Abstract
The ubiquitin-proteasome pathway plays an important role in DNA damage signaling and repair by facilitating the recruitment and activation of DNA repair factors and signaling proteins at sites of damaged chromatin. Proteasome activity is generally not thought to be required for activation of apical signaling kinases including the PI3K-related kinases (PIKKs) ATM, ATR, and DNA-PK that orchestrate downstream signaling cascades in response to diverse genotoxic stimuli. In a previous work, we showed that inhibition of the proteasome by MG-132 suppressed 53BP1 (p53 binding protein1) phosphorylation as well as RPA2 (replication protein A2) phosphorylation in response to the topoisomerase I (TopI) poison camptothecin (CPT). To address the mechanism of proteasome-dependent RPA2 phosphorylation, we investigated the effects of proteasome inhibitors on the upstream PIKKs. MG-132 sharply suppressed CPT-induced DNA-PKcs autophosphorylation, a marker of the activation, whereas the phosphorylation of ATM and ATR substrates were only slightly suppressed by MG-132, suggesting that DNA-PK among the PIKKs is specifically regulated by the proteasome in response to CPT. On the other hand, MG-132 did not suppress DNA-PK activation in reponse to UV or IR. MG-132 blocked the interaction between DNA-PKcs and Ku heterodimer enhanced by CPT, and hydroxyurea pre-treatment completely abolished CPT-induced DNA-PKcs autophosphorylation, indicating a requirement for ongoing DNA replication. CPT-induced TopI degradation occurred independent of DNA-PK activation, suggesting that DNA-PK activation does not require degradation of trapped TopI complexes. The combined results suggest that CPT-dependent replication fork collapse activates DNA-PK signaling through a proteasome dependent, TopI degradation-independent pathway. The implications of DNA-PK activation in the context of TopI poison-based therapies are discussed.
Keywords: DNA-PK, DNA damage, camptothecin, proteasome, topoisomerase I
1. Introduction
DNA damage responses, including signaling and repair, are enormously important for the maintenance of genome integrity. In response to DNA damages such as a DNA double-strand breaks (DSBs) and DNA replication stress, members of the phosphatidylinositol 3-kinase related protein kinase (PIKK) family, including ataxia telangiectasia mutated (ATM), ATM and Rad3-related (ATR), and DNA-dependent protein kinase (DNA-PK) are rapidly activated [1]. Activation of PIKKs triggers coordinated signaling pathways leading to cell cycle checkpoint arrest, DNA repair, and apoptosis. As widely accepted, ATM and ATR respond to DSB and replication stress, respectively, and are involved in DNA damage checkpoint, whereas DNA-PK is activated by DSBs for non-homologous end joining (NHEJ) repair with Ku70 and Ku80 proteins [1].
Replication protein A2 (RPA2) is a 32 kDa subunit of the heterotrimeric RPA complex, which binds single-strand DNA and is essential for DNA replication and DNA repair. RPA2 has a serine/threonine cluster in its N-terminus, which is phosphorylated in response to DNA damage [2]. In particular, the topoisomerase I (TopI) poison camptothecin (CPT) induces a highly phosphorylated form of RPA2 that is detected as a slower mobility species on SDS-PAGE gels [3, 4]. Hyperphosphorylation of RPA2 is dependent on PIKKs including ATR and DNA-PK [5, 6]. Cyclin-dependent kianses (CDKs) also contribute to RPA2 hyperphosphorylation in a cell cycle-specific manner. CPT causes DNA single-strand breaks (SSBs) by preventing the resolution step of the TopI cleavage reaction. TopI-DNA cleavage complexes (TopI-cc) are converted to DSBs following collision with DNA replication forks [7, 8]. Induction of DSBs is thought to underlie the anti-cancer properties of CPT derivatives, such as topotecan and irinotecan.
Jacquemont and Taniguchi [9] have reported that proteasome activity regulates DNA damage-responsive proteins including FANCD2, 53BP1, NBS1, and BRCA1 in response to ionizing radiation (IR). Several studies have shown that UBC13, an E2 ubiquitin (Ub) conjugating enzyme, and the E3 ubiquitin ligases RNF8/RNF168 mediate IR-induced 53BP1 and BRCA1 foci formation mediated by an ATM-γH2AX-MDC1 pathway, as well as homologous recombination [10–15]. Proteasome activity is also essential for 53BP1 recruitment to damage sites in response to DNA replication stress [16]. UBC13 makes RNF8 ubiquitinate histone H2AX and contributes to BRCA1 and 53BP1 recruitment through lysine63-mediated poly ubiquitin chain formation [17, 18]. From the combined findings, it is clear that protein ubiquitination and proteolysis are critical for processing chromatin prior to DNA repair.
Although the critical importance of Ub-dependent steps for the recruitment of mediator proteins to damage sites is now well established, the role of Ub pathways in apical PIKK activation is less clear. Here we used the CPT-induced phosphorylation of RPA as a paradigm to show that the activation of DNA-PK is critically dependent on proteasome activity in mammalian cells. Our studies suggest that proteasome-dependent chromatin modifications are required for DNA-PK-association with regulatory subunits at collapsed DNA replication forks.
2. Materials and Methods
2.1. Cell Culture and Drug Treatment
HeLa, U2OS, and HCT116 cells were maintained in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum. Cells were UV irradiated without medium using a UVP CL-1000 ultraviolet cross-linker. A JL Shepherd Model JL-109 irradiator with a 137Cs source was used for gamma irradiation. Camptothecin (CPT) was dissolved in DMSO at 10 mM and used at a final concentration of 2 μM. Hydroxyurea (HU) was dissolved in water at 1 M and used at a final concentration of 2 mM. MG-132 and N-acetyl-Leu-Leu-Nle-CHO (ALLN) (Calbiochem) dissolved in DMSO at 10 mM were added to the medium at a final concentration of 25 and 50 μM, respectively, 1 h prior to DNA damage. Bortezomib (commercially obtained from Millennium Pharmaceuticals, Inc. (Cambridge, MA) for experimental purposes) was dissolved in water at a concentration of 100 μM and used at a final concentration of 100 nM. For 53BP1 knockdown, pooled siRNAs (Dharmacon) were transfected by the phospho-calcium method.
2.2. Western Blotting
Cells were lysed with SDS sample buffer (60 mM Tris-HCl, pH 6.8, 2% SDS, 5% glycerol, 5% 2-mercaptoethanol, and bromphenol blue) and separated by SDS-polyacrylamide gel electrophoresis (PAGE). Separated proteins were transferred to polyvinylidene difluoride membrane (Millipore), and the membrane was blocked with 5% nonfat dry milk. The membrane was incubated with primary antibodies against phospho-DNA-PKcs (S2056, catalog no. ab18192, abcam), DNA-PKcs (Ab-4, Thermo SCIENTIFIC), phospho-ATM (S1981, catalog no. AF-1655, R&D Systems), ATM (catalog no. GTX70103, GeneTex), phospho-CHK1 (S317, catalog no. AF-2054, R&D Systems), CHK1 (G-4, Santa Cruz Biotechnology), RPA2 (Ab-3, Oncogene), and 53BP1 (catalog no. A300-272A, Bethyl Laboratories, Inc.). After primary antibody incubation, the membranes were incubated with secondary antibodies and visualized using SuperSignal chemiluminescent substrate (Pierce). For detection of immunoprecipitated proteins, HeLa cells treated with CPT with or without MG-132 were lysed with IP buffer (50 mM Tris pH7.5, 100 mM NaCl, 1.5 mM MgCl2, 0.2% Nonidet P-40) containing protease inhibitors (10 μg/ml leupeptin, 5 μg/ml aprotinin, 5 μg/ml pepstatin A, 0.2 mM phenylmethylsulfonyl fluoride) and soluble fraction was incubated with anti-DNA-PKcs antibody. Protein-antibody complexes were precipitated by protein G Sepharose (GE Healthcare) and co-precipitated Ku70 was detected by Western blotting using specific antibody.
2.3. Immunofluorescence Microscopy
Cells grown on 15-mm coverslips were fixed with 4% paraformaldehyde in PBS for 15 min and then permeabilized with PBS containing 0.2% Triton X-100 for 5 min. For RPA2 staining, cells were pre-extracted with PBS containing 0.1% Triton X-100. The fixed cells were incubated with primary antibodies specific for 53BP1, RPA2, and phosphorylated RPA2 (catalog no. A300-245A, Bethyl Laboratories, Inc.), which were diluted in PBS containing 5% bovine serum albumin. For bromodeoxyuridine (BrdU) staining, cells were treated with 20 μM BrdUrd for 20 min and treated with 2 N HCl for 20 min at 37 °C. Cells were stained with anti-BrdU antibody (Ab-2, Oncogene), followed by secondary antibodies conjugated to fluorescein isothiocyanate. The cells were stained with 4′, 6-diamidino-2-phenylindole in PBS (2 μg/ml) and mounted using Vectashield (Vector Laboratories). A Carl Zeiss Axiovert 200 fluorescence microscope was used to visualize samples.
2.4. Cell Cycle Synchronization and Flow Cytometry
To synchronize cells in S phase, cells were treated with thymidine at 2.5 mM for 18 h and incubated with a fresh medium for 3 h. For cell cycle analysis, cells were harvested and stained with propidium iodide (PI) after fixing with 70% ethanol. PI stained cells were analyzed using a FACSCalibur (BD Biosciences).
3. Results
3.1. MG-132 suppresses CPT-induced RPA2 phosphorylation by PIKKs, but not CDK
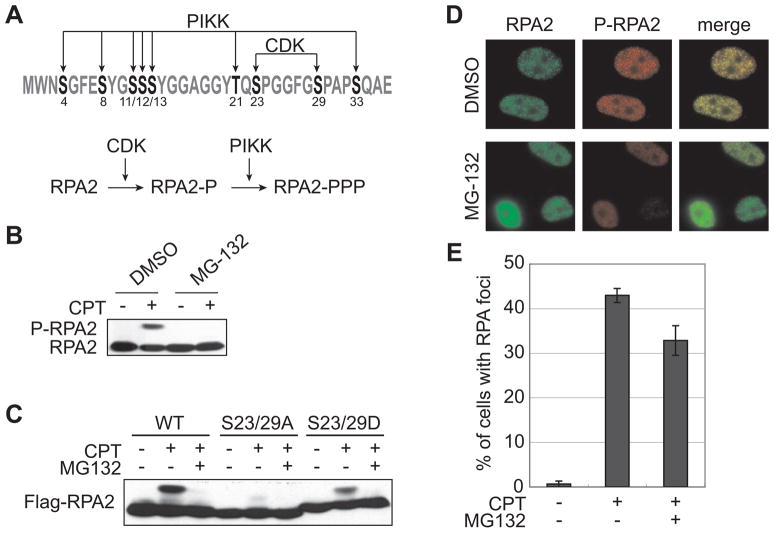
We previously reported that proteasome inhibitors suppress 53BP1 phosphorylation and foci formation in response to certain types of DNA replication stress [16]. In that study, we also showed that CPT-induced RPA2 hyperphosphorylation was suppressed by MG-132 (Fig. 1B). RPA2 has eight serines and a threonine in the N-terminus, which are sequentially phosphorylated by CDK and PIKKs in response to DNA-damaging agents, including CPT (Fig. 1A) [6]. One possible explanation for the inhibition of CPT-induced RPA2 phosphorylation was that MG-132 prevented CDK-dependent priming of RPA2. To test this idea, we introduced non-phosphorylatable (Ala) or phospho-mimetic (Asp) amino acid substitutions at the CDK sites (Ser23 and Ser29) and tested the phosphorylation profiles of the mutant proteins in transiently transfected HEK 293T cells. The RPA2 S23/29A double mutant showed a defect in CPT-induced hyperphosphorylation as already reported (Fig. 1C) [6]. On the other hand, the phospho-mimetic RPA2 substituted S23/29D mutant exhibited robust CPT-induced hyperphosphorylation that retained sensitivity to MG-132 (Fig. 1C). Because phospho-mimetic substitutions should bypass the CDK requirement, this finding suggests that the blocking of RPA2 phosphorylation caused by MG-132 is unlikely to be at the level of CDK priming phosphorylation. Instead, MG-132 appears to block PIKK-dependent RPA2 phosphorylation.
Fig 1.
Suppression of RPA2 hyperphosphorylation by MG-132. (A) The N-terminus sequence of RPA2. Eight serines and a threonine are the target of PIKKs and CDK. (B) MG-132 suppressed CPT-induced hyperphosphorylation of RPA2. RPA2 hyperphosphorylation was analyzed by Western blotting in CPT (2 μM, 1 h)-treated HeLa cells with or without MG-132 (25 μM, 1 h) pretreatment. (C) Bypass of CDK-mediated phosphorylation does not reverse MG-132 suppression of RPA2 phosphorylation. RPA2 proteins mutated on CDK phosphorylation sites (S23/29A and S23/29D) were expressed in 293T cells and analyzed phosphorylation by Western blotting using Flag antibody after CPT (2 μM, 1 h) treatment. (D, E) MG-132 does not affect RPA foci formation. HeLa cells were treated with CPT (2 μM, 1 h) after mock or MG-132 (25 μM, 1 h) treatment and stained with RPA2 antibody and phospho-specific antibody against Ser4/8 of RPA2 (D). Graphical data of the percentage of cells with RPA2 foci (E). Error bars show S.E. calculated from three independent experiments.
The targeting of RPA to nuclear foci precedes and is thought to be required for its phosphorylation in response to DNA damage [5, 6]. However, RPA2 foci formation induced by CPT was not significantly affected by MG-132 (Fig. 1D, E). These results suggest that MG-132 specifically affects the phosphorylation of RPA2 by PIKKs after foci formation.
3.2. CPT-induced DNA-PK activation is suppressed by MG-132
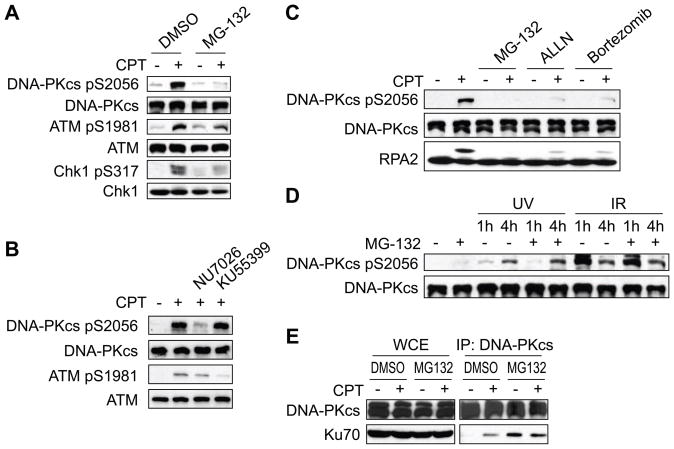
It has been reported that RPA2 phosphorylation requires PIKKs including ATR and DNA-PK [5, 6]. Thus, we set out to measure the effects of MG-132 on the activities of DNA-PK, ATM, and ATR following CPT treatment of HeLa cells. MG-132 partially suppressed CPT-induced ATM autophosphorylation on Ser1981 as well as ATR-dependent Chk1 phosphorylation on Ser317, whereas phosphorylation of DNA-PK catalytic subunit (DNA-PKcs) on Ser2056 was dramatically suppressed by MG-132 treatment (Fig. 2A). Phosphorylation of DNA-PKcs on Ser2056 is known as autophopshorylation in response to IR [19]. In response to CPT, DNA-PKcs phosphorylation at Ser2056 was suppressed by the DNA-PK inhibitor NU7026, but not the ATM inhibitor KU55399 (Fig. 2B). This result indicates that CPT-induced Ser2056 phosphorylation of DNA-PK is autophosphorylation. To further confirm this finding, DNA-PKcs was immunoprecipitated and immunoblotted with phospho-DNA-PKcs antibody. DNA-PKcs immunoprecipitated from cells treated with MG-132 showed a strong suppression in autophosphorylation caused by CPT (Fig. S1). Other proteasome inhibitors, ALLN and Bortezomib also suppressed CPT-induced DNA-PKcs autophosphorylation on Ser2056 and RPA2 hyperphosphorylation (Fig. 2C). On the other hand, UV- and IR-induced DNA-PKcs autophosphorylation was resistant to MG-132 (Fig. 2D). Together, these results suggest that the proteasome specifically regulates DNA-PK activation in response to CPT and that inhibition of DNA-PK is responsible for the loss of RPA2 phosphorylation.
Fig 2.
CPT-induced DNA-PK activation is abrogated by proteasome inhibitors. (A) Suppression of DNA-PK activation by MG-132. HeLa cells were pretreated with vehicle or MG-132 (25 μM, 1 h) and CPT (2 μM, 1 h)-induced DNA-PKcs, ATM, and Chk1 phosphorylation were analyzed by Western blotting using the indicated antibodies. (B) DNA-PKcs Ser2056 is an autophosphorylation site in response to CPT. HeLa cells were pretreated with DNA-PK inhibitor (NU7026, 10 μM) or ATM inhibitor (KU55399, 10 μM) for 1 h followed by CPT (2 μM, 1 h) treatment. DNA-PKcs and ATM phosphorylation were analyzed by Western blotting. (C) Suppression of DNA-PK by ALLN and Bortezomib. DNA-PK and RPA2 phosphorylation were analyzed by Western blotting in cells treated with MG-132 (25 μM, 1 h), ALLN (50 μM, 1 h), or Bortezomib (100 nM, 2 h). (D) MG-132 does not affect UV- or IR-induced DNA-PK activation. DNA-PKcs autophosphorylation was analyzed by Western blotting using extracts prepared from cells UV irradiated (20 J/m2) or exposed to 5 Gy IR. (E) MG-132 blocks CPT-induced association of DNA-PKcs and Ku70. HeLa cells were treated with CPT (2 μM, 1 h) and whole cell extracts were prepared. DNA-PKcs was immunoprecipitated and coimmunoprecipitated Ku70 was detected by Western blotting.
3.3. MG-132 blocks DNA-PK activation at the level of DNA-PK recruitment
Catalytic activation of DNA-PK requires its association with DNA-binding Ku70/Ku80 heterodimer. To test whether MG-132 blocked DNA-PK activation at the level of recruitment, we coimmunoprecipitated DNA-PKcs and Ku70 from HeLa cells before and after CPT treatment in the absence or presence of MG-132. As expected, CPT induced DNA-PKcs-Ku heterodimer association in the absence of MG-132 (Fig. 2E). Interestingly, MG-132 treated cells showed an elevated level of Ku70 coimmunoprecipitation in the absence of CPT treatment that was not further induced upon CPT exposure (Fig. 2E). The inability of CPT to induce DNA-PKcs-Ku heterodimer complexes in the presence of MG-132 suggests that defective DNA-PK recruitment underlies the activation defect.
3.4. Slight effect of MG-132 on DNA replication is not critical for DNA-PK activation
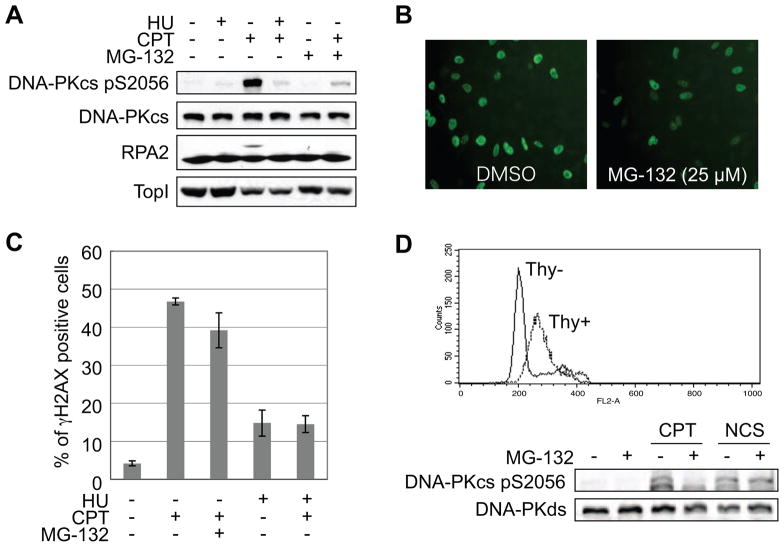
CPT-induced Top I-cc impedes DNA replication and transcription. To test whether DNA replication is required for DNA-PK activation by CPT, HeLa cells were pre-treated with the replication inhibitor HU 10 min prior to CPT addition. HU pre-treatment strongly suppressed DNA-PKcs autophosphorylation as well as MG-132 treatment (Fig. 3A). This indicates that CPT-induced DNA-PK activation is dependent on replication fork progression, and raises the possibility that MG-132 also suppresses DNA replication. To check the effect of MG-132 on DNA replication, cells were labeled with BrdU during exposure to several concentrations of MG-132 and the percentages of BrdU-positive cells were compared with the level of DNA-PKcs autophosphorylation. Whereas 5 μM MG-132 was sufficient for the suppression of DNA-PK activation, this concentration of MG-132 did not reduce the percentage of BrdU positive cells (Fig. S2). Although higher concentrations of MG-132 (25 μM) caused partial suppression of DNA replication (Fig. 3B), it did not block CPT-induced accumulation of γH2AX foci, a surrogate marker of DSBs (Fig. 3C). The fact that CPT-induced γH2AX foci were suppressed by HU pre-treatment suggests that the DNA replication rate is sufficient for DSBs to occur following CPT treatment in the presence of high MG-132 concentrations. From these results we conclude that the suppression of DNA-PK activation by MG-132 is not caused by the repression of DNA replication. On the other hand, CPT-induced TopI degradation was not affected by HU pre-treatment even though DNA-PK activation was clearly suppressed (Fig. 3A). This result is consistent with the report that TopI degradation caused by CPT treatment is dependent on transcription [20], and indicates that TopI is not the relevant proteasome target required for DNA-PK activation in response to CPT.
Fig 3.
DNA-PK activation is dependent on DNA replication, but not S phase specific regulation. (A) DNA replication-dependent activation of DNA-PK. CPT (2 μM), HU (4 mM), and MG-132 (10 μM) were added to the media of HeLa cells in the indicated combinations. DNA-PKcs and RPA2 phosphorylation and Top1 protein were analyzed by Western blotting. (B) DNA replication in the presence of MG-132. BrdU-incorporated cells were stained with specific antibody after 1 h treatment of MG-132 (25 μM). (C) Replication-sensitive and MG-132-resistant γH2AX induced by CPT. Cells were treated with CPT (2 μM, 1 h) following mock, MG-132 (10 μM, 1 h), or HU (4 mM, 15 min) pretreatment and stained with γH2AX antibody. The percentage of cells with γH2AX was averaged from three independent experiments. Error bars are the S.E. (D) MG-132 does not affect NCS-induced DNA-PK activation in S phase. U2OS cells were released from thymidine block to synchronize in S phase and analyzed for DNA-PKcs autophosphorylation induced by CPT (2 μM, 1 h) or NCS (100 ng/ml, 1 h). Thy− and Thy+ indicate cells without and with thymidine treatment, respectively, to synchronize in S phase.
The DNA replication dependence of DNA-PK activation by CPT raised the possibility that activation of DNA-PK in S phase requires proteasome activity irrespective of the damage insult. To test this, cells were synchronized in S phase by thymidine block and treated with CPT or the radiomimetic Neocarzinostatin (NCS). S phase synchronization itself did not affect DNA-PKcs autophosphorylation. CPT-induced DNA-PKcs autophosphorylation was suppressed by MG-132 in the synchronized cells, whereas NCS-induced DNA-PKcs autophosphorylation was not (Fig. 3D). This result suggests that DNA-PK activation in S phase is not universally dependent on proteasome activity and that stalled replication forks require proteasome-dependent processing to activate DNA-PK.
3.5. DNA-PK and 53BP1 function in the distinct pathways in response to CPT
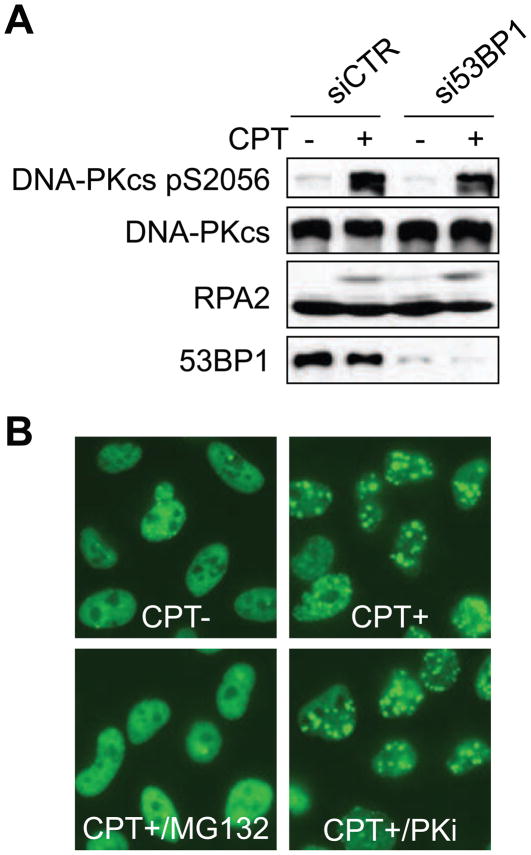
In our previous report, we showed that proteasome inhibitors suppress 53BP1 phosphorylation and foci formation caused by DNA replication stress. Therefore, we considered the possibility that DNA-PK and 53BP1 lie in a linear, MG-132 sensitive pathway activated by CPT. To test this possibility, 53BP1 expression was suppressed by siRNA and the response to CPT was analyzed by Western blotting. CPT-induced DNA-PKcs autophosphorylation and RPA2 hyperphosphorylation were not affected by 53BP1 knockdown (Fig. 4A). We also tested the effects of DNA-PK inhibition on 53BP1 foci formation. Cells treated with a DNA-PK inhibitor, NU7026, exhibited normal levels of CPT-induced 53BP1 foci formation (Fig. 4B), suggesting that 53BP1 and DNA-PK do not function in a linear, proteasome-dependent pathway in response to CPT.
Fig 4.
DNA-PK and 53BP1 are independently regulated by the proteasome. (A) 53BP1-independent DNA-PK activation in response to CPT. HeLa cells were transfected with control or 53BP1 siRNA, and CPT (2 μM, 1 h)-induced DNA-PKcs autophosphorylation was analyzed by Western blotting. (B) DNA-PK-independent foci formation of 53BP1. HeLa cells were treated with vehicle or the DNA-PK inhibitor NU7026 (10 μM, 1 h) prior to treatment with CPT (2 μM, 1 h). 53BP1 foci formation was examined by immunofluorescence microscopy.
4. Discussion
In this study we have demonstrated proteasome-dependent activation of DNA-PK in response to the TopI poison CPT. CPT belongs to a clinically relevant class of TopI poisons used in cancer chemotherapy. CPT-induced signaling has been extensively characterized and this study provides insights into how CPT engages DNA-PK, a critical regulator of DSB repair.
The activation of DNA-PK by CPT is strictly S-phase specific, suggesting that DNA-PK might promote NHEJ repair of CPT-induced damage in S phase. However, the DNA damage induced by CPT through DNA replication is considered not to be suitable for NHEJ because it has one free DNA-end and large gapped DNA. Although NHEJ occurs during all phases of the cell cycle for DSB repair, homologous recombination (HR) repair is thought to be the dominant mechanism of the repair of DSB and stalled replication fork in S and G2 phases [21, 22]. Although the overall contribution of NHEJ to genomic stability in S phase is still not clear, several reports have shown that DNA-PKcs deficient cells are sensitive to UV and CPT [4, 23], suggesting that DNA-PK might be required for stalled fork repair through HR or checkpoint activation, which is necessary for cell survival. Since phosphorylated RPA2 binds to Rad51 [24], it is conceivable that DNA-PK participates in HR repair through phosphorylation of RPA2, which is both DNA-PK and proteasome dependent in CPT-treated cells.
The molecular details of proteasome-dependent DNA-PK activation remain to be elucidated. MG-132 treatment suppressed CPT-induced enhancement of DNA-PKcs-Ku heterodimer association, providing a possibility that the DNA-PK complex cannot be recruited onto DNA damage sites in the presence of MG-132. Interestingly, MG-132 treatment itself promoted the association between DNA-PKcs and Ku heterodimer as shown in Fig 2E. Dissociation of DNA-PKcs-Ku heterodimer complex is considered to require DNA-PKcs autophosphorylation [25]. The turnover of DNA-PK complex responding to spontaneous DSBs might be blocked by MG-132 through the DNA-PK inhibition, resulting in accumulation of DNA-PKcs associated with Ku heterodimer. Available evidence suggests that MG-132 prevents CPT-induced DNA-PK activation at the level of recruitment. On the other hand, Ku heterodimer itself, a targets of the proteasome, is ubiquitinated and degraded when Ku is displaced from chromatin [26]. However, we did not observe CPT-induced degradation of Ku70/80, suggesting that Ku heterodimer degradation is not involved in DNA-PK activation. Another candidate is TopI, because its degradation in response to CPT has been reported [27]. Evidence that TopI degradation is not required for DNA-PK activation comes from the finding that CPT-induced TopI degradation occurs in the presence of HU, even though DNA-PKcs autophosphorylation was dramatically suppressed. This is consistent with the report that TopI degradation caused by CPT is dependent on transcription, but not DNA replication [20].
Altogether our data suggest that degradation of a chromatin factor(s) following CPT-induced DNA damage is required for DNA-PK activation and DNA-PK-dependent phosphorylation of RPA2 in S-phase cells. Murakawa et al. [28] have also reported that proteasome inhibition suppresses DSB repair by HR. In this model a stalled replication fork is transiently protected from DNA damage-responsive proteins including 53BP1, DNA-PK, and Rad51. Degradation of the putative factor(s) allows recruitment of these factors and initiation of DNA repair. The operative DNA-PK activation mechanism is apparently distinct from that induced by radiation and radiomimetic drugs, as these agents activate DNA-PK independent of the proteasome, even in S-phase cells. In addition, MG-132 appears to inhibit DNA-PK activation and 53BP1 recruitment through different mechanisms, as 53BP1 knockdown did not affect DNA-PK activation, and a DNA-PK inhibitor did not block 53BP1 targeting to damage sites. Our findings are therefore not congruent with a report that 53BP1 is required for CPT-induced RPA2 hyperphosphorylation [29].
It is also possible that ubiquitination itself, but not protein degradation, is required for DNA-PK activation. Xu et al. [30] have reported that proteasome inhibition causes the starvation of free Ub leading to defects in regulatory ubiquitination of histones H2A and H2B. γH2AX ubiquitination was suppressed by MG-132 in response to either CPT or NCS (Fig. S3). This suggests that Ub starvation is responsible for the suppression of γH2AX ubiquitination in MG-132-treated cells and that DNA-PK can be activated in the absence of Ub in response to NCS. Furthermore, knockdown of RNF8, the responsible enzyme for H2AX ubiquitination [12, 13], did not block CPT-induced DNA-PKcs autophosphorylation (data not shown). Combined these data reveal that γH2AX ubiquitination is not required for DNA-PK activation. Instead, the ubiquitination of other chromatin-associated proteins may facilitate access of DNA-PK to CPT-induced DNA damage. Recently, Lin et al. [31] have published that proteasome inhibition suppresses the generation of DSBs in response to CPT. In contrast to their data, MG-132 did not significantly decrease the percentage of γH2AX positive cells in our hands, which was suppressed by HU, suggesting that replication-dependent DSBs are generated even in the presence of MG-132. In addition, the effect of MG-132 on ATM and Chk1 phosphorylation was partial as shown by Lin et al., whereas MG-132 sharply abolished DNA-PKcs and RPA2 phosphorylation, suggesting that MG-132 effect on DNA-PK activation is definitely different from the effect on ATM/ATR activation.
It has been reported that co-treatment with proteasome inhibitor improves the effect of CPT against a colorectal cancer cell line [32]. This synergistic effect of proteasome inhibition has been thought to be caused by the suppression of NF-κB activation. CPT activates NF-κB through the proteasome-dependent degradation of I Ba which is inhibitory factor of NF-κB [33]. Given that proteasome inhibition blocks several steps in DNA damage signaling, including 53BP1 recruitment and DNA-PK activation, it is plausible that a DNA repair defect may contribute to enhanced cytotoxicity of TopI poisons.
Supplementary Material
Fig S1. MG-132 suppresses CPT-induced DNA-PKcs autophosphorylation. DNA-PKcs was immunoprecipitated with anti-DNA-PKcs antibody from extracts of HeLa cells treated with the indicated compounds. Immunoprecipitated DNA-PKcs was resolved by SDS-PAGE and immunoblotted with anti-DNA-PKcs antibody.
Fig S2. Concentrations of MG-132 that ablate DNA-PKcs autophosphorylation marginally affect DNA replication in HeLa cells. (A) MG-132 dose response. DNA-PKcs, Chk1, and RPA2 phosphorylation was analyzed in HeLa cells treated with CPT (2 μM, 1 h) following MG-132 treatment at the indicated concentrations for 1 h. (B) Effects of MG-132 on BrdU incorporation. After treatment with MG-132 at the indicated concentration for 2 h, HeLa cells were labeled with BrdU (20 μM, 20 min) and immunostained with anti-BrdU antibodies. The percentage of BrdU-positive cells were counted using a fluorescence microscope. Error bars show S.E. calculated in three independent experiments.
Fig S3. MG-132 suppresses ubiquitination of γH2AX caused by distinct types of DNA damage. HeLa cells were treated with CPT (2 μM, 1 h) or NCS (100 ng/ml, 1 h) following exposure to vehicle or MG-132 (10 μM, 1 h). After cell lysis and sonication, γH2AX ubiquitination was analyzed by Western blotting. The higher molecular mass ubiquitinated form of γH2AX is denoted. The lower panel shows accumulation of poly-ubquitinated proteins in cells treated with MG-132.
Acknowledgments
This work was supported by grants from the NIH (CA124722), The American Cancer Society, and a Shaw Scientist Award to R.S.T. from the Greater Milwaukee Foundation.
Footnotes
Conflict of interest
The authors declare that there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bakkenist CJ, Kastan MB. Initiating cellular stress responses. Cell. 2004;118:9–17. doi: 10.1016/j.cell.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 2.Binz SK, Sheehan AM, Wold MS. Replication protein A phosphorylation and the cellular response to DNA damage. DNA Repair. 2004;3:1015–1024. doi: 10.1016/j.dnarep.2004.03.028. [DOI] [PubMed] [Google Scholar]
- 3.Wang Y, Perrault AR, Iliakis G. Down-regulation of DNA replication in extracts of camptothecin-treated cells: activation of an S-phase checkpoint? Cancer Res. 1997;57:1654–1659. [PubMed] [Google Scholar]
- 4.Shao RG, Cao CX, Zhang H, Kohn KW, Wold MS, Pommier Y. Replication-mediated DNA damage by camptothecin induces phosphorylation of RPA by DNA-dependent protein kinase and dissociates RPA:DNA-PK complexes. EMBO J. 1999;18:1397–1406. doi: 10.1093/emboj/18.5.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sakasai R, Shinohe K, Ichijima Y, Okita N, Shibata A, Asahina K, Teraoka H. Differential involvement of phosphatidylinositol 3-kinase-related protein kinases in hyperphosphorylation of replication protein A2 in response to replication-mediated DNA double-strand breaks. Genes Cells. 2006;11:237–246. doi: 10.1111/j.1365-2443.2006.00942.x. [DOI] [PubMed] [Google Scholar]
- 6.Anantha RW, Vassin VM, Borowiec JA. Sequential and synergistic modification of human RPA stimulates chromosomal DNA repair. J Biol Chem. 2007;282:35910–35923. doi: 10.1074/jbc.M704645200. [DOI] [PubMed] [Google Scholar]
- 7.Liu LF, Desai SD, Li TK, Mao Y, Sun M, Sim SP. Mechanism of action of camptothecin. Ann N Y Acad Sci. 2000;922:1–10. doi: 10.1111/j.1749-6632.2000.tb07020.x. [DOI] [PubMed] [Google Scholar]
- 8.Pommier Y. Topoisomerase I inhibitors: camptothecins and beyond. Nat Rev Cancer. 2006;6:789–802. doi: 10.1038/nrc1977. [DOI] [PubMed] [Google Scholar]
- 9.Jacquemont C, Taniguchi T. Proteasome function is required for DNA damage response and fanconi anemia pathway activation. Cancer Res. 2007;67:7395–7405. doi: 10.1158/0008-5472.CAN-07-1015. [DOI] [PubMed] [Google Scholar]
- 10.Zhao GY, Sonoda E, Barber LJ, Oka H, Murakawa Y, Yamada K, Ikura T, Wang X, Kobayashi M, Yamamoto K, Boulton SJ, Takeda S. A critical role for the ubiquitin-conjugating enzyme Ubc13 in initiating homologous recombination. Mol Cell. 2007;25:663–675. doi: 10.1016/j.molcel.2007.01.029. [DOI] [PubMed] [Google Scholar]
- 11.Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, Panier S, Mendez M, Wildenhain J, Thomson TM, Pelletier L, Jackson SP, Durocher D. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 2007;318:1637–1640. doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 14.Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, Ellenberg J, Panier S, Durocher D, Bartek J, Lukas J, Lukas C. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009;136:435–446. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 15.Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, Nakada S, Ylanko J, Olivarius S, Mendez M, Oldreive C, Wildenhain J, Tagliaferro A, Pelletier L, Taubenheim N, Durandy A, Byrd PJ, Stankovic T, Taylor AM, Durocher D. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 2009;136:420–434. doi: 10.1016/j.cell.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 16.Sakasai R, Tibbetts R. RNF8-dependent and RNF8-independent regulation of 53BP1 in response to DNA damage. J Biol Chem. 2008;283:13549–13555. doi: 10.1074/jbc.M710197200. [DOI] [PubMed] [Google Scholar]
- 17.Hofmann RM, Pickart CM. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell. 1999;96:645–653. doi: 10.1016/s0092-8674(00)80575-9. [DOI] [PubMed] [Google Scholar]
- 18.Plans V, Scheper J, Soler M, Loukili N, Okano Y, Thomson TM. The RING finger protein RNF8 recruits UBC13 for lysine 63-based self polyubiquitylation. J Cell Biochem. 2006;97:572–582. doi: 10.1002/jcb.20587. [DOI] [PubMed] [Google Scholar]
- 19.Chen BP, Chan DW, Kobayashi J, Burma S, Asaithamby A, Morotomi-Yano K, Botvinick E, Qin J, Chen DJ. Cell cycle dependence of DNA-dependent protein kinase phosphorylation in response to DNA double strand breaks. J Biol Chem. 2005;280:14709–14715. doi: 10.1074/jbc.M408827200. [DOI] [PubMed] [Google Scholar]
- 20.Desai SD, Zhang H, Rodriguez-Bauman A, Yang JM, Wu X, Gounder MK, Rubin EH, Liu LF. Transcription-dependent degradation of topoisomerase I-DNA covalent complexes. Mol Cell Biol. 2003;23:2341–2350. doi: 10.1128/MCB.23.7.2341-2350.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takata M, Sasaki MS, Sonoda E, Morrison C, Hashimoto M, Utsumi H, Yamaguchi-Iwai Y, Shinohara A, Takeda S. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 1998;17:5497–5508. doi: 10.1093/emboj/17.18.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rothkamm K, Kruger I, Thompson LH, Lobrich M. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol Cell Biol. 2003;23:5706–5715. doi: 10.1128/MCB.23.16.5706-5715.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muller C, Calsou P, Frit P, Cayrol C, Carter T, Salles B. UV sensitivity and impaired nucleotide excision repair in DNA-dependent protein kinase mutant cells. Nucleic Acids Res. 1998;26:1382–1389. doi: 10.1093/nar/26.6.1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu X, Yang Z, Liu Y, Zou Y. Preferential localization of hyperphosphorylated replication protein A to double-strand break repair and checkpoint complexes upon DNA damage. Biochem J. 2005;391:473–480. doi: 10.1042/BJ20050379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chan DW, Lees-Miller SP. The DNA-dependent protein kinase is inactivated by autophosphorylation of the catalytic subunit. J Biol Chem. 1996;271:8936–8941. doi: 10.1074/jbc.271.15.8936. [DOI] [PubMed] [Google Scholar]
- 26.Postow L, Ghenoiu C, Woo EM, Krutchinsky AN, Chait BT, Funabiki H. Ku80 removal from DNA through double strand break-induced ubiquitylation. J Cell Biol. 2008;182:467–479. doi: 10.1083/jcb.200802146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Desai SD, Liu LF, Vazquez-Abad D, D’Arpa P. Ubiquitin-dependent destruction of topoisomerase I is stimulated by the antitumor drug camptothecin. J Biol Chem. 1997;272:24159–24164. doi: 10.1074/jbc.272.39.24159. [DOI] [PubMed] [Google Scholar]
- 28.Murakawa Y, Sonoda E, Barber LJ, Zeng W, Yokomori K, Kimura H, Niimi A, Lehmann A, Zhao GY, Hochegger H, Boulton SJ, Takeda S. Inhibitors of the proteasome suppress homologous DNA recombination in mammalian cells. Cancer Res. 2007;67:8536–8543. doi: 10.1158/0008-5472.CAN-07-1166. [DOI] [PubMed] [Google Scholar]
- 29.Yoo E, Kim BU, Lee SY, Cho CH, Chung JH, Lee CH. 53BP1 is associated with replication protein A and is required for RPA2 hyperphosphorylation following DNA damage. Oncogene. 2005;24:5423–5430. doi: 10.1038/sj.onc.1208710. [DOI] [PubMed] [Google Scholar]
- 30.Xu Q, Farah M, Webster JM, Wojcikiewicz RJ. Bortezomib rapidly suppresses ubiquitin thiolesterification to ubiquitin-conjugating enzymes and inhibits ubiquitination of histones and type I inositol 1,4,5-trisphosphate receptor. Mol Cancer Ther. 2004;3:1263–1269. [PubMed] [Google Scholar]
- 31.Lin CP, Ban Y, Lyu YL, Liu LF. Proteasome-dependent processing of topoisomerase I-DNA adducts into DNA double-strand breaks at arrested replication forks. J Biol Chem. 2009;284:28084–28092. doi: 10.1074/jbc.M109.030601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cusack JC, Jr, Liu R, Houston M, Abendroth K, Elliott PJ, Adams J, Baldwin AS., Jr Enhanced chemosensitivity to CPT-11 with proteasome inhibitor PS-341: implications for systemic nuclear factor-kappaB inhibition. Cancer Res. 2001;61:3535–3540. [PubMed] [Google Scholar]
- 33.Huang TT, Wuerzberger-Davis SM, Seufzer BJ, Shumway SD, Kurama T, Boothman DA, Miyamoto S. NF-kappaB activation by camptothecin. A linkage between nuclear DNA damage and cytoplasmic signaling events. J Biol Chem. 2000;275:9501–9509. doi: 10.1074/jbc.275.13.9501. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1. MG-132 suppresses CPT-induced DNA-PKcs autophosphorylation. DNA-PKcs was immunoprecipitated with anti-DNA-PKcs antibody from extracts of HeLa cells treated with the indicated compounds. Immunoprecipitated DNA-PKcs was resolved by SDS-PAGE and immunoblotted with anti-DNA-PKcs antibody.
Fig S2. Concentrations of MG-132 that ablate DNA-PKcs autophosphorylation marginally affect DNA replication in HeLa cells. (A) MG-132 dose response. DNA-PKcs, Chk1, and RPA2 phosphorylation was analyzed in HeLa cells treated with CPT (2 μM, 1 h) following MG-132 treatment at the indicated concentrations for 1 h. (B) Effects of MG-132 on BrdU incorporation. After treatment with MG-132 at the indicated concentration for 2 h, HeLa cells were labeled with BrdU (20 μM, 20 min) and immunostained with anti-BrdU antibodies. The percentage of BrdU-positive cells were counted using a fluorescence microscope. Error bars show S.E. calculated in three independent experiments.
Fig S3. MG-132 suppresses ubiquitination of γH2AX caused by distinct types of DNA damage. HeLa cells were treated with CPT (2 μM, 1 h) or NCS (100 ng/ml, 1 h) following exposure to vehicle or MG-132 (10 μM, 1 h). After cell lysis and sonication, γH2AX ubiquitination was analyzed by Western blotting. The higher molecular mass ubiquitinated form of γH2AX is denoted. The lower panel shows accumulation of poly-ubquitinated proteins in cells treated with MG-132.