Abstract
Background
Endogenous internal controls (‘reference’ or ‘housekeeping’ genes) are widely used in real-time PCR (RT-PCR) analyses. Their use relies on the premise of consistently stable expression across studied experimental conditions. Unfortunately, none of these controls fulfills this premise across a wide range of experimental conditions; consequently, none of them can be recommended for universal use.
Methods
To determine which endogenous RT-PCR controls are suitable for analyses of renal tissues altered by kidney disease, we studied the expression of 16 commonly used ‘reference genes’ in 7 mildly and 7 severely affected whole kidney tissues from a well-characterized cystic kidney disease model. Expression levels of these 16 genes, determined by TaqMan® RT-PCR analyses and Affymetrix GeneChip® arrays, were normalized and tested for overall variance and equivalence of the means.
Results
Both statistical approaches and both TaqMan- and GeneChip-based methods converged on 3 out of the 4 top-ranked genes (Ppia, Gapdh and Pgk1) that had the most constant expression levels across the studied phenotypes.
Conclusion
A combination of the top-ranked genes will provide a suitable endogenous internal control for similar studies of kidney tissues across a wide range of disease severity.
Key Words: Real-time PCR, Kidney disease, Endogenous internal control, Reference gene, Housekeeping gene
Introduction
Real-time PCR (RT-PCR) has become a new standard for quantitative gene expression analyses. However, the accuracy of this technique depends on the quality of internal controls. Endogenous internal controls, often referred to as ‘housekeeping’ or ‘reference’ genes, offer multiple practical advantages and are therefore widely used in research and clinical applications. Their use, however, relies on the premise that these genes are expressed at consistently stable levels across all experimental conditions under investigation. Unfortunately, none of the endogenous controls was found to be constantly expressed across different tissues, developmental stages, or pathological and study conditions [1, 2].
The suitability of individual ‘reference genes’ as internal RT-PCR controls for a specific experimental design can be tested with various approaches, including methods of overall variance [3], ANOVA models [4], Bayesian models [5] and equivalence testing [6, 7]. More recently, genome-wide expression technologies have been recognized as an effective tool for the identification and evaluation of endogenous RT-PCR controls [8, 9]. Even more accurate RT-PCR controls could be obtained by accounting for the expression of multiple genes, e.g. using a method of a geometric mean that is the basis of the GeNorm [10] and BestKeeper programs [11].
In the case of renal tissues, 18S rRNA and the cyclophilin A-encoding gene Ppia (but not Gapdh) have been recommended as the preferred RT-PCR controls for studies of the renal tubulointerstitial compartment, based on analyses of these 3 genes in renal biopsies [12]. In the current study, we used TaqMan® and Affymetrix GeneChip® assays to compare the expression stability of 16 commonly used RT-PCR controls in whole kidneys affected by mild and severe cystic kidney disease from an extensively characterized animal model [13, 14].
Materials and Methods
Gene Expression Profiling
RNA and cDNA were prepared previously [15] from whole kidneys harvested from 7 mildly and 7 severely affected 10-day-old F2 mice generated in a (C57BL/6J-cpk/+ × CAST)F1 intercross (cystic disease severity was defined by kidney length, weight and volume; e.g. average kidney volume was more than 8 times higher among the 7 highly vs. 7 mildly affected mice) [16]. These 14 samples were used in triplicates for gene expression analyses using 16 TaqMan assays arranged into a mouse endogenous control low-density array [Applied Biosystems, Foster City, Calif., USA (online suppl. table 1, for online supplementary material, see www.karger.com/doi/10.1159/000235993)]. CT values were determined with the 7900 HT Real-Time PCR System using SDS 2.1 RQ software. All statistical analyses were based on CT values.
The genome-wide expression data were generated previously with 14 Affymetrix GeneChip® Mouse Genome 430 2.0 arrays (Affymetrix Inc., Santa Clara, Calif., USA; 7 biological replicates for mild and 7 for severe cystic kidney phenotype) [15]. Large-scale validation of these gene expression data with 14 low-density Affymetrix U74Av2 arrays provided a formal technical validation of the 430 2.0 data (gene expression correlated strongly across these 2 array platforms; r = 0.72 for all genes, r = 0.90 for differentially expressed genes with p < 0.05 after adjusting for multiple testing) [15].
Statistical Analyses
Normalization of CT values was performed by subtracting the sample mean from each value. This mean-centering adjustment removes potential RNA loading differences from sample to sample. The assumption for this normalization is that the total signals from all studied ‘reference genes’ are the same across all samples.
The preprocessing of the microarray data analysis was performed as previously described [15]. Data from all probe sets corresponding to the 16 endogenous controls were examined for appropriateness of inclusion in this study. The probe sets were excluded from further analyses if they matched alternative transcripts or represented ‘rare EST events’ according to the UCSC Genome Browser. Each of the remaining probe sets (online suppl. table 1) was used for all subsequent analyses.
Gene expression equivalence of the 16 genes was tested with the two one-sided t tests (TOST) procedure [17] that has been used in bioequivalence testing of RT-PCR data [6, 7]. The p values were derived based on the normal distribution assumption. However, the ranking was not affected by the normal assumption. For the RT-PCR data, 1 technical replicate was arbitrarily chosen from each sample for the equivalence test. For the microarray data, each probe set was analyzed separately. The median p value from each gene was used for overall gene ranking. In contrast to the previously utilized 2-fold change threshold for the equivalence test [6], we used a more stringent 1.5-fold change threshold since a 2-fold change is often assumed to be differentially expressed and not appropriate to consider as a ‘reference gene’.
The overall variance analysis was conducted on the same data in the same fashion as the equivalence test except that the overall variances of all samples were computed for each gene.
Results
Renal expression levels of 16 commonly used endogenous controls for RT-PCR analyses were determined in 2 groups of age-matched cpk mice generated in a (C57BL/6J-cpk × CAST/Ei)F1 intercross [15]: 7 mice with the mildest cystic kidney phenotype and 7 mice with the most severe cystic kidney phenotype. The expression levels were determined in triplicates by TaqMan®-based RT-PCR assays (online suppl. table 1). These 16 genes were also included in our previously conducted microarray experiments with Affymetrix GeneChip® arrays in these renal tissues [15]. Comparisons of data generated by these 2 distinct gene expression platforms allowed an independent validation of these 2 approaches.
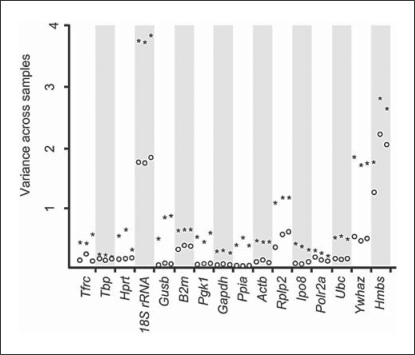
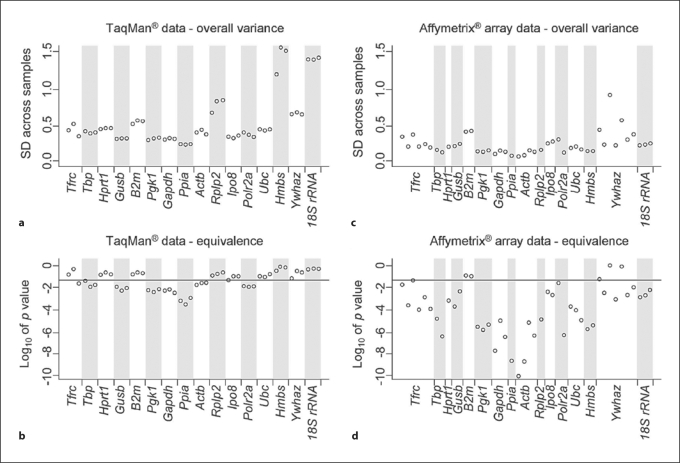
First, normalization of the TaqMan®-based RT-PCR data was performed for all samples by subtracting the mean of the CT values from each CT value obtained from a specific sample. This normalization was applied to remove RNA loading differences across samples. Compared to original CT values, this approach successfully reduced variances in the expression of all studied genes, although to various extents (fig. 1). Then, we ranked the appropriateness of the 16 ‘reference genes’ as endogenous internal RT-PCR controls based on their overall variance, with smaller variance indicating higher gene expression stability. The overall variance, a basis of a simple, yet highly effective, tool for the identification of ‘reference genes’ [3], was calculated for the normalized CT values of each of the 3 technical replicates. The overall variances were compared across the 16 ‘reference genes’ to identify optimal RT-PCR controls with the most stable gene expression (smallest overall variance) across all kidney samples (fig. 2a). For example, 18S rRNA and Hmbs showed the largest variation for all 3 replicates, indicating that these genes represented suboptimal endogenous controls in this experimental setup. In contrast, Ppia, Gapdh and Pgk1 showed the smallest variation and would serve as the more appropriate endogenous internal RT-PCR controls. However, the overall variance method does not separate for the between-group variance (i.e. severe and mild kidney disease) from the within-group variance. In other words, a small overall variance does not necessarily imply small fold changes between the 2 groups. A small overall variance could mean extremely small within-group variance with relatively large fold changes between the groups. Since our goal is to identify ‘reference genes’ that do not change expression levels between study groups, it is desirable to test for the equivalence of the means of the gene expressions between groups for each gene. We applied the TOST [17] to determine gene expression equivalence of our TaqMan® data and ranked the studied genes according to the test statistics of the TOST. The outcomes of these analyses (fig. 2b) closely resembled the ranking of optimal endogenous RT-PCR controls based on overall variance analysis (fig. 2a). However, only 7 of the 16 studied ‘reference genes’ (Ppia, Gapdh, Pgk1, Gusb, Polr2a, Actb, Tbp) remained significantly within the 1.5-fold limit for expression difference between the studied groups (i.e. mildly and severely affected kidneys), with p < 0.05.
Fig. 1.
Normalization reduces overall variance of CT values. This mean-centering adjustment was performed to remove potential RNA loading differences across samples. For each gene, CTvariance across all studied samples was calculated for each of the 3 technical replicates before (★) and after (○) normalization (each point represents a single replicate of data from 7 mildly and 7 severely affected kidneys). The normalization reduced overall variance of CT values for all genes, although at different rates for specific genes.
Fig. 2.
Gene expression stability of commonly used endogenous RT-PCR controls. Gene expression stability based on TaqMan® RT-PCR data (a, b) and Affymetrix GeneChip® data (c, d) was evaluated with the method of ‘total variance’ that is expressed as the standard deviation (SD) across all samples (a, c) and the TOST method that measures gene expression equivalence across all samples (b, d). For the TaqMan® analyses (a, b), each point represents data from a single technical replicate from all studied kidneys (3 replicates per gene). For the GeneChip® analyses (c, d), each point designates data from all studied kidneys corresponding to one of the multiple probe sets that represent a specific gene on the GeneChip® array. The horizontal line in the lower panels represents gene-specific p values (0.05) for a 1.5-fold equivalence threshold. Genes with favorable endogenous control profiles have consistently low overall variance and high significance of equivalence (e.g. Ppia, Gapdh and Pgk1), unlike controls with less suitable expression (e.g. B2m and Ywhaz).
While both Affymetrix GeneChip® and TaqMan® analyses used the same 7 biological replicas for mild phenotypes and the same 7 for severe phenotypes, true technical replicates were generated only for the TaqMan® analyses (fig. 2a, b; each gene is represented by 3 technical replicates). In contrast, genes on Affymetrix GeneChip® arrays are represented by a variable number of probe sets (fig. 2c, d), each consisting of eleven 25-mer probe pairs. The evaluation outcomes and ranking of the Affymetrix GeneChip array data closely resemble the TaqMan®-based analyses (fig. 2). Importantly, the top-ranked endogenous internal RT-PCR controls (e.g. Ppia, Gapdh and Pgk1) were top-ranked by both the TaqMan® and Affymetrix GeneChip® platforms. However, some of the lower-ranked genes differed between these platforms. For example, 18S rRNA, Hmbs and Rplp2 showed large variation in the TaqMan® data, but were relatively constant in the Affymetrix GeneChip® data.
Discussion
Our analyses of mildly and severely affected whole kidney tissues using 2 different statistical methods and 2 gene expression platforms identified consistently 3 ‘reference genes’ with the highest equivalence and lowest variability in expression: Ppia, Gapdh and Pgk1. In the case of Ppia, a cyclophilin A-encoding gene, our data are consistent with a previous recommendation of Ppia as an endogenous control for expression studies involving the renal tubulointerstitial compartment [12]. In contrast, Gapdh, one of the most commonly used endogenous controls, was recommended as a control for dissected glomeruli [18], but not for a microdissected renal tubulointerstitial compartment [12]. A comprehensive evaluation of the appropriateness of Pgk1 as an internal RT-PCR control for renal tissues has not been described before.
We also show that expression of 18S rRNA, one of the two recommended endogenous expression controls for the renal tubulointerstitial compartment [12], is relatively constant, although poorly ranked, in GeneChip® analyses. However, this gene was highly variably expressed in TaqMan® analyses. Although this discrepancy could be specific to our experiment design or to assay-specific factors, it casts doubt on the appropriateness of 18S rRNA as a universal endogenous control for RT-PCR studies involving whole cystic kidneys.
Although the use of a gene encoding β-actin (Actb) as an endogenous internal control for RT-PCR analyses is controversial [19, 20], it is commonly used as such a control in studies of renal tissues. In the current study, the expression of Actb was moderately stable among the 16 endogenous controls in TaqMan® analyses and it was the second most stable gene in GeneChip® analyses (fig. 2 and online suppl. table 2).
Similar to Actb, expression stability of other moderately-ranked genes was very similar and any of these genes would perform well as an endogenous RT-PCR control in this setting. However, genes in the bottom half of the rankings showed large expression variability and should be avoided as RT-PCR controls in affected kidney tissues.
It is important to recognize that the highest accuracy of RT-PCR analyses can be achieved with multiple internal control genes [10, 11]. However, the use of several genes that belong to the same functional class (e.g. glycolysis in cases of Gapdh and Pgk1) should be avoided to decrease potential effects of their co-regulation.
In summary, to evaluate the appropriateness of the 16 commonly used ‘reference genes’ as endogenous internal RT-PCR controls in mildly and severely affected kidney tissues, we determined expression levels of these genes in age-matched cpk kidneys with extreme rates of cystic kidney disease progression. Analyses of overall gene expression variability and gene expression equivalence based on TaqMan® as well as Affymetrix GeneChip® data, yielded 3 top-ranked genes (Ppia, Gapdh and Pgk1). The top-ranked genes, or indexes reflecting their geometric means, may represent a suitable endogenous internal control for gene expression studies involving similarly affected kidney tissues. However, this study requires further validation in other kidney disease models.
Supplementary Material
Endogenous RT-PCR controls and their corresponding TaqMan© assay and Affymetrix© probeset IDs
Ranking of variance and equivalence of RT-PCR controls
Acknowledgements
We thank Dr. Lisa M. Guay-Woodford for providing partial resources for this study. This work was supported by the Polycystic Kidney Disease Foundation Grant-In-Aid (M.M.), and Dr. Cui was supported in part by the UAB-UCSD O’Brien Center (1P30 DK079337).
References
- 1.Huggett J, Dheda K, Bustin S, Zumla A. Real-time RT-PCR normalisation: strategies and considerations. Genes Immun. 2005;6:279–284. doi: 10.1038/sj.gene.6364190. [DOI] [PubMed] [Google Scholar]
- 2.Blanquicett C, Johnson MR, Heslin M, Diasio RB. Housekeeping gene variability in normal and carcinomatous colorectal and liver tissues: applications in pharmacogenomic gene expression studies. Anal Biochem. 2002;303:209–214. doi: 10.1006/abio.2001.5570. [DOI] [PubMed] [Google Scholar]
- 3.Szabo A, Perou CM, Karaca M, et al. Statistical modeling for selecting housekeeper genes. Genome Biol. 2004;5:R59. doi: 10.1186/gb-2004-5-8-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brunner AM, Yakovlev IA, Strauss SH. Validating internal controls for quantitative plant gene expression studies. BMC Plant Biol. 2004;4:14. doi: 10.1186/1471-2229-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 6.Haller F, Kulle B, Schwager S, et al. Equivalence test in quantitative reverse transcription polymerase chain reaction: confirmation of reference genes suitable for normalization. Anal Biochem. 2004;335:1–9. doi: 10.1016/j.ab.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 7.Huang Y, Hsu JC, Peruggia M, Scott AA. Statistical selection of maintenance genes for normalization of gene expressions. Stat Appl Genet Mol Biol. 2006;5 doi: 10.2202/1544-6115.1122. Article4. [DOI] [PubMed] [Google Scholar]
- 8.Maccoux LJ, Clements DN, Salway F, Day PJ. Identification of new reference genes for the normalisation of canine osteoarthritic joint tissue transcripts from microarray data. BMC Mol Biol. 2007;8:62. doi: 10.1186/1471-2199-8-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodriguez-Lanetty M, Phillips WS, Dove S, Hoegh-Guldberg O, Weis VM. Analytical approach for selecting normalizing genes from a cDNA microarray platform to be used in q-RT-PCR assays: a cnidarian case study. J Biochem Biophys Methods. 2008;70:985–991. doi: 10.1016/j.jbbm.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 10.Vandesompele J, De Preter K, Pattyn F, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper – Excel-based tool using pair-wise correlations. Biotechnol Lett. 2004;26:509–515. doi: 10.1023/b:bile.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- 12.Schmid H, Cohen CD, Henger A, Irrgang S, Schlondorff D, Kretzler M. Validation of endogenous controls for gene expression analysis in microdissected human renal biopsies. Kidney Int. 2003;64:356–360. doi: 10.1046/j.1523-1755.2003.00074.x. [DOI] [PubMed] [Google Scholar]
- 13.Fry J, Koch W, Jennette J, McFarland E, Fried F, Mandell J. A genetically determined murine model of infantile polycystic kidney disease. J Urol. 1985;134:828–833. doi: 10.1016/s0022-5347(17)47448-9. [DOI] [PubMed] [Google Scholar]
- 14.Preminger G, Koch W, Fried F, McFarland E, Murphy E, Mandell J. Murine congenital polycystic kidney disease: a model for studying development of cystic disease. J Urol. 1982;127:556–560. doi: 10.1016/s0022-5347(17)53911-7. [DOI] [PubMed] [Google Scholar]
- 15.Mrug M, Zhou J, Woo Y, et al. Overexpression of innate immune response genes in a model of recessive polycystic kidney disease. Kidney Int. 2008;73:63–76. doi: 10.1038/sj.ki.5002627. [DOI] [PubMed] [Google Scholar]
- 16.Mrug M, Li R, Cui X, Schoeb TR, Churchill GA, Guay-Woodford LM. Kinesin family member 12 is a candidate polycystic kidney disease modifier in the cpk mouse. J Am Soc Nephrol. 2005;16:905–916. doi: 10.1681/ASN.2004121083. [DOI] [PubMed] [Google Scholar]
- 17.Schuirmann DJ. A comparison of the two one-sided tests procedure and the power approach for assessing the equivalence of average bioavailability. J Pharmacokinet Biopharm. 1987;15:657–680. doi: 10.1007/BF01068419. [DOI] [PubMed] [Google Scholar]
- 18.Ceol M, Del Prete D, Tosetto E, et al. GAPDH as housekeeping gene at renal level. Kidney Int. 2004;65:1972–1973. doi: 10.1111/j.1523-1755.2004.607_7.x. author reply 1973–1974. [DOI] [PubMed] [Google Scholar]
- 19.Schmittgen TD, Zakrajsek BA. Effect of experimental treatment on housekeeping gene expression: validation by real-time, quantitative RT-PCR. J Biochem Biophys Methods. 2000;46:69–81. doi: 10.1016/s0165-022x(00)00129-9. [DOI] [PubMed] [Google Scholar]
- 20.Selvey S, Thompson EW, Matthaei K, Lea RA, Irving MG, Griffiths LR. Beta-actin – an unsuitable internal control for RT-PCR. Mol Cell Probes. 2001;15:307–311. doi: 10.1006/mcpr.2001.0376. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Endogenous RT-PCR controls and their corresponding TaqMan© assay and Affymetrix© probeset IDs
Ranking of variance and equivalence of RT-PCR controls