Abstract
Background/Aims
Patients with anti-glomerular basement membrane diseases produce pathogenic autoantibodies (autoAb) that deposit in the kidney and initiate severe inflammation. Restricted antigenic specificity of the autoAb against 2 regions (with related sequences) within α3(IV)NC1, along with shared idiotypes (i.e. structural determinants), among pathogenic human autoAb suggested that common genetic elements encode the autoAb. The aim of this study was to determine whether the idiotypic relatedness of the autoAb was due to the fact that unique and similar genes were used to encode them, divergent genes were used to produce Ab with similar Ag-binding properties and conformation, or if other mechanisms were operative.
Methods
The encoding V gene sequences of pathogenic human anti-α3(IV)NC1 Ab, derived following immunization of XenoMice® which produce human but not murine IgG, with α3(IV)NC1 were determined. Predicted conformations of autoAb–α3(IV)NC1 interactions were derived using the Ab sequences and molecularmodels of the α3(IV)NC1 structure.
Results
The pathogenic Ab were encoded by multiple, common VH and VL gene families indicating that they were not encoded by a unique subset of genes and that normal individuals have the capacity to produce them. However, modeling of the Ag–Ab interactions suggested that although the contact regions varied for individual Ab, the optimized energy constraints facilitate interaction of both Ab-binding regions with pathogenically relevant epitopes on α3(IV)NC1.
Conclusions
The results suggest that the repetitive nature and relatedness of the α3(IV)NC1 antigenic epitopes facilitate cross-linking of pathogenic Ab, in vivo, by allowing both IgG Fab to bind to the basement membrane. This most likely accounts for the high-affinity Ab binding we and others observed among human anti-α3(IV)NC1 Ab. Based on these observations, we postulate that this interaction provides for the stability of the Ab interaction, resulting in a high-affinity interaction that serves as an ideal scaffold for optimal FcR engagement and complement activation, thereby accelerating inflammation and contributing to the rapidly progressive nature of this disease.
Key Words: Glomerulonephritis, Anti-glomerular basement membrane disease, α3(IV) collagen
Background/Aims
Spontaneous production of human anti-glomerular basement membrane (anti-GBM) Ab often results in rapidly progressive glomerulonephritis and renal failure [1]. A unique feature of this disease is that pathogenic Ab bind to specific regions within the GBM (the noncollagenous domain (NC1) of α3(IV) collagen). Two major epitopes, termed EA (C2) and EB (C6), have been defined and are variably recognized by all patient sera, with primary sequence homology among them [1]. Anti-GBM Ab derived from the sera of different patients share a common idiotype (termed Id-GBM) [2] and they are uniformly of high affinity [3]. Taken together, the results raise the possibility that similar and/or unique genes encode them. Herein, the basis for these observations was examined. The encoding (primary) V gene sequences of pathogenic, human monoclonal anti-α3(IV) Ab, derived following immunization of XenoMice® (an engineered strain that produces human but not murine Ab), were determined, with the sequences used to generate best fit models of Ab–Ag (antigen) interactions with α3(IV). We found that different genes were used to encode the autoantibodies (autoAb); however, best fit conformations of the interactions suggested that multiple contact regions of individual Ab contribute to high affinity.
Methods
Production of Human mAb and Evaluation of mAb Activity
XenoMouse® animals (Abgenix Inc., Fremont, Calif., USA), hyperimmunized with either Escherichia coli-expressed, human 293 cell-expressed or bovine-derived α3(IV)NC1 collagen [2, 3, 7], were sacrificed, and single cell suspensions of splenocytes were fused to the myeloma fusion partner Sp2 mIL6 [10]. Supernatants were tested (ELISA and Western blot) for Ab activity, and positive wells were subcloned (>2×) and selected. mAb were purified using Protein A/G (Pierce, Rockford, Ill., USA). To approximate the epitope-specificity of the mAb, a competitive ELISA was utilized by evaluating the capacity of soluble peptides: C2 corresponding to epitope EA (a.a. sequence: TAIPSCPEGTVPLYS), soluble peptide C6 corresponding to EB (a.a. sequence: TDIPPCPHGWISLWK) or an irrelevant control peptide (a.a. sequence RRVPHFYHFAPVIAR) to inhibit Ab binding [9]. Similarly, a competition ELISA was used to determine if human anti-GBM Ab and XenoMouse II mAb bound to similar epitopes on human α3(IV)NC1, using purified and biotinylated mAb.
In vivo Analysis of Human Anti-α3(IV) mAb
Previously described methods were utilized; briefly, affinity-purified, pathogen-free, human mAb (1–2 mg) were administered intravenously to either XenoMouse II or C57Bl/6 mice. After 5 days, 24-hour Upr, BUN and serum creatinine were determined, and kidneys were examined and graded by light microscopy for severity of nephritis and direct immunoflourescence for human IgG deposition [10].
Sequence Analysis
All sequencing reactions were performed using ABI 377 and 373A automated sequencers with Taq FS Big Dye™ Terminator or Dye Primer chemistry. The PCR heavy and light chain products of the mAb were ligated into the TOPO vector (Invitrogen, Carlsbad, Calif., USA), transformed into chemically competent TOP10 One Shot® cells and grown on LB agar/kanamycin plates. Individual colonies were picked, grown overnight and plasmid DNA was prepared. After restriction enzyme digestion, the DNA was sequenced, using T3 and T7 primers. The variable region sequences were then compared and aligned using the V-Base germline databank (DNAPLOT 2.0.1 using V BASE Version 1.0).
3D Modeling of Human Monoclonal Anti-α3(IV) Ab–α3(IV) Interactions
Three of the mAb anti-α3(IV) were modeled using the elucidated structure of the human α3(IV)NC1 antigenic epitopes C2 and C6 [6, 8, 9] and the UCSF Chimera program (http://www.cgl.ucsf.edu/chimera/). The latter is a highly extensible, interactive molecular graphics program, funded by the NIH National Center for Research Resources (grant No. P41-RR01081) [8, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20]. The contact residues were inferred from the interaction between the heavy and light chain complementary regions of the mAb and the putative epitopes [14, 15, 16] using the BndLst contact residue software. Bndlst reads a PDB format file and prints a list of covalent and H-bonded neighboring atoms, along with several atomic properties, for each atom. It is useful in building scripts that process atomic structures permitting development of homology-based models (http://kinemage.biochem.duke.edu/software/utilities.php).
Results
Human Anti-α3(IV) mAb Are Pathogenic
As previously reported for F1.1, F2.1 and F3.1 were pathogenic. After injection of soluble Ig to naïve mice, they produced immune deposits and nephritis (table 1). By contrast, irrelevant human mAb neither produced immune deposits nor nephritis (not shown). Like human patient serum autoAb, the mAb bound to α3(IV)NC1 and exhibited preferential binding to native Ag, with minimal binding to denatured α3(IV)NC1 (fig. 1). Furthermore, the XII mAb competed with patient-derived anti-GBM Ab for binding to α3(IV) (fig. 2), and human mAb and patient serum-derived anti-GBM Ab were inhibited to human anti-α3(IV)NC1 by preincubation with cyclic peptides corresponding to the major α3(IV) epitopes, EA (C2) and EB (C6) (table 1), providing additional support for the relatedness of the human mAb and the patient serum Ab.
Table 1.
Pathogenicity and epitope specificity of human monoclonal anti-α3(IV)NC1 Ab after administration to XenoMouse II®
| mAb | n | BUN (SD) creatinine for F1.1∗ |
Albuminuria/proteinuria∗ |
GN |
C2 | C6 | |||
|---|---|---|---|---|---|---|---|---|---|
| pre |
post |
pre |
post |
H&E |
IF |
||||
| F2.1 | 4 | NA | 32 (7) | 31 (23) | 37 (10) | 1–2+ | 2+ | + | +++ |
| F3.1 | 8 | 32 (8) | 43 (10)∗ | 41 (22) | 47 (28) | 2–3+1 | 3+ | + | ++ |
| F1.1 | 8 | 0.007 | 0.424 | 1.6∗ | 340 (314)∗ | 2+ | 2+ | + | ++ |
| Polyclonal human | 2 | ND | ND | ND | ND | 2+ | 2+ | ND | ND |
In some short-tem experiments (5 days), normal mice were used, because of the limited availability of XenoMouse II. Grading of histopathologic findings: 0–1+ = normal or mild increase in cellularity; 2+ = glomerular enlargement and focal hypercellu-larity (increased matrix may be present in some); 3+ = focal hy-percellularity/proliferation in >50% of glomeruli with <30% diffuse proliferative changes and/or crescents; 4+ = diffuse prolif-erative changes with crescents/sclerosis in >50% of glomeruli. Epitope specificity was defined by competitive inhibition, using cyclic peptides corresponding to the C2 and C6 regions of α3(IV)NC1 (1 μg/well for mAb and 10 μg/well for serum-derived polyclonal anti-GBM Ab): + = 10–25%; ++ = 25–50%; +++ = >50%.
p < 0.05.
Early crescents present.
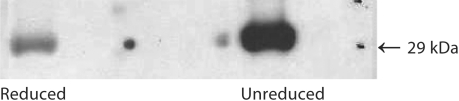
Fig. 1.
Anti-α3(IV)NC1 mAb bind to human α3(IV)NC1. Human mAb produced from XenoMouse II® after immunization with α3(IV)NC1 preparations bound to human α3(IV)NC1 by Western blot. Greater binding of m anti-α3(IV) to intact human α3(IV)NC1 with lesser binding to reduced human α3(IV)NC1, similar to that observed for human polyclonal serum-derived Ab. 4–6 μg Ag were used in these experiments.
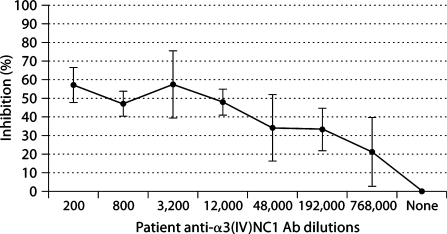
Fig. 2.
XenoMouse II pathogenic mAb compete for binding with human-derived polyclonal anti-GBM Ab to α3(IV). Negative control mAb showed less than 5% inhibition at any time and was equivalent to ‘none’ as shown in the figure(+/– error bars = SD).
Human Anti-α3(IV) mAb Are Encoded by Multiple VH and VL Genes
Given their similar Ag-binding properties, we questioned whether pathogenic Ab were encoded by similar genes. However, V gene sequence analysis of the human anti-α3(IV)NC1 mAb (table 2), demonstrated that the encoding VH and VL gene sequences were different (i.e. they were not even derived from the same V gene families). Additionally, multiple base changes from germline were found in each case, suggesting that the autoAb arose from an Ag-driven response, leading to B cell clonal expansion, somatic mutation, etc., and contributing to additional differences in their primary heavy and light chain sequences.
Table 2.
XenoMouse®-derived human anti-α3(IV)NC1 mAb
| mAb |
VH (gene) |
CDR1 |
CDR2 |
CDR3 |
JH |
|---|---|---|---|---|---|
| Heavy chain analysis demonstrating multiple VH genes may be used to encode pathogenic antibodies | |||||
| F1.1 | VH4 (DP-70) | Δ 3 bp | Δ 3 bp | NI | Δ 5b (6 bp) |
| F2.1 | VH3 (DP-50) | Δ 14 bp | germline | NI | NI |
| F3.1 |
VH3 (DP-47) |
germline |
germline |
Δ D3-22 (6 bp) |
Δ 6b (germline) |
| Light chain analysis demonstrating multiple VK genes may be used to encode pathogenic antibodies | |||||
| F1.1 | VK2 (DPk-12) | Δ 2 bp | germline | germline | Δ 5 |
| F2.1 | VK3 (DPk-22) | germline | germline | Δ 1 bp | Δ 2 (3 bp) |
| F3.1 | VK3 (DPk-21) | germline | germline | germline | Δ 1 (2 bp) |
Blast search alignment for F1.1 was closest to Macaca fasicu-laris Ab derived from a phage display library vs. hepatitis A (not shown), but the others were not. Δ = Change from germline; VH = variable heavy chain; CDR 1, 2 and 3 = complement determining regions 1, 2 and 3; JH = junction heavy variable to heavy constant region; NI = not identifiable; bp = base pairs.
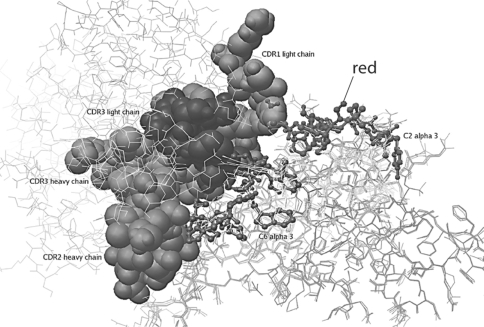
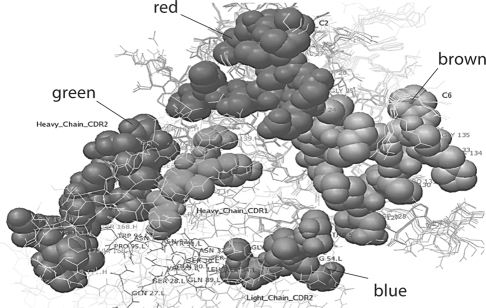
Multiple Contact Regions on the Heavy and Light Chains React with the Major Repeating Pathogenic Epitopes of α3(IV)NC1 (fig. 3, 4, 5)
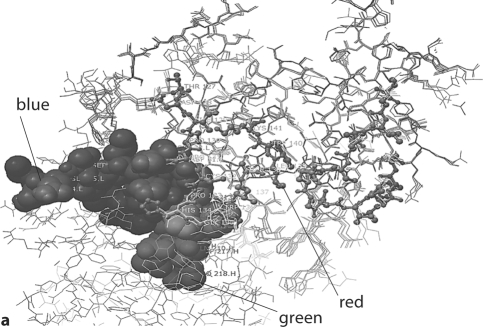
Fig. 3.
Model of human m anti-α3(IV)NC1 F1.1 binding to α3(IV)NC1. a In this model, the heavy chain CDR1 (shown in blue) and light chain CDR1 (shown in green) are shown interacting with the C2 epitope of α3(IV)NC1 (shown in red). b In this model, the heavy chain CDR1 (shown in blue) and light chain CDR1 (shown in green) are shown interacting with the C6 epi- tope C6 of α3(IV)NC1 (shown in red). Note the multiple contact Ab contact regions of the Ab with the repeating epitopes on α3(IV)NC1.
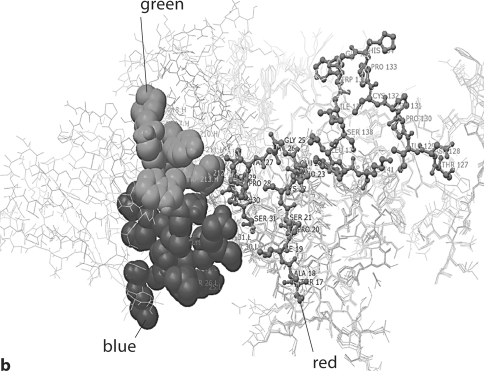
Fig. 4.
Model of human monoclonal anti-α3(IV)NC1 F2.1 binding to α3(IV)NC1. F2.1 is shown interacting at multiple sites with both the C2 and C6 epitopes α3(IV)NC1 (shown in red). Note the multiple contact Ab contact regions of the Ab with the repeating epitopes on α3(IV)NC1. The contact regions, however, are different from those of either F1.1. or F3.1.
Fig. 5.
Model of human m anti-α3(IV)NC1 F3.1 binding to α3(IV)NC1. F3.1 is shown interacting through heavy chain CDR2 (shown in green) and light chain CDR2 (shown in blue) sites with both the C2 (shown in red) and C6 (shown in brown) epitopes. Note the multiple contact Ab contact regions of the Ab with the repeating epitopes on α3(IV)NC1. The contact regions, however, are different from those of either F1.1. or F2.1.
Although different V gene sequences were used to encode the mAb, the possibility that their 3D structures were similar was suggested by a shared idiotype among patient serum Ab. To address whether the mAb were structurally related, homology-based models were employed using the sequences of the pathogenic mAb. It was noteworthy that their Ag-binding regions were different. However, modeling of the mAb and the putative structure of the α3(IV)NC1 region [6] to approximate Ag–Ab interactions provided intriguing results. Contact residues inferred from the interaction between the heavy chain and light chain complementary determining regions of the mAb and the relevant α3(IV)NC1 epitopes predicted the best energy constraints. It was particularly noteworthy that although the predicted 3D mAb structures and Ag contact regions differed for each mAb (as expected from the primary sequence differences), in each case the heavy chain binding region was predicted to provide contact, whereas the light chains either provided stability or did not interfere with the interactions. Furthermore, the close spatial proximity of related epitopes within α3(IV)NC1 was predicted to facilitate binding of both Ab heavy chains, thus providing the basis for Ab cross-linking in each case. It was also noteworthy that the best energy constraints were predicted among the mAb and the major epitopes (i.e. C2 and C6), as indicated by epitope mapping studies by Hudson et al. [1], using serum Ab. These latter observations also support the validity of the models.
Discussion
Human anti-α3(IV) autoAb are uniformly of high affinity, leading to the notion that this property contributes to the aggressive nature of disease [3]. Herein, we explore a possible molecular basis for high affinity, and, by inference, for pathogenicity. From the sequence analysis of multiple pathogenic autoAb, the results indicate that multiple heavy and light chain gene sequences may encode them. Nevertheless, from the modeling studies, it was predicted that the heavy and light chains mediate binding largely through one of the chains, with the other acting permissively. More importantly, the repetitive nature and spatial relationship of α3(IV) epitopes provides an ideal scaffold for interaction of both mAb chain arms. We postulate that this provides the basis for Ag cross-linking, leading to stability of the Ab–Ag interactions. Whether or not other possible combinations may contribute to similar-type, high-affinity interactions is unclear, although it seems reasonable to presume they would. In any event, the common idiotype shared by patients likely results from polyclonal Ab (i.e. anti-Id) reactive with multiple sites within the Ag-binding region of the anti-GBM Ab, although common genetic origins among some of the Ab is also feasible.
From the results, it is predicted that juxtapositions and stability of multiple autoAb on the GBM provides an ideal platform for FcR engagement and activation of inflammatory cells, especially since multiple Fc–FcR interactions are required for cellular recruitment and infiltration. Similarly, complement activation would be enhanced by multiplicity and proximity of the individual Fc heavy chains by providing the scaffolding necessary for engagement and stabilization of complement components. For the mAb examined, the light chains were predicted to stabilize the interaction, although we cannot exclude a minor contact role for them. Light chain-swapping experiments could resolve this issue. In this regard, it is likely that some light chains will inhibit binding or/and lower affinity, thereby limiting pathogenicity.
Multiple stable high-affinity interactions will likely reduce the anti-inflammatory properties of α3(IV) collagen by limiting access to regions within α3(IV) to circulating inflammatory cells [4], either by directly binding or through steric hindrance, thus preventing cellular engagement and subsequent dampening of activity. In either scenario, disease will be exacerbated, since the normal processes that promote disease resolution will be limited. As importantly, it is predictable that immunosuppressive therapy will be relatively ineffective, with reduced turnover of deposited pathogenic Ab tightly bound to these sites.
These high-affinity interactions may also contribute to autoimmunity. During inflammation with local release of proteases and other enzymes, detachment of Ag-Ab complexes (instead of release of Ab alone) will allow for the efficient delivery of immune complexes to activated APC, through interactions of Ag-Fc with FcR/DC. This will, in turn, serve as a highly efficient means of further activation of autoreactive T cells and stimulation of autoreactive B cells, etc., to further target immunity to the kidney. By contrast, these findings may also explain why some patients with circulating autoAb do not develop disease, as insufficient clustering of available epitopes for circulating Ab will limit complex stability, thereby limiting the half-life of bound Ab and/or providing an unstable platform for FcR engagement. On the other hand, increased epitope exposure (e.g. as with reactive oxygen species) will enhance binding and stability, leading to greater pathogenicity.
Multiple V genes were used to encode the human anti-α3(IV) Ab, and they were mutated from germline, consistent with the conclusion that they arise from an Ag-driven response. Thus, most individuals have the capacity to produce these autoAb; however, the immune response to this particular Ag is tightly regulated. In this context, exposure of α3(IV) with repeating epitopes to the surface receptors of B or T cells (i.e. binding with high affinity to either surface Ig or TcR) during normal development (e.g. in the thymus or bone marrow) should be particularly effective in receptor cross-linking, leading to long-lasting immune tolerance. The findings that the immune response is very restricted [i.e. to α3(IV) in patients with anti-GBM disease, and that anti-α3(IV) Ab are unusual in normal individuals, but common after renal transplantation of Alport's patients] is consistent with this conclusion [21, 22]. It is also consistent with the conclusion that α3(IV) may be the driving Ag in this disease [23], although molecular mimicry with other Ag may also be operative.
Acknowledgements
We thank Kelvin Gustilo and Mengshu Wang for providing technical support. We thank Eric Neilson and Raghu Kalluri for their advice and providing various forms and means of expression of α3(IV)NC1 and α1(IV)NC1. This work was supported by NIH grants NIDDK: DK53088 (M.P.M.) and KO827205 (M.C.).
References
- 1.Hudson BG, Tryggvason K, Sundaramoorthy M, Neilson EG. Alport's syndrome, Goodpasture's syndrome, and type IV collagen. N Eng J Med. 2003;348:2543–2556. doi: 10.1056/NEJMra022296. [DOI] [PubMed] [Google Scholar]
- 2.Meyers KE, Kinniry PA, Kalluri R, Neilson EG, Madaio MP. Human Goodpasture anti-alpha3(IV)NC1 autoantibodies share structural determinants. Kidney Int. 1998;53:402–407. doi: 10.1046/j.1523-1755.1998.00827.x. [DOI] [PubMed] [Google Scholar]
- 3.Rutgers A, Meyers KE, Canziani G, Kalluri R, Lin J, Madaio MP. High affinity of anti-GBM antibodies from Goodpasture and transplanted Alport patients to alpha3(IV)NC1 collagen. Kidney Int. 2000;58:115–122. doi: 10.1046/j.1523-1755.2000.00146.x. [DOI] [PubMed] [Google Scholar]
- 4.Fawzi A, Robinet A, Monboisse JC, Ziaie Z, Kefalides NA, Bellon G. A peptide of the alpha 3(IV) chain of type IV collagen modulates stimulated neutrophil function via activation of cAMP-dependent protein kinase and Ser/Thr protein phosphatase. Cell Signal. 2000;12:327–335. doi: 10.1016/s0898-6568(00)00074-7. [DOI] [PubMed] [Google Scholar]
- 5.Kalluri R, Weber M, Netzer KO, Sun MJ, Neilson EG, Hudson BG. COL4A5 gene deletion and production of post-transplant anti-alpha 3(IV) collagen alloantibodies in Alport syndrome. Kidney Int. 1994;45:721–726. doi: 10.1038/ki.1994.96. [DOI] [PubMed] [Google Scholar]
- 6.Netzer KO, Leinonen A, Boutaud A, Borza DB, Todd P, Gunwar S, Langeveld JP, Hudson BG. The goodpasture autoantigen. Mapping the major conformational epitope(s) of alpha3(IV) collagen to residues 17–31 and 127–141 of the NC1 domain. J Biol Chem. 1999;274:11267–11274. doi: 10.1074/jbc.274.16.11267. [DOI] [PubMed] [Google Scholar]
- 7.Kalluri R, Torre A, Shield CF, 3rd, Zamborsky ED, Werner MC, Suchin E, Wolf G, Helmchen UM, LP van den Heuvel, Grossman R, Aradhye S, Neilson EG. Identification of alpha3, alpha4, and alpha5 chains of type IV collagen as alloantigens for Alport posttransplant anti-glomerular basement membrane antibodies. Transplantation. 2000;69:679–683. doi: 10.1097/00007890-200002270-00038. [DOI] [PubMed] [Google Scholar]
- 8.Cleveland DW, Fischer SG, Kirschner MW, Laemmli UK. Peptide mapping by limited proteolysis in sodium dodecyl sulfate and analysis by gel electrophoresis. J Biol Chem. 1977;252:1102–1106. [PubMed] [Google Scholar]
- 9.Borza DB, Bondar O, Todd P, Sundaramoorthy M, Sado Y, Ninomiya Y, Hudson BG. Quaternary organization of the goodpasture autoantigen, the alpha 3(IV) collagen chain. Sequestration of two cryptic autoepitopes by intrapromoter interactions with the alpha4 and alpha5 NC1 domains. J Biol Chem. 2002;277:40075–40083. doi: 10.1074/jbc.M207769200. [DOI] [PubMed] [Google Scholar]
- 10.Meyers KE, Allen J, Gehret J, Jacobovits A, Gallo M, Neilson EG, Hopfer H, Kalluri R, Madaio MP. Human antiglomerular basement membrane autoantibody disease in XenoMouse II. Kidney Int. 2002;61:1666–1673. doi: 10.1046/j.1523-1755.2002.00312.x. comment 1905–1906. [DOI] [PubMed] [Google Scholar]
- 11.Lin J, Yanase K, Rutgers A, Madaio MP, Meyers KE. Selection of specific phage from display libraries: monoclonal antibody against VCS M13 helper phage coat protein III (gIIIp) Hybridoma. 1999;18:257–261. doi: 10.1089/027245799315916. [DOI] [PubMed] [Google Scholar]
- 12.Harrison JL, Williams SC, Winter G, Nissim A. Screening of phage antibody libraries. Methods Enzymol. 1996;267:83–109. doi: 10.1016/s0076-6879(96)67007-4. [DOI] [PubMed] [Google Scholar]
- 13.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 15.Peitsch MC. Protein modeling by E-mail. Bio/Technology. 1995;13:658–660. [Google Scholar]
- 16.Martin AC, Cheetham JC, Rees AR. Modelling antibody hypervariable loops: a combined algorithm. Proc Natl Acad Sci USA. 1989;86:9268–9272. doi: 10.1073/pnas.86.23.9268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin ACR, Cheetham JC, Rees AR. Molecular modelling of antibody combining sites. Methods Enzymol. 1991;203:121–153. doi: 10.1016/0076-6879(91)03008-5. [DOI] [PubMed] [Google Scholar]
- 18.Pedersen T, et al. Antibody modelling: beyond homology. Immunomethods. 1992;1:126–136. [Google Scholar]
- 19.Rees AR, et al. Antibody combining sites: structure and prediction. In: Sternberg MJE, editor. Protein Structure Prediction. New York: Oxford University Press; 1996. pp. 141–172. [Google Scholar]
- 20.Whitelegg NRJ, Rees AR. WAM – an improved algorithm for modelling antibodies on the Web. Protein Eng. 2000;13:819–824. doi: 10.1093/protein/13.12.819. [DOI] [PubMed] [Google Scholar]
- 21.Kalluri R, et al. COL4A5 gene deletion and production of post-transplant anti-alpha 3(IV) collagen alloantibodies in Alport syndrome. Kidney Int. 1994;45:721–726. doi: 10.1038/ki.1994.96. [DOI] [PubMed] [Google Scholar]
- 22.Kalluri R, et al. Identification of alpha3, alpha4, and alpha5 chains of type IV collagen as alloantigens for Alport posttransplant anti-glomerular basement membrane antibodies. Transplantation. 2000;69:679–683. doi: 10.1097/00007890-200002270-00038. [DOI] [PubMed] [Google Scholar]
- 23.Ooi JD, Holdsworth SR, Kitching AR. Advances in the pathogenesis of Goodpasture's disease: from epitopes to autoantibodies to effector T cells. J Autoimmun. 2008;31:295–300. doi: 10.1016/j.jaut.2008.04.005. [DOI] [PubMed] [Google Scholar]