate samples in three independent experiments. (± SEM, * P < 0.05, ** P < 0.01).
Transfection of shRNA constructs targeting Rac1, Rac3 or RhoA inhibits the expression of their respective targets in hippocampal neurons. Hippocampal neurons were transfected on DIV7 with Myc-Rac1, Myc-Rac3 or HA-RhoA ± sh-Rac1, ± sh-Rac3 ± sh-RhoA or a scrambled control. On DIV8 the expression of Myc-Rac1 and Myc-Rac3 was analyzed by Western blot. Representative Western blots of the knockdown and Erk2 as a loading control are shown.