Abstract
TGFβ and BMP receptor kinases activate Smad transcription factors by C-terminal phosphorylation. We have identified a subsequent agonist-induced phosphorylation that plays a central dual role in Smad transcriptional activation and turnover. As receptor-activated Smads form transcriptional complexes, they are phosphorylated at an interdomain linker region by CDK8 and CDK9, which are components of transcriptional mediator and elongation complexes. These phosphorylations promote Smad transcriptional action, which in the case of Smad1, is mediated by the recruitment of YAP to the phosphorylated linker sites. An effector of the highly conserved Hippo organ size control pathway, YAP supports Smad1-dependent transcription and is required for BMP suppression of neural differentiation of mouse embryonic stem cells. The phosphorylated linker is ultimately recognized by specific ubiquitin ligases, leading to proteasome-mediated turnover of activated Smad proteins. Thus, nuclear CDK8/9 drive a cycle of Smad utilization and disposal that is an integral part of canonical BMP and TGFβ pathways.
Introduction
The transforming growth factor β (TGFβ) family of cytokines are key regulators of metazoan embryo development and adult tissue homeostasis. In the canonical pathway ligands of both the TGFβ and the BMP (bone morphogenetic protein) branches of this family, bind to heteromeric serine/threonine kinase receptor complexes, which in turn phosphorylate Smad transcription factors at their C-terminal tail. This phosphorylation induces Smads 1, 5 and 8 in the BMP pathway and Smads 2 and 3 in the TGFβ pathway to accumulate in the nucleus and assemble transcriptional complexes that regulate hundreds of target genes (Feng and Derynck, 2005; Massagué, 1998).
The TGFβ and BMP pathways are intensely regulated by inputs that adjust pathway activity according to contextual status. Antagonists such as FGF and EGF, and cell stress signals act through mitogen-activated protein kinases (MAPKs), to cause phosphorylation of a region that links the DNA-binding and transcriptional domains of the Smads (Aubin et al., 2004; Grimm and Gurdon, 2002; Kretzschmar et al., 1997; Pera et al., 2003). The Smad linker is also phosphorylated by G1 cyclin-dependent kinases during the cell cycle (Matsuura et al., 2004) and by GSK3β complementing MAPK action (Fuentealba et al., 2007; Sapkota et al., 2007). Linker phosphorylation of Smads in the basal state leads to their cytoplasmic retention and ubiquitin ligase-driven, proteasomal degradation (Gao et al., 2009; Sapkota et al., 2007), with an attendant decrease in the responsiveness of cells to BMP and TGFβ signals (Grimm and Gurdon, 2002; Kretzschmar et al., 1997; Kretzschmar et al., 1999; Pera et al., 2003).
Smad linker phosphorylation by antagonists provides a critical counterbalance to TGFβ and BMP signaling. This has led to postulates that in the canonical pathways C-tail phosphorylation activates Smad signaling and linker-phosphorylation inhibits it (Fuentealba et al., 2007; Sapkota et al., 2007). However, this dichotomy is not so tidy. Our present investigation of the BMP-induced Smad1 linker phosphorylation we had reported previously (Sapkota et al., 2007), reveals unexpected new facets of the canonical TGFβ and BMP pathways. Unlike linker phosphorylation by antagonistic signals, which is cytoplasmic and MAPK-mediated, agonist-induced linker phosphorylation (abbreviated from here on as ALP) occurs during or directly prior to the assembly of Smad proteins into transcriptional complexes and is mediated by CDK8 and CDK9. CDK8 is part of Mediator, a multi-subunit complex that couples transcription factors to RNA polymerase II (RNAP II) (Malik and Roeder, 2000). CDK8 phosphorylates the C-terminal domain (CTD) of RNAP II and certain enhancer-binding transcription factors (Donner et al., 2007; Firestein et al., 2008; Morris et al., 2008). CDK9 phosphorylates the RNAP II CTD at distinct sites to enhance transcriptional elongation (Durand et al., 2005; Komarnitsky et al., 2000).
The present work further reveals that the CDK8/9 mediated Smad ALP results in full activation of Smad-dependent transcription, while at the same time priming Smad proteins for eventual degradation. We show that ALP activation of Smad1 involves YAP (Yes-associated protein, also known as YAP1 or YAP65), the end target of the Hippo pathway (Huang et al., 2005), which mediates cell-contact growth inhibition, organ size control, and tumor suppression (Dong et al., 2007; Overholtzer et al., 2006; Zhao et al., 2007). Thus the present findings reveal a dual role for ALP and shed light on previously unrecognized events of the canonical BMP and TGFβ pathways.
Results
Agonist-induced Smad linker phosphorylation
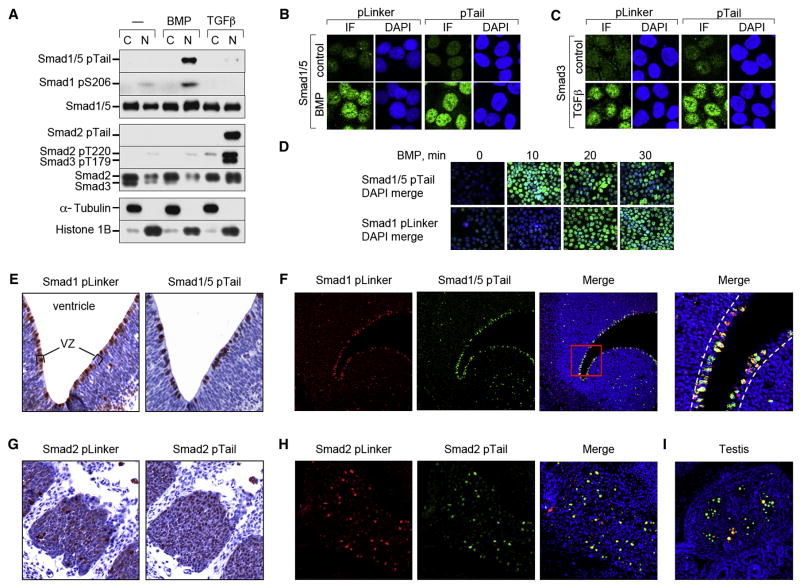
Phosphorylation of the Smad1 linker region is induced not only by antagonists acting through MAPKs, but also by the pathway agonist BMP2 (Sapkota et al., 2007). To determine the generality of Smad ALP, BMP2 or TGFβ1 treated HaCaT cell extracts were probed with Smad phosphopeptide antibodies against phospho-Ser206 (pS206) in Smad1 (Sapkota et al., 2007), which does not appear to cross-react with Smad5 (data not shown); and phospho-Thr220/179 (pT220/179) in Smad2/3 (Gao et al., 2009)(refer to Figure S1). BMP induced the phosphorylation of the Smad1 linker region and C-tail of Smad1/5, and TGFβ did the same to Smads 2 and 3 (Figure 1A). Cell fractionation (Figure 1A) and immunofluorescence staining (Figure 1B, C) showed that linker-phosphorylated Smads accumulate in the nucleus. ALP occurred 10 minutes after receptor-mediated tail-phosphorylation (Figure 1D).
Figure 1. TGFβ and BMP induce Smad C-tail and linker phosphorylation.
(A) Nuclear and cytoplasmic fractions of BMP or TGFβ treated (1h) HaCaT cells were analyzed by western blot with the indicated antibodies. (B) and (C) HaCaT cells treated with BMP, TGFβ or no addition (control) for 1 h were fixed and analyzed by immunofluorescence (IF) with antibodies against Smad1 (pLinker, Smad1 pS206; pTail, Smad1/5 pTail) and Smad3 (pLinker, Smad3 pS213; pTail, Smad3 pTail) including DAPI-staining of nuclei. (D) As in (B), with BMP treatment for the indicated times. (E–I) Images of E13.5 mouse embryo sections. (E) Adjacent sections through the telencephalic ventricular zone immunohistochemically stained with anti-Smad1 antibodies described in (B and C), with Hematoxylin counterstaining. Images at 40x magnification. Ventricle and ventricular zone (VZ) are indicated. (F) Confocal images of a section similar to (A) with double-immunofluorescence staining using the indicated antibodies and DAPI counterstaining. Images are at 20x magnification, with the far right panel at 200x. The ventricular zone is indicated by white dashed lines. (G) Immunohistochemistry on adjacent sections through dorsal root ganglia stained as in (E), with antibodies against Smad2 (pLinker Smad2, Smad2 pS245/250/245; pTail Smad2). Images at 80x magnification. (H) Confocal images of dorsal root ganglia with double-immunofluorescence staining with the antibodies used in (G). Magnification was 80x. (I) Merged confocal image of E13.5 testis from the same section as in (H).
In E13.5 mouse embryos the immunostaining pattern of both linker-phosphorylated Smad1 and tail-phosphorylated Smad1/5 was mostly nuclear (Figure 1E; Figure S2A, B) and showed a high degree of co-localization (Figure 1F; Figure S2B). Phospho-linker Smad1 and phospho-tail Smad1/5 were detected in the ventricular zones of the brain ventricles (Figure 1E); in tooth buds (Figure S2A); and in the spinal cord canal and dorsal root ganglia (data not shown). Moderate levels were seen in the gastric wall (Figure S2A), in developing heart valves, epithelial cells of lung bronchioles and kidney tubules (data not shown). Phospho-linker and phospho-tail Smad2 staining overlapped in nuclei of dorsal root ganglia (Figure 1G, H), and only partially co-localized in male germ cells (Figure 1I), and in brain and spinal cord ventricular zones (data not shown). Phospho-tail Smad2 with little or no phospho-linker staining was observed in tooth buds, mesenchymal cells surrounding large airways (Figure S2C, D), and in heart valves, the aortic wall, and vertebral ossification centers (data not shown). In sum, Smad linker phosphorylation accompanying C-tail phosphorylation is a general feature of the BMP and TGFβ pathways.
ALP occurs during transcriptional complex assembly
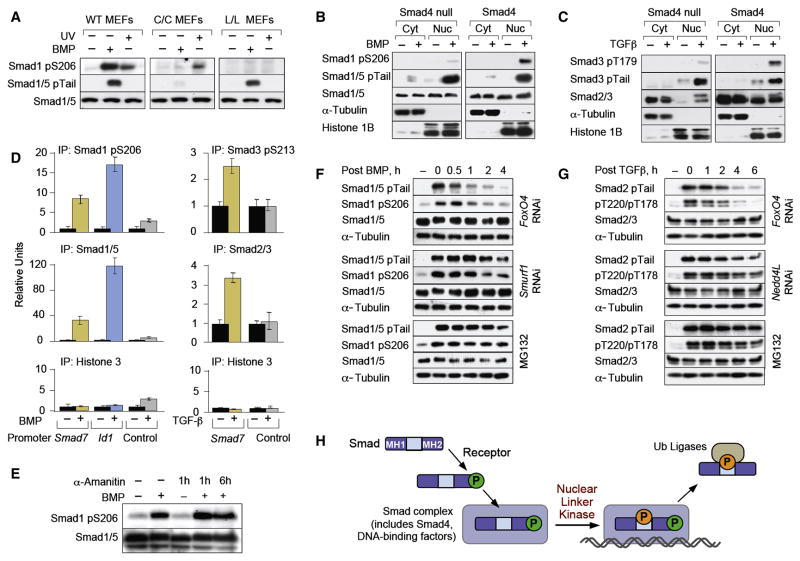
To determine the requirements for ALP we used mouse embryonic fibroblasts (MEFs) derived from wild-type (WT) embryos and embryos homozygous for knocked-in Smad1 alleles with alanine mutations of C-tail (Smad1C) or linker (Smad1L) phosphorylation sites (Aubin et al., 2004). BMP failed to induce ALP of Smad1C, despite the presence in this mutant of intact linker sites, in contrast to UV cell irradiation (Figure 2A), which induces cytoplasmic Smad1 linker phosphorylation through JNK and p38 MAPKs (Sapkota et al., 2007). This suggested that Smad1 C-tail phosphorylation is not required for linker phosphorylation by antagonistic MAPKs, but is essential in vivo for linker phosphorylation by agonist-dependent kinases.
Figure 2. Requirements for Smad ALP.
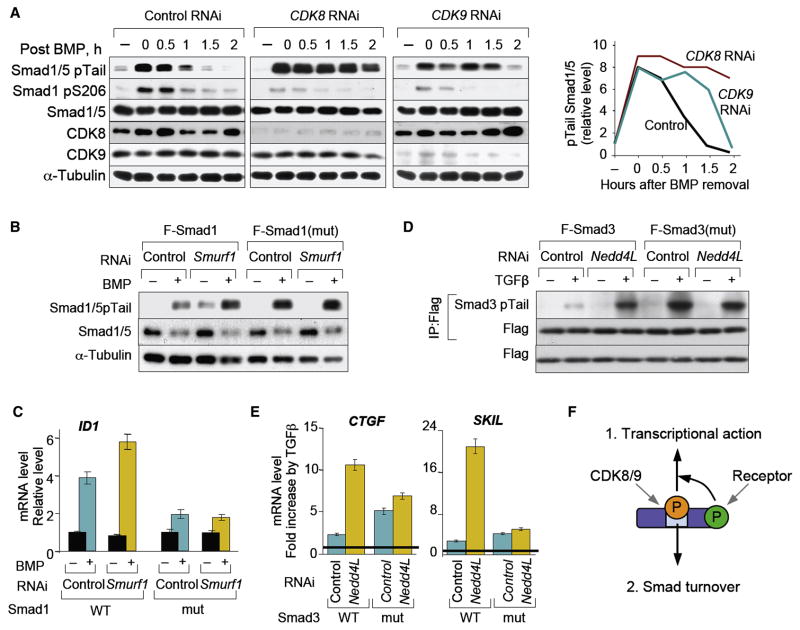
(A) Immunoblot analysis of wild-type, Smad1C and Smad1L mutant knock-in MEFs treated with BMP or UV, with antibodies against the indicated proteins. (B, C) SW480 Smad4-null cells, or cells stably expressing Smad4, were treated with BMP (B) or TGFβ (C). Nuclear and cytoplasmic fractions were analyzed using the indicated antibodies. (D) Chromatin immunoprecipitation of BMP and TGFβ stimulated C2C12 cells with the indicated antibodies. Precipitates were subjected to qRT-PCR of the BMP and TGFβ responsive regions of the indicated genes with unresponsive regions of the same genes serving as negative controls. Data show the mean ± S.D. of triplicates and are representaive of at least two independent expreriments. (E) Immunoblot analysis of BMP treated HaCaT cells in the absence or after addition of α-amanitin for the indicated times. Total cell extracts were analyzed using the indicated antibodies. (F) HaCaT cells were stimulated with BMP for 1 h in the absence or presence of MG132 or of a siRNA against Smurf1. Cells were harvested at the indicated times after BMP removal and total cell extracts were analyzed by immunoblot. (G) As in (F) but cells were stimulated with TGFβ and siRNA against Nedd4L was used. (H) Schematic model of the sequential steps leading to Smad-ALP and binding of ubiquitin ligases.
Smad ALP was observed in all cell lines tested except in cells lacking Smad4, a general partner of receptor-activated Smads (R-Smads) which binds to their phosphorylated C-tail and nucleates transcriptional complexes (Massagué et al., 2005). In the Smad4-defective human colon cancer line SW480 and pancreatic cancer line BxPC3 BMP induced tail phosphorylation and nuclear accumulation of Smad1/5, but only minimal Smad1 linker phosphorylation (Figure 2B; Figure S3). Similar results were obtained with Smad3 in response to TGFβ (Figure 2C). Restoration of Smad4 expression rescued the ability of Smad1 and Smad3 to undergo ALP (Figure 2B, C; Figure S3). These results suggested that Smads undergo ALP as a result of phosphotail-driven incorporation into Smad4-containing transcriptional complexes.
To determine whether the ALP-Smads are present on the regulatory regions of target genes, we performed chromatin immunoprecipitation (ChIP) assays. In BMP-treated cells, but not in controls, both an anti-Smad1/5 antibody and an antibody against phospho-Ser206 of Smad1 pulled down DNA that included the BMP responsive regions of Inhibitor of DNA binding 1 (Id1) and Smad7 (Figure 2D). Similarly, in TGFβ treated cells, an antibody against the linker phosphorylated Smad3 and an anti-Smad2/3 antibody pulled down DNA containing the TGFβ responsive element of the Smad7 gene (Figure 2D). Treating cells with the RNAP II inhibitor α-amanitin did not affect Smad1 ALP (Figure 2E), indicating that this event accompanies, but is not a consequence of active transcription.
ALP primes Smads for turnover
Linker-phosphorylated Smad1 is recognized by Smurf1 (Sapkota et al., 2007) and linker-phosphorylated Smad2/3 by Nedd4L (Gao et al., 2009), both of which belong to the HECT family of E3 ubiquitin ligases. Members of this family bind their substrates via WW domains that interact with PPXY sequences (PY motifs), normally without requiring supporting contacts with phosphorylated sites (Ingham et al., 2004). However, the PY motifs in the linker regions of Smads 1, 2 and 3 are not sufficient for productive interactions with Smurf1 or Nedd4L. Smurf1 binding requires phosphorylation of at least one serine residue in a SerPro cluster of the Smad1 linker region, preferably S206 and S214 (Sapkota et al., 2007). Nedd4L binding to Smads 2 and 3 requires phosphorylation of a Thr residue (T220 in Smad2, T179 in Smad3) located immediately upstream of the PY motif (Gao et al., 2009). Since ALP prominently targeted these residues (Figure 1A; Figure S1), we postulated that Smurf1 and Nedd4L mediate proteasome degradation of activated Smad proteins.
Cells were treated with BMP or TGFβ for 1 h to achieve peak Smad tail phosphorylation, followed by removal of agonist to determine the decay of tail-phosphorylated Smads. Depletion of Smurf1 by RNAi delayed the decay of activated Smad1/5 as effectively as addition of a proteasome inhibitor MG132 (Figure 2F), and the same was seen for activated Smad2/3 after Nedd4L depletion (Figure 2G). RNAi-mediated depletion of FoxO4, which is ubiquitously coexpressed and functionally redundant with FoxO1 and FoxO3 (Gomis et al., 2006), was used as a negative control.
Proteasome inhibition with MG132 led to accumulation of tail-phosphorylated Smad1/5 and linker-phosphorylated Smad1 both in the nucleus and in the cytoplasm (Figure S4). MG132 did not fully block the decay of tail-phosphorylated Smads, consistent with the participation of Smad C-terminal phosphatases as an alternative mechanism for Smad deactivation (Inman et al., 2002; Knockaert et al., 2006; Lin et al., 2006). Furthermore the CRM1 inhibitor leptomycin B, which had been previously reported to block Smad1 nuclear export (Xiao et al., 2001), resulted in increased levels of tail-phosphorylated Smad1/5 and linker-phosphorylated Smad1 (Figure S4). Taken together these results indicate that ALP is a consequence of Smad assembly into transcriptional complexes in the nucleus, occurs during or just prior to Smad binding to chromatin, and targets Smads to specific ubiquitin ligases for proteosomal turnover (Figure 2H).
CDK8 and CDK9 mediate Smad ALP
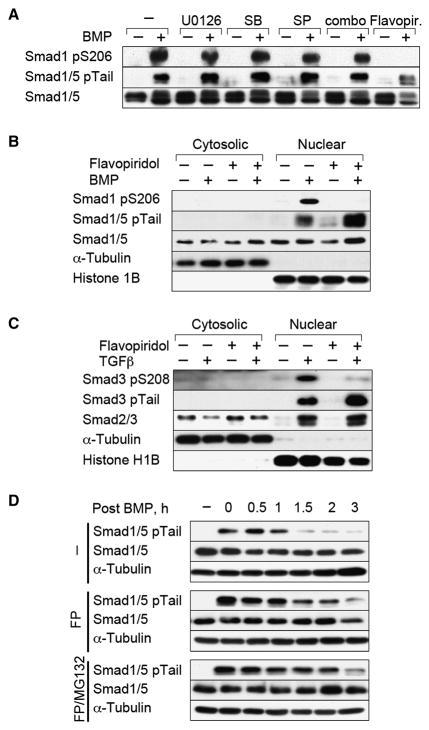
BMP-induced Smad1 linker phosphorylation was not suppressed by inhibitors of MEK (U0126), p38 (SB203580), or JNK (SP600125) tested individually; in double; or triple combinations (Figure 3A; data not shown). Of all the protein kinase inhibitors screened, only the semi-synthetic flavonoid flavopiridol (Shapiro, 2004) effectively inhibited Smad ALP (Figure 3A; Figure S5), by preventing ALP of nuclear Smad1 in BMP treated cells (Figure 3B) and of nuclear Smad3 in TGFβ treated cells (Figure 3C). This was accompanied by an increase in the level of tail-phosphorylated Smad1 and Smad3 (Figure 3B,C). Indeed, flavopiridol extended the half-life of BMP-activated Smad1/5 as much as MG132 (Figure 3D), and a similar effect was observed with TGFβ-activated Smad3 (data not shown).
Figure 3. Flavopiridol inhibits Smad ALP.
(A) Cell extracts of BMP treated HaCaT cells with or without kinase inhibitors were analyzed with antibodies against the indicated proteins. SB, SB203580; SP, SP60125; combo, combination of the three MAPK inhibitors; Flavopir., Flavopiridol. (B, C) Nuclear and cytoplasmic fractions of HaCaT cells stimulated with BMP or TGFβ in the absence or presence of Flavopiridol were analyzed by immunoblotting. (D) HaCaT cells were stimulated with BMP for 1 h in the absence or presence of Flavopiridol (FP), or of Flavopiridol plus MG132. Incubations were continued for the indicated periods with media without BMP.
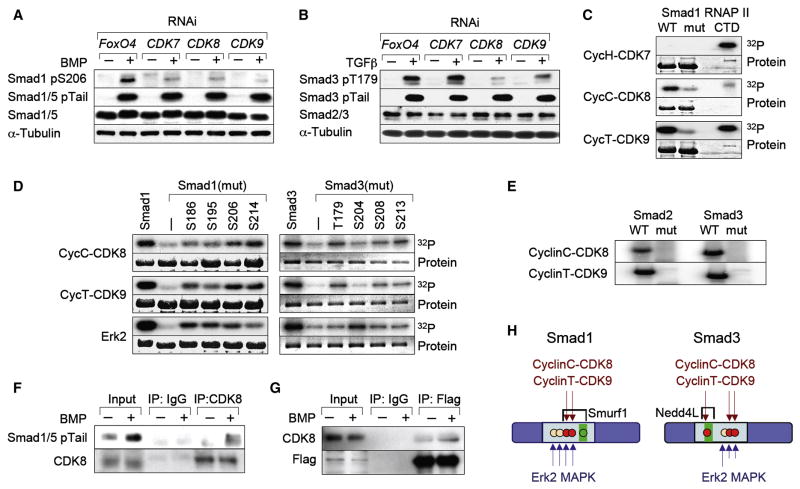
Reducing the list of flavopiridol-sensitive kinases (Fabian et al., 2005) by using inhibitors of partially overlapping specificity (Figure S5), led us to cyclin-dependent kinases (CDKs) as potential Smad-ALP mediators. Various inhibitors of CDKs that function in the cell cycle did not inhibit BMP-induced Smad1 linker phosphorylation. These included roscovitine, purvalanol A, and UCN01 (Figure S5), which inhibit CDKs 1, 2, 4, 5 and 6 (McClue et al., 2002; Omura-Minamisawa et al., 2000). The inducible overexpression of p27Kip1 or p15Ink4b, which inhibit CDKs 2, 4 and 6 and their phosphorylation of the retinoblastoma protein pRb (Reynisdottir and Massagué, 1997) (Figure S6A), as well as RNAi-mediated knockdown of CDK1, CDK2, CDK4 or CDK5 (Figure S6B-D) also had no effect. These results left as candidates the transcription-regulatory CDKs 7, 8 and 9. RNAi-mediated knockdown of CDK8 or CDK9 inhibited the BMP-induced phosphorylation of S206 in Smad1 and the TGFβ induced phosphorylation of T179 in Smad3 (Figure 4A,B). RNAi inhibition of both CDK8 and CDK9 resulted in greater reduction of Smad1-ALP (Figure S6F) suggesting that these kinases act redundantly, while knockdown of CDK7 inhibited the ALP of S206 in Smad1 but not that of T179 in Smad3 (Figure 4A, B). Knockdown of one CDK did not affect the levels of the others (Figure S6E).
Figure 4. CDK8 and CDK9 as mediators of Smad ALP.
(A, B) HaCaT cells were transfected with siRNAs against CDK7, CDK8, CDK9 or against FoxO4 as a negative control. Total extracts from cells stimulated with BMP or TGFβ for 1 h were analyzed by immunoblot. (C) Autoradiograph of bacterially expressed Smad1 wild-type or linker mutant, phosphorylated in vitro by the indicated CDK/Cyclin complexes with γ32P-ATP as substrate. Purified CTD domain of RNA Pol II was used as a positive control for CDK activity. (D) In vitro kinase assay of the indicated CDK9/CyclinT, CDK8/CyclinC complexes or ERK, on bacterially expressed Smad1 and Smad3 wild-type or linker mutant. The numbers indicate the residue that was left as Ser/Thr in the linker region. (E) As in (C) but with Smad2 and Smad3 wild-type or linker mutants. (F) Co-immunoprecipitation of endogenous Smad1/5 and CDK8. Cell extracts from untreated or BMP-treated HaCaT cells were immunoprecipitated using an anti-CDK8 antibody and analyzed by western immunoblotting with antibodies against Smad1/5 pTail, or CDK8 as a loading control. (G) Flag immunoprecipitates of BMP-treated HaCaT cells stably expressing Flag-Smad1 were analyzed using antibodies against CDK8, or Flag as a loading control. (H) Schematic summary of CDK8/9 phosphorylation sites in Smad linker regions and their relationship to ubiquitin ligase recognition sites (Gao et al., 2009; Sapkota et al., 2007). Red dots, principal phosphorylation sites; green boxes, PY motifs.
In vitro, recombinant cyclinC-CDK8 and cyclinT1-CDK9 phosphorylated Smads 1, 2 and 3 but induced much reduced phosphorylation of Smad proteins with mutated linker sites (Figure 4C–E; see Supplementary Experimental Procedures for description of the Smad linker mutants). Using as substrates Smad1 and Smad3 proteins with valine or alanine mutations in all but one of the four Ser/Thr residues of interest, cyclinC-CDK8 and cyclinT-CDK9 showed a preference for S206 and S214 but also phosphorylated S186 and S195 in the case of Smad1; and T179, S208 and S213 in the case of Smad3. In contrast, ERK2 phosphorylated all four Smad1 residues almost evenly, while showing a preference for S204 over S208 and S213 in Smad3 (Figure 4D). Activated, tail-phosphorylated Smad1 could be co-immunoprecipitated with endogenous CDK8 (Figure 4F), and endogenous CDK8 with stably expressed Flag-tagged Smad1 in response to BMP (Figure 4G). CyclinH-CDK7 did not phosphorylate Smads in vitro, even though it was active at phosphorylating RNAPII CTD (Figure 4C, and data not shown), and thus does not appear to be a direct Smad linker kinase. Collectively these results identified CDK8 and CDK9 as mediators of agonist-dependent linker phosphorylation of Smads (Figure 4H).
Dual role of CDK8/9 and linker phosphorylation in Smad function and turnover
Since Smad phosphorylation by CDK8 and CDK9 creates ubiquitin ligase-binding sites, we asked whether interfering with CDK8/9 function would stabilize the pool of activated, C-tail phosphorylated Smads. CDK8- or CDK9-depleted cells were treated with BMP for 1 h, followed by incubation without the agonist to track the decay of tail-phosphorylated Smad1. CDK8 or CDK9 knockdown delayed the decay of activated Smad1 (Figure 5A) and Smad3 (data not shown), thus mimicking the effects of flavopiridol addition (refer to Figure 3B–D) and of Smad ubiqutin ligase depletion (refer to Figure 2F).
Figure 5. Role of CDK8/9 and linker phosphorylation in Smad turnover.
(A) Time course of Smad1/5 C-tail phosphorylation after 1 h of BMP stimulation as in Figure 2F. HaCaT cells transfected with siRNA against CDK8, CDK9 or the control FoxO4 were stimulated 48 h later cells and total lysates were analyzed by immunoblotting. The fourth panel shows a quantitation of the Smad1/5 pTail bands. (B) HaCaT cells with stable shRNA Smad1 knockdown and stably transduced with vectors encoding Smad1 wild-type or linker mutant were treated with BMP in the presence of Smurf1 or control siRNA and analyzed by immunoblotting. (C) The cell lines described in (B) were treated with BMP for 1 h and cells were harvested 2 h after BMP removal to test the expression of ID1 by qRT-PCR. Data show the mean ± S.D of quadruplicates and are representative of two independent experiments. (D) HeLa-S3 cells stably expressing Flag-tagged Smad3 (wild-type or linker mutant) were retrovirally infected with vectors encoding control shRNA or shRNA against Nedd4L. After treating cells with or without TGFβ for 3 h whole cell lysates were subjected to anti-Flag immunoprecipitation and analyzed by immunoblotting. (E) The cell lines used in (D) were treated with TGFβ for 3 h, and total RNA was isolated for qRT-PCR analysis of CTGF or SKIL levels. Values are normalized to the untreated levels, and shown as the fold induction by TGFβ in each cell line. Data are the mean ± S.D of quadruplicates and are representative of two independent experiments. (F) Schematic representation of the dual role of linker phopshorylation.
To assess the effect of ALP on the transcriptional function of Smad proteins we compared cells expressing wild-type or mutant Smad lacking the linker phosphorylation sites. Knocking down CDK8 and CDK9 was ruled out, since the effects of these protein kinases on general transcription would confound our results. We generated HaCaT cell lines in which endogenous Smad1 has been depleted and which stably overexpress either wild-type Smad1 or the mutant Smad1(mut) with alanines replacing all four serines in the linker SerPro cluster. Additional Smurf1 depletion increased the BMP-dependent accumulation of tail-phosphorylated Smad1/5 in these cells (Figure 5B). This effect was accompanied by a stronger induction of the typical BMP/Smad1 target gene ID1 (Figure 5C). The absence of linker phosphorylation sites led to a constitutive increase in BMP-dependent accumulation of tail-phosphorylated Smad1(mut), and this increase was not expanded by Smurf1 depletion (Figure 5B). This result was consistent with a role of ALP in Smurf1-dependent turnover of activated Smad1. Surprisingly, the ID1 response in Smad1(mut) cells was weaker than in Smad1 cells (Figure 5C), suggesting that lack of ALP makes Smad1 not only resistant to Smurf1-dependent turnover, but also inefficient as a mediator of transcriptional responses.
A similar pattern was observed with HeLa-S3 cells expressing Smad3 or a linker phosphorylation site mutant Smad3(mut), while retaining endogenous Smad3 expression. Nedd4L depletion strongly increased the TGFβ-dependent accumulation of activated Smad3 (Figure 5D) and the expression of the typical TGFβ target genes CTGF (connective tissue growth factor) and SKIL (SKI-like, also known as SnoN) (Figure 5E). Tail-phosphorylated Smad3(mut) accumulated to high levels in response to TGFβ (Figure 5D), but although the presence of endogenous Smad3 supported target gene induction, Nedd4L depletion failed to significantly increase these responses (Figure 5E). These results suggest that ALP promotes Smad transcriptional function while marking Smads for turnover (Figure 5F).
Smad1 ALP recruits YAP
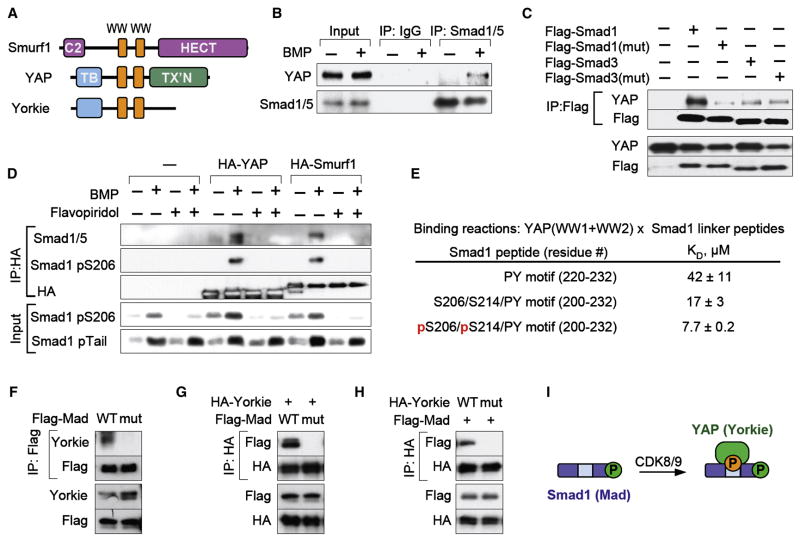
We hypothesized that this dual role of Smad ALP might be based on the recruitment of different partners at different stages of the signal transduction cycle. In light of the highly selective interaction between linker-phosphorylated Smads and different ubiquitin ligases, we further postulated that the Smad transcriptional function depends on the recruitment to the same phosphorylated sites of transcription cofactors containing WW domains similar to those of the corresponding Smad ubiquitin ligase. Focusing on Smad1 we conducted a genome-wide blastp search for proteins that contain Smurf1-like WW domains but are not ubiquitin ligases. The top-scoring hit was YAP (Figure 6A), a transcriptional coactivator that binds PY motifs of target proteins (Edgar, 2006; Pan, 2007; Zeng and Hong, 2008).
Figure 6. Smad1 linker phosphorylation mediates binding of YAP.
(A) Schematic representation of Smurf1 and YAP. Orange boxes, WW domains; C2, calcium and lipid binding domain; HECT, ubiquitin ligase domain; TB, TEAD bindng domain, TX’N, transcription activation domain. (B) Co-immunoprecipitation of endogenous YAP and Smad1 in BMP-treated HaCaT cells. Complexes were analyzed by immunoblotting. (C) HEK293T cells transfected with vectors encoding Flag-tagged Smad1 or Smad3 (wild-type or linker mutant) were subjected to Flag immunoprecipitation and analyzed by immunoblotting. (D) HEK293T cells were transfected with vectors encoding HA-tagged YAP or Smurf1 and after BMP stimulation in the presence or absence of flavopiridol, HA immunoprecipitates were analyzed by immunoblotting using the indicated anti-Smad1 antibodies. (E) Dissociation constants of the interaction of a YAP (WW1+WW2) protein fragment with the indicated Smad1 linker peptides, as measured by isothermal titration calorimetry. (F) Drosophila S2 cells were transfected with Flag-Mad wild-type or linker mutant and Flag-immunoprecipitates were tested for the presence of endogenous Yorkie by immunoblot. (G) As in (E), with cotransfection of HA-Yorkie and immunoprecipitation of the HA-species. (H) As in (F), with transfection and immunoprecipitation of wild-type or WW domain mutant Yorkie. (I) Schematic representation of the recruitment of YAP upon CDK8/9-mediated linker phosphorylation of Smad1.
Endogenous YAP and Smad1/5 in HaCaT cells could be co-immunoprecipitated in a BMP dependent manner (Figure 6B). Using epitope-tagged Smad expression vectors, showed that YAP binding to Smad1 requires the phosphorylation sites of the SerPro cluster (Figure 6C), but not T222, the residue directly adjacent to the PY motif (Figure S7). Moreover, flavopiridol abolished the BMP induced interaction between endogenous Smad1 and epitope-tagged YAP or Smurf1 (Figure 6D), confirming the importance of Smad1-ALP for YAP and Smurf binding. Isothermal titration calorimetry experiments with a recombinant 104-amino acid polypeptide that includes the two YAP WW domains, and three Smad1 peptides, also showed that the YAP WW construct had low affinity for a Smad1 peptide containing only the PY motif (Figure 6E). This interaction was increased by 2.5-fold by extending the Smad1 peptide to include the two principal CDK8/9 sitesS206 and S214, and was further increased by 2.2-fold when these sites were phosphorylated. An interaction was observed between YAP and Flag-tagged Smad3 in transfected cells, but this was weak and independent of Smad3 linker phosphorylation (Figure 6C).
To investigate the conservation of the Smad1-YAP interaction through species we tested the ability of their Drosophila orthologs, Mad and Yorkie, to interact in S2 Drosophila cells. Endogenous or transfected epitope-tagged Yorkie could be co-immunoprecipitated with wild-type Flag-Mad, but not with a linker phosphorylation site mutant (Figure 6F, G). Conversely no interaction was detected between wild-type Flag-Mad and a WW domain Yorkie mutant (Figure 6H). The loss of interaction of Yorkie with the Mad linker mutant, indicates that overexpression of wild-type Mad leads to linker hyperphosphorylation, as seen with overexpression of mammalian Smads (Sapkota et al., 2007). The lack of Mad phospholinker antibodies precluded corroboration of this interpretation. Taken together these results show that YAP interacts with Smad1 with the same binding requirements and selectivity as Smurf1 and that this interaction is evolutionarily conserved from flies to mammals.
YAP enhances Smad1 function
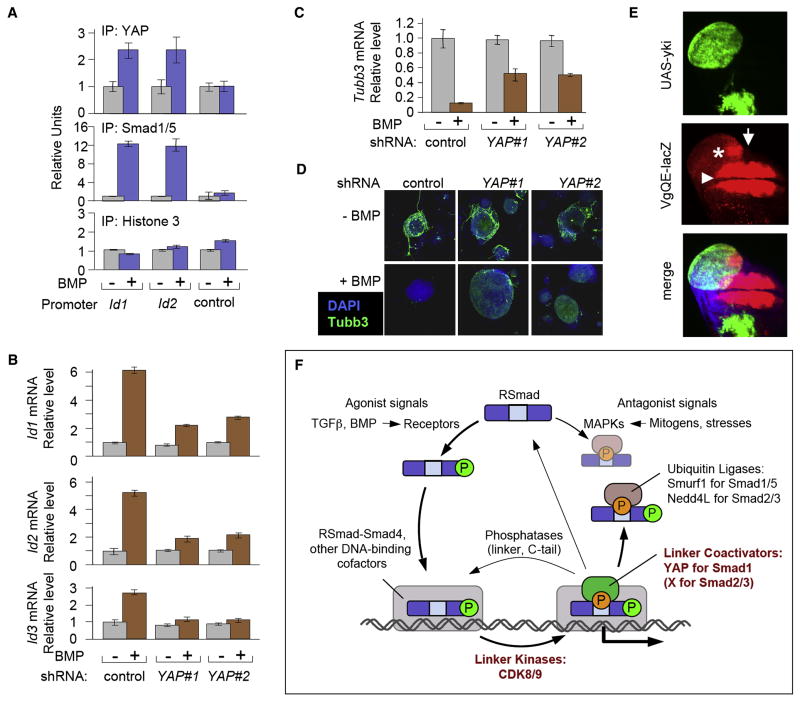
Given that BMP has roles in mouse embryonic stem cell (mESC) self-renewal and differentiation (Varga and Wrana, 2005) we chose mESCs to analyze the impact of YAP on BMP mediated gene responses. Transcriptomic analysis of BMP-stimulated mESCs, identified a limited number of BMP responsive genes (Table S1). The top scoring genes on this list belonged to the Id family (Id3, Id1 and Id4; with Id2 also among the top-ten targets), which had been previously identified as prominent BMP targets in undifferentiated and differentiating mESC cultures (Hollnagel et al., 1999; Ying et al., 2003a). Chromatin immunoprecipitation showed that YAP and Smad1/5 were bound to the BMP responsive region of Id1 and Id2 when these genes were actively transcribed in response to BMP (Figure 7A). To test the effect of YAP on BMP-dependent gene responses, we depleted YAP from mESCs by stable shRNA transduction, generating two independent cell lines, which exhibited ~80% YAP knockdown without significantly altering Smad1/5 levels (Figure S8A, B). The effect of BMP on the expression of Id1, Id2 and Id3 was sensitive to depletion of YAP (Figure 7B).
Figure 7. YAP enhances BMP-Smad responses in different biological contexts.
(A) Chromatin immunoprecipitation of BMP-stimulated mESCs using antibodies against YAP and Smad1/5. BMP-responsive regions of Id1 and Id2 and an unresponsive control region of Id1 (control) were analyzed by qRT-PCR as in Figure 2D. (B) Wild-type YAP-knockdown mESCs were stimulated with BMP4 for 1 h. Total RNA was analyzed by qRT-PCR. Data show the mean ± S.D of quadruplicates and are representative of two independent experiments. (C) Gene expression analysis of the neural differentiation marker β-III tubulin (Tubb3), in differentiating wild-type and YAP-knockdown mESCs. Cells were cultured in N2B27 supplemented media in the presence or absence of BMP, harvested five days later and total RNA was subjected to qRT-PCR analysis for Tubb3 expression as in (B). (D) Confocal images of mESCs treated as in (C) and subjected to immunofluorescence staining with an anti-Tubb3 antibody (green) and DAPI counterstaining (blue). Tubb3 staining detects neuronal differentiation. (E) vgQE-lacZ expression (red) in Drosophila wing imaginal disc containing Yorkie-overexpressing clones (marked by GFP+). Confocal z-stack through the wing imaginal disc was projected into a single plane. Note that the ectopic vgQE-lacZ expression (asterisk) is discontinuous with the endogenous vgQE-lacZ expression domain (arrowhead) and only observed close to the anterior-posterior compartment boundary (arrow). (F) The Smad signaling cycle. Receptor-bound BMP and TGFβ ligands induce C-tail phosphorylation of R-Smads, which then accumulate in the nucleus. Nuclear R-Smads complexed with Smad4, bind to the regulatory elements of target genes and interact with other DNA binding cofactors (shown as a grey box), becoming linker-phosphorylated by CDK8/9 at some point during this process. This facilitates Smad1 binding to YAP, which is required for efficient transcription of BMP target genes. Linker phosphorylated Smad2/3 is thought to activate transcription of TGFβ target genes in a similar manner, but through as yet unknown cofactors. After fulfilling their transcriptional role, Smads may be targeted by linker and tail phosphatases that reset them to their ground state or linker phosphatases only that may allow further Smad activity. Alternatively, Smads can be recognized in a phospho-linker dependent manner, by ubiquitin ligases, such as Smurf1 in the case of Smad1, and Nedd4L in the case of Smad2/3, resulting in their eventual degradation. The R-Smad linker regions can also be phosphorylated by MAPK pathway kinases in response to antagonists, such as FGF, EGF, or stress signals, which ultimately leads to Smad degradation. By providing a platform for the regulation of the different and often opposing fates of the Smads, linker phosphorylation represents a focal point in the Smad signaling cycle.
BMP inhibits neural differentiation of mouse ES cells through the induction of Id proteins (Ying et al., 2003a). Moreover, activated Smad1/5 is abundant in the subventricular zone of the mouse telencephalon (Figure 1C), which is rich in neural stem and progenitor cells (Alvarez-Buylla and Lim, 2004). When incubated in LIF- and serum-free media supplemented with N2/B27, mESCs commit to neural cell lineages as shown by the expression of the neuronal marker β-III tubulin (Tubb3), and this effect is drastically inhibited by BMP (Figure 7C) (Ying et al., 2003b). YAP depletion attenuated this effect of BMP, as determined by qRT-PCR analysis of Tubb3 mRNA levels (Figure 7C) and immunofluorescence staining of the cells with anti-tubb3 antibodies (Figure 7D; Figure S8). Collectively, these results suggest that BMP-induced linker phosphorylation of Smad1 serves to recruit YAP to Id genes for enhanced transcription.
To further probe the significance of the Smad-YAP interaction, we investigated whether their Drosophila counterparts Mad and Yorkie (Yki) cooperate to affect Drosophila biological processes in vivo. In the wing imaginal disc a gradient of the BMP ortholog Dpp activates Mad to achieve induction of target genes such as vestigial (vg), for correct patterning and growth (Kim et al., 1997). Overexpression of Yorkie in wing imaginal disc clones induced ectopic expression of the vgQE-lacZ reporter (Figure 7E), which contains a previously described Mad binding element (Kim et al., 1997). Yorkie-induced ectopic vgQE-lacZ expression is discontinuous with the endogenous expression domain of the reporter and is detected near the AP boundary, where the Dpp signal is at its highest. Thus, the ectopic vgQE-lacZ expression reflects an intrinsic response of the cells to high levels of Yorkie and Dpp at those positions, rather than being a result of clone overproliferation. The fact that this ectopic expression is only observed at positions with the highest level of Dpp further suggests that the cooperation between Mad and Yorkie might be important for achieving maximum level Dpp signaling. Thus Mad and Yorkie parallel in Drosophila the role established in the mammalian ES cell system for the Smad1-YAP interaction and the induction of BMP target genes.
Discussion
The present findings reveal a remarkable integration of Smad regulatory functions by agonist-induced, CDK8/9-mediated phosphorylation of the linker region and highlight this previously unrecognized event as an integral feature of the transcriptional action and turnover of receptor-activated Smad proteins (Figure 7).
Smad linker phosphorylation by CDK8/9 in canonical BMP and TGFβ pathways
Agonist-induced linker phosphorylation of R-Smads is a general feature of BMP and TGFβ pathways, occurring in all the responsive cell types examined, shortly after Smad tail phosphorylation. Our evidence identifies CDK8 and CDK9 as the kinases involved and does not support a major role for MAPKs or cell cycle-regulatory CDKs in this process. CDK8 and cyclin C are components of the Mediator complex that couples enhancer-binding transcriptional activators to RNAPII for transcription initiation (Malik and Roeder, 2005). CDK9 and cyclin T1 constitute the P-TEFb complex, which promotes transcriptional elongation (Bres et al., 2008). CDK8 and CDK9 phosphorylate overlapping serine clusters in the C-terminal domain of RNAPII (Phatnani and Greenleaf, 2006), a region which integrates regulatory inputs by binding proteins involved in mRNA biogenesis (Phatnani and Greenleaf, 2006). Thus, CDK8 and CDK9 may provide coordinated regulation of Smad transcriptional activators and RNAPII.
Precedent exists for the ability of CDK8 to phosphorylate enhancer-binding transcription factors. The CDK8 ortholog Srb10 in budding yeast phosphorylates Gcn4 marking this transcriptional activator of amino acid biosynthesis for recognition by the SCF(Cdc4) ubiquitin ligase (Chi et al., 2001). In mammalian cells, CDK8 phosphorylates the ICD signal transduction component of Notch, targeting it to the Fbw7/Sel10 ubiquitin ligase (Fryer et al., 2004). However, whereas CDK8-mediated phosphorylation inhibits Gcn4 and Notch activity, we show here that phosphorylation of agonist-activated Smads by CDK8/9 enables Smad-dependent transcription before triggering Smad turnover.
One structure, two binding partners and two opposite functions
Activated Smads undergo proteasome-mediated degradation (Lo and Massagué, 1999) as well as phosphatase-mediated tail dephosphorylation (Inman et al., 2002) to keep signal transduction closely tied to receptor activation. We show that BMP-induced Smad1-ALP generates binding sites for Smurf1, accomplishing in the nucleus what MAPK-mediated phosphorylation of basal-state Smad1 accomplishes in the cytoplasm (Sapkota et al., 2007) (Figure 7). Similarly, TGFβ-induced linker phosphorylation of Smad2/3 provides a binding site for Nedd4L (Gao et al., 2009).
Our results also reveal a positive role for ALP in Smad-dependent transcription. Smad proteins with phosphorylation-resistant linker mutations are more stable as receptor-activated signal transducers than their wild-type counterparts, yet they are transcriptionally less active. Indeed, mutation of Smad1 linker phosphorylation sites (in a wild-type Smad5 background) does not result in a straight BMP gain-of-function phenotype but rather in an unforeseen gastric epithelial phenotype (Aubin et al., 2004). Although the interpretation of this phenotype is confounded by the contribution of MAPK signaling to linker phosphorylation, it is consistent with the present evidence that Smad1 linker phosphorylation plays an active role in BMP signaling.
Focusing on Smad1 to define this dual role, we have found that the phosphorylated linker sites, together with a neighboring PY motif, are recognized also by the transcriptional coactivator YAP. Smurf1 and YAP present closely related WW domains with a similar selectivity towards linker-phosphorylated Smad1. YAP is recruited with Smad1 to BMP responsive enhancers and knockdown of YAP inhibits BMP-induced Id gene responses in mouse embryonic stem cells. Both BMP and YAP act as suppressors of neural differentiation in specific contexts (Cao et al., 2008; Varga and Wrana, 2005). As we show here YAP supports the ability of BMP to block neural lineage commitment through the induction of Id family members (Ying et al., 2003a), creating a link between YAP-dependent BMP transcriptional output and ES cell fate determination.
Thus, a common structure fulfills two opposite functions –Smad1 transcriptional action and turnover – by recruiting different proteins, YAP and Smurf1 – at different stages of the signal transduction cycle (Figure 7). The cyclic recruitment and continuous turnover of transcription factors on target enhancers is required for the proper response of cells to developmental and homeostatic cues. We propose that Smad activation by TGFβ family agonists accomplishes this important requirement through linker phosphorylation that triggers transcriptional action and messenger turnover in one stroke.
A link to the Hippo pathway
Activation of the Hippo pathway by cell density cues triggers a kinase cascade that culminates in the inactivation of YAP (Yorkie in Drosophila), a transcriptional co-activator which acts through interactions with enhancer-binding factors, including TEAD/scalloped, Runx, p73 and others (Edgar, 2006; Pan, 2007; Zeng and Hong, 2008). Yorkie/YAP promotes cell proliferation and survival and organ growth, whereas the upstream components of the inhibitory kinase cascade constrain organ size and act as tumor suppressors (Dong et al., 2007; Overholtzer et al., 2006; Zhao et al., 2007). Elucidating the links between the Hippo pathway and other signaling cascades is an important open question (Edgar, 2006; Pan, 2007; Zeng and Hong, 2008). Our evidence that YAP is recruited to BMP-activated Smad1 reveals a previously unknown link between the BMP and the Hippo pathways. Both these signaling cascades have the capacity to control organ size, and do so in a manner suggestive of interactions with other patterned signals (Affolter and Basler, 2007; Martin-Castellanos and Edgar, 2002). An example is the regulation of imaginal disc growth by Dpp via cell competition, a process by which slow proliferating cells are eliminated in favor of their higher proliferating neighbors (Moreno et al., 2002). A genetic screen for negative regulators of Dpp signaling that protect cells from being outcompeted, identified upstream components of the Hippo pathway (Tyler et al., 2007). Inactivation of these factors elevated Dpp target gene expression, presumably by failing to inhibit Yorkie, and allowed cells to outcompete their neighbors, suggesting a functional convergence of the Hippo and BMP pathways that foreshadowed our findings.
Although ALP is a general event in Smad activation, YAP may not be a universal partner of linker-phosphorylated Smad1. Smad ALP likely plays a wider role potentially acting to recruit co-activators other than YAP, depending on the cellular context or the target gene. Also of interest is the identity of factors that may play an analogous role in linker-phosphorylated Smad2/3 in the TGFβ pathway. The linker phosphorylation sites and PY motifs of Smad1 and Smad2/3 are conserved in the otherwise divergent linker regions of the Drosophila orthologs, Mad/dSmad1 and Smox/dSmad2, respectively (Figure S1C). Although the contribution of the MAPK pathway in linker phosphorylation precludes a clearcut genetic investigation of these functions, they are probably conserved across metazoans. A concerted search for Smad phospho-linker interacting factors would answer many of these questions and would fully elucidate the role of the Smad linker region as a centerpiece in the function, regulation and connectivity of Smad transcription factors.
Experimental Procedures
Cell culture and transfections
HaCaT keratinocytes, HEK293T cells, SW480 colorectal adenocarcinoma cells and wild-type, Smad1 L/L and Smad1 c/c MEFs were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% FBS. Mouse C2C12 cells were maintained in DMEM with 20% FBS. Mv1Lu tetracycline-inducible cells were cultured as described (Blain and Massagué, 2000). BxPC3 cells were maintained in RPMI1640 media with 10% FBS. The SW480 and BxPC3 Smad4 stable cell lines were generated previously (He et al., 2006) and Hela-S3 cells stably expressing Flag-tagged Smad3 with shRNA mediated Nedd4L stable knockdown are described elsewhere (Gao et al., 2009). Mouse embryonic stem cells E14Tg2a.IV were maintained in feeder-layer free LIF-supplemented medium (Keller, 1995). Prior to total RNA extraction ES cells were treated with BMP4 (25 ng/ml, R&D Systems) for 1 h. Differentiation assays were carried out as described (Ying et al., 2003b) in the presence or absence of BMP2 (25 ng/ml, R&D Systems)..
Prior to treatment with BMP2 (25ng/ml, R&D systems), TGFβ1 (100 pM, R&D Systems), or UV radiation (200 mJ/m2), cells were serum-starved for 16 h. The chemical inhibitors U0126 (10 μM, Promega), SP600125 (10 μM, Calbiochem), SB203580 (10 μM, Calbiochem), MG132 (10 μM, Calbiochem), and LY294002 (100 μM, Calbiochem), Flavopiridol (0.3 μM, National Cancer Institute), Roscovitine (50 μM, Calbiochem), SU11248 (5 μM), CGP57380 (5 μM, Sigma-Aldrich), TG003 (10 μM, Calbiochem), Ro318220 (5 μM, Calbiochem), CHIR 99021 and CHIR 98014 (both 2 μM, Division of Signal Transduction Therapy, School of Life Sciences, Dundee University), UCN01 (2 μM, Calbiochem), DRB (50 μM, Calbiochem), Purvalanol A (2 μM, Calbiochem) were added to cells 30 min prior to BMP2 or TGFβ1 addition. Transfections of mammalian and Drosophila S2 cells and siRNA oligonucleotides were as described (Sapkota et al., 2007) (Wu et al., 2003) (Knockaert et al., 2006). Nuclear and cytosolic fractionations were performed with a Nuclear and Cytoplasmic Extraction Kit (Pierce) following the manufacturer’s instructions.
Immunohistochemistry and immunofluorescence
Immunohistochemistry (IHC) and immunofluorescence (IF) of mouse embryo tissue sections were processed at the Molecular Cytology Core Facility of MSKCC using a Discovery XT processor (Ventana Medical Systems). Tissue sections were blocked for 30 min in 10% normal goat serum, 2% BSA in PBS, followed by avidin/biotin block (12 min for Smad1 and 16 min for Smad2 antibodies). The 3 h primary antibody incubation was followed by 1 h incubation with biotinylated goat anti-rabbit IgG (ABC kit; Vector labs; 1:200 dilution). For IHC, detection was performed with the DAB detection kit (Ventana Medical Systems) according to the manufacturer’s instructions. For IF, detection was performed with Streptavidin-HRP D (Ventana Medical Systems), followed by incubation with Tyramide-Alexa Fluor 488 (Invitrogen) or Tyramide Alexa Fluor 568 (Invitrogen). The double IF was carried out sequentially.
For IF of Smad1 and Smad3 phospho-tail and phopsho-linker in cell lines, HaCaT cells were fixed in 4% Paraformaldehyde and immunostained with the indicated antibodies as described previously (Xu et al., 2002).
Drosophila genetics
Flies of the genotype y w hs-Flp; vgQE-lacZ/+; Act>CD2>Gal4, UAS-GFP/UAS-Yorkie were heat shocked at 48–60 hr after egg deposition to induce Yorkie-overexpressing clones in imaginal discs. UAS-Yorkie and vgQE-lacZ were described in (Huang et al., 2005; Kim et al., 1997), respectively. Confocal images were collected on a Zeiss 510 microscope and processed using the Zeiss LSM Image software.
Other assays
Immunoprecipitations, western immunoblotting and kinase assays were done as previously described (Sapkota et al., 2007). Chromatin immunoprecipitations were performed with a ChIP kit (Upstate Biotechnology Inc.) following the manufacturer’s protocol with modifications and details described in the Supplementary Methods. CDK9/CyclinT and CDK8/CyclinC complexes were purchased from Invitrogen and CDK7/CyclinH was a gift from Dr. R. P. Fisher (MSKCC). Total RNA extraction, reverse transcription and quantitative real-time PCR to detect gene transcript levels, were performed as previously described (Knockaert et al., 2006). Primers used in qRT-PCR analysis are listed in Table S2. For microarray analysis duplicate RNA samples were extracted from E14Tg2a.IV cells treated with BMP2 for 1 h and untreated control cells (Accession #: GSE17896). Array hybridization, and data analysis was performed as previously described (Xi et al., 2008) using the MOE 430 2.0 mouse platform.
Supplementary Material
Acknowledgments
We thank M. Yaffe, R.P. Fisher and F. Liu for reagents; S. Malik, S. Tavazoie, D. Nguyen, S. Keeney and R. Benezra for helpful discussions; X.H.-F. Zhang for valuable help with microarray analysis; E. Lai for providing the S2 Cells; and M. Squatrito, M. Tavazoie, E. Montalvo, P.Y. Chen and N. Fan for technical support. This work was supported by NIH grants CA34610 (JM), EY015708 (DP) and BFU2008-02795 (MJM). JM and DP are investigators of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Affolter M, Basler K. The Decapentaplegic morphogen gradient: from pattern formation to growth regulation. Nat Rev Genet. 2007;8:663–674. doi: 10.1038/nrg2166. [DOI] [PubMed] [Google Scholar]
- Alvarez-Buylla A, Lim DA. For the long run: maintaining germinal niches in the adult brain. Neuron. 2004;41:683–686. doi: 10.1016/s0896-6273(04)00111-4. [DOI] [PubMed] [Google Scholar]
- Aubin J, Davy A, Soriano P. In vivo convergence of BMP and MAPK signaling pathways: impact of differential Smad1 phosphorylation on development and homeostasis. Genes Dev. 2004;18:1482–1494. doi: 10.1101/gad.1202604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blain SW, Massagué J. Different sensitivity of the transforming growth factor-beta cell cycle arrest pathway to c-Myc and MDM-2. J Biol Chem. 2000;275:32066–32070. doi: 10.1074/jbc.M006496200. [DOI] [PubMed] [Google Scholar]
- Bres V, Yoh SM, Jones KA. The multi-tasking P-TEFb complex. Curr Opin Cell Biol. 2008;20:334–340. doi: 10.1016/j.ceb.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Pfaff SL, Gage FH. YAP regulates neural progenitor cell number via the TEA domain transcription factor. Genes Dev. 2008 doi: 10.1101/gad.1726608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi Y, Huddleston MJ, Zhang X, Young RA, Annan RS, Carr SA, Deshaies RJ. Negative regulation of Gcn4 and Msn2 transcription factors by Srb10 cyclin-dependent kinase. Genes Dev. 2001;15:1078–1092. doi: 10.1101/gad.867501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donner AJ, Szostek S, Hoover JM, Espinosa JM. CDK8 is a stimulus-specific positive coregulator of p53 target genes. Mol Cell. 2007;27:121–133. doi: 10.1016/j.molcel.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand LO, Advani SJ, Poon AP, Roizman B. The carboxyl-terminal domain of RNA polymerase II is phosphorylated by a complex containing cdk9 and infected-cell protein 22 of herpes simplex virus 1. J Virol. 2005;79:6757–6762. doi: 10.1128/JVI.79.11.6757-6762.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA. From cell structure to transcription: Hippo forges a new path. Cell. 2006;124:267–273. doi: 10.1016/j.cell.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Fabian MA, Biggs WH, 3rd, Treiber DK, Atteridge CE, Azimioara MD, Benedetti MG, Carter TA, Ciceri P, Edeen PT, Floyd M, et al. A small molecule-kinase interaction map for clinical kinase inhibitors. Nat Biotechnol. 2005;23:329–336. doi: 10.1038/nbt1068. [DOI] [PubMed] [Google Scholar]
- Feng XH, Derynck R. Specificity and versatility in TGF-β signaling through Smads. Annu Rev Cell Dev Biol. 2005;21:659–693. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- Firestein R, Bass AJ, Kim SY, Dunn IF, Silver SJ, Guney I, Freed E, Ligon AH, Vena N, Ogino S, et al. CDK8 is a colorectal cancer oncogene that regulates beta-catenin activity. Nature. 2008;455:547–551. doi: 10.1038/nature07179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryer CJ, White JB, Jones KA. Mastermind recruits CycC:CDK8 to phosphorylate the Notch ICD and coordinate activation with turnover. Mol Cell. 2004;16:509–520. doi: 10.1016/j.molcel.2004.10.014. [DOI] [PubMed] [Google Scholar]
- Fuentealba LC, Eivers E, Ikeda A, Hurtado C, Kuroda H, Pera EM, De Robertis EM. Integrating patterning signals: Wnt/GSK3 regulates the duration of the BMP/Smad1 signal. Cell. 2007;131:980–993. doi: 10.1016/j.cell.2007.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S, Alarcón C, Sapkota G, Rahman S, Chen P-Y, Goerner N, Macias M, Erdjument-Bromage H, Tempst P, Massagué J. Ubiquitin ligase Nedd4L targets CDK8/9-activated Smad2/3 to end TGFβ signaling. 2009 doi: 10.1016/j.molcel.2009.09.043. (submitted for publication) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomis RR, Alarcón C, He W, Wang Q, Seoane J, Lash A, Massagué J. A FoxO-Smad synexpression group in human keratinocytes. Proc Natl Acad Sci USA. 2006;103:12747–12752. doi: 10.1073/pnas.0605333103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm OH, Gurdon JB. Nuclear exclusion of Smad2 is a mechanism leading to loss of competence. Nat Cell Biol. 2002;4:519–522. doi: 10.1038/ncb812. [DOI] [PubMed] [Google Scholar]
- He W, Dorn DC, Erdjument-Bromage H, Tempst P, Moore MA, Massagué J. Hematopoiesis controlled by distinct TIF1gamma and Smad4 branches of the TGFβ pathway. Cell. 2006;125:929–941. doi: 10.1016/j.cell.2006.03.045. [DOI] [PubMed] [Google Scholar]
- Hollnagel A, Oehlmann V, Heymer J, Ruther U, Nordheim A. Id genes are direct targets of bone morphogenetic protein induction in embryonic stem cells. J Biol Chem. 1999;274:19838–19845. doi: 10.1074/jbc.274.28.19838. [DOI] [PubMed] [Google Scholar]
- Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Ingham RJ, Gish G, Pawson T. The Nedd4 family of E3 ubiquitin ligases: functional diversity within a common modular architecture. Oncogene. 2004;23:1972–1984. doi: 10.1038/sj.onc.1207436. [DOI] [PubMed] [Google Scholar]
- Inman GJ, Nicolas FJ, Hill CS. Nucleocytoplasmic shuttling of Smads 2, 3, and 4 permits sensing of TGF-β receptor activity. Mol Cell. 2002;10:283–294. doi: 10.1016/s1097-2765(02)00585-3. [DOI] [PubMed] [Google Scholar]
- Keller GM. In vitro differentiation of embryonic stem cells. Curr Opin Cell Biol. 1995;7:862–869. doi: 10.1016/0955-0674(95)80071-9. [DOI] [PubMed] [Google Scholar]
- Kim J, Johnson K, Chen HJ, Carroll S, Laughon A. Drosophila Mad binds to DNA and directly mediates activation of vestigial by Decapentaplegic. Nature. 1997;388:304–308. doi: 10.1038/40906. [DOI] [PubMed] [Google Scholar]
- Knockaert M, Sapkota G, Alarcón C, Massagué J, Brivanlou AH. Unique players in the BMP pathway: small C-terminal domain phosphatases dephosphorylate Smad1 to attenuate BMP signaling. Proc Natl Acad Sci USA. 2006;103:11940–11945. doi: 10.1073/pnas.0605133103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar M, Doody J, Massagué J. Opposing BMP and EGF signalling pathways converge on the TGF-β family mediator Smad1. Nature. 1997;389:618–622. doi: 10.1038/39348. [DOI] [PubMed] [Google Scholar]
- Kretzschmar M, Doody J, Timokhina I, Massagué J. A mechanism of repression of TGFβ/Smad signaling by oncogenic Ras. Genes Dev. 1999;13:804–816. doi: 10.1101/gad.13.7.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Duan X, Liang YY, Su Y, Wrighton KH, Long J, Hu M, Davis CM, Wang J, Brunicardi FC, et al. PPM1A functions as a Smad phosphatase to terminate TGFβ signaling. Cell. 2006;125:915–928. doi: 10.1016/j.cell.2006.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo RS, Massagué J. Ubiquitin-dependent degradation of TGF-β-activated smad2. Nat Cell Biol. 1999;1:472–478. doi: 10.1038/70258. [DOI] [PubMed] [Google Scholar]
- Malik S, Roeder RG. Transcriptional regulation through Mediator-like coactivators in yeast and metazoan cells. Trends Biochem Sci. 2000;25:277–283. doi: 10.1016/s0968-0004(00)01596-6. [DOI] [PubMed] [Google Scholar]
- Malik S, Roeder RG. Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem Sci. 2005;30:256–263. doi: 10.1016/j.tibs.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Martin-Castellanos C, Edgar BA. A characterization of the effects of Dpp signaling on cell growth and proliferation in the Drosophila wing. Development. 2002;129:1003–1013. doi: 10.1242/dev.129.4.1003. [DOI] [PubMed] [Google Scholar]
- Massagué J. TGF-β signal transduction. Annu Rev Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- Massagué J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- Matsuura I, Denissova NG, Wang G, He D, Long J, Liu F. Cyclin-dependent kinases regulate the antiproliferative function of Smads. Nature. 2004;430:226–231. doi: 10.1038/nature02650. [DOI] [PubMed] [Google Scholar]
- McClue SJ, Blake D, Clarke R, Cowan A, Cummings L, Fischer PM, MacKenzie M, Melville J, Stewart K, Wang S, et al. In vitro and in vivo antitumor properties of the cyclin dependent kinase inhibitor CYC202 (R-roscovitine) Int J Cancer. 2002;102:463–468. doi: 10.1002/ijc.10738. [DOI] [PubMed] [Google Scholar]
- Moreno E, Basler K, Morata G. Cells compete for decapentaplegic survival factor to prevent apoptosis in Drosophila wing development. Nature. 2002;416:755–759. doi: 10.1038/416755a. [DOI] [PubMed] [Google Scholar]
- Morris EJ, Ji JY, Yang F, Di Stefano L, Herr A, Moon NS, Kwon EJ, Haigis KM, Naar AM, Dyson NJ. E2F1 represses beta-catenin transcription and is antagonized by both pRB and CDK8. Nature. 2008;455:552–556. doi: 10.1038/nature07310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omura-Minamisawa M, Diccianni MB, Batova A, Chang RC, Bridgeman LJ, Yu J, de Wit E, Kung FH, Pullen JD, Yu AL. In vitro sensitivity of T-cell lymphoblastic leukemia to UCN-01 (7-hydroxystaurosporine) is dependent on p16 protein status: a Pediatric Oncology Group study. Cancer Res. 2000;60:6573–6576. [PubMed] [Google Scholar]
- Overholtzer M, Zhang J, Smolen GA, Muir B, Li W, Sgroi DC, Deng CX, Brugge JS, Haber DA. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc Natl Acad Sci USA. 2006;103:12405–12410. doi: 10.1073/pnas.0605579103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D. Hippo signaling in organ size control. Genes Dev. 2007;21:886–897. doi: 10.1101/gad.1536007. [DOI] [PubMed] [Google Scholar]
- Pera EM, Ikeda A, Eivers E, De Robertis EM. Integration of IGF, FGF, and anti-BMP signals via Smad1 phosphorylation in neural induction. Genes Dev. 2003;17:3023–3028. doi: 10.1101/gad.1153603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- Reynisdottir I, Massagué J. The subcellular locations of p15(Ink4b) and p27(Kip1) coordinate their inhibitory interactions with cdk4 and cdk2. Genes Dev. 1997;11:492–503. doi: 10.1101/gad.11.4.492. [DOI] [PubMed] [Google Scholar]
- Sapkota G, Alarcón C, Spagnoli FM, Brivanlou AH, Massagué J. Balancing BMP signaling through integrated inputs into the Smad1 linker. Mol Cell. 2007;25:441–454. doi: 10.1016/j.molcel.2007.01.006. [DOI] [PubMed] [Google Scholar]
- Shapiro GI. Preclinical and clinical development of the cyclin-dependent kinase inhibitor flavopiridol. Clin Cancer Res. 2004;10:4270s–4275s. doi: 10.1158/1078-0432.CCR-040020. [DOI] [PubMed] [Google Scholar]
- Tyler DM, Li W, Zhuo N, Pellock B, Baker NE. Genes affecting cell competition in Drosophila. Genetics. 2007;175:643–657. doi: 10.1534/genetics.106.061929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga AC, Wrana JL. The disparate role of BMP in stem cell biology. Oncogene. 2005;24:5713–5721. doi: 10.1038/sj.onc.1208919. [DOI] [PubMed] [Google Scholar]
- Wu S, Huang J, Dong J, Pan D. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell. 2003;114:445–456. doi: 10.1016/s0092-8674(03)00549-x. [DOI] [PubMed] [Google Scholar]
- Xi Q, He W, Zhang XH, Le HV, Massagué J. Genome-wide impact of the BRG1 SWI/SNF chromatin remodeler on the transforming growth factor beta transcriptional program. J Biol Chem. 2008;283:1146–1155. doi: 10.1074/jbc.M707479200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z, Watson N, Rodriguez C, Lodish HF. Nucleocytoplasmic shuttling of Smad1 conferred by its nuclear localization and nuclear export signals. J Biol Chem. 2001;276:39404–39410. doi: 10.1074/jbc.M103117200. [DOI] [PubMed] [Google Scholar]
- Xu L, Kang Y, Col S, Massagué J. Smad2 nucleocytoplasmic shuttling by nucleoporins CAN/Nup214 and Nup153 feeds TGFβ signaling complexes in the cytoplasm and nucleus. Mol Cell. 2002;10:271–282. doi: 10.1016/s1097-2765(02)00586-5. [DOI] [PubMed] [Google Scholar]
- Ying QL, Nichols J, Chambers I, Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003a;115:281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- Ying QL, Stavridis M, Griffiths D, Li M, Smith A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat Biotechnol. 2003b;21:183–186. doi: 10.1038/nbt780. [DOI] [PubMed] [Google Scholar]
- Zeng Q, Hong W. The emerging role of the hippo pathway in cell contact inhibition, organ size control, and cancer development in mammals. Cancer Cell. 2008;13:188–192. doi: 10.1016/j.ccr.2008.02.011. [DOI] [PubMed] [Google Scholar]
- Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.