Abstract
Dendritic spine morphology is thought to play important roles in synaptic development and plasticity, and morphological derangements in spines are correlated with several neurological disorders. Here, we identified an interaction between Spine-Associated RapGAP (SPAR), a postsynaptic protein that reorganizes actin cytoskeleton and drives dendritic spine head growth, and PDLIM5 / Enigma Homolog (ENH), a PDZ-LIM (postsynaptic density-95/Discs large/zona occludens 1-Lin11/Isl-1/Mec3) family member. PDLIM5 has been implicated in susceptibility to bipolar disorder, major depression and schizophrenia but its function in neurological disease is poorly understood. We show that PDLIM5 is present in the postsynaptic density, where it promotes decreased dendritic spine head size and longer, filopodia-like morphology. Conversely, RNA interference against PDLIM5 or loss of PDLIM5 interaction with SPAR caused increased spine head diameter. Furthermore, PKC activation promoted delivery of PDLIM5 into dendritic spines and increased its spine colocalization with SPAR. These data reveal new postsynaptic functions for PDLIM5 in shrinkage of dendritic spines that may be relevant to its association with psychiatric illness.
Keywords: PDLIM5, ENH, PDZ-LIM, Enigma, SPAR, dendritic spines, postsynaptic density
Introduction
Dendritic spines are small protrusions that serve as the primary sites of excitatory synaptic transmission in the CNS (Bourne and Harris, 2008). Spines have received enormous interest because of their dynamic morphology, which is highly responsive to various patterns of synaptic activity (De Roo et al., 2008). Spine morphology is also developmentally regulated, progressing from thin filopodia-like structures in immature neurons to bulbous mushroom headed spines as neurons age (Ethell and Pasquale, 2005). Functionally, larger spines possess greater synaptic strength and stability and have been suggested to play the role of ‘memory’ spines, while thin spines are more dynamic and responsive to potentiating stimulation and are thus proposed to act as ‘plasticity’ spines (Bourne and Harris, 2007). However, several neurological conditions associated with mental retardation are characterized by unusually thin spines (Newey et al., 2005; Purpura, 1974) suggesting that inability to form or maintain appropriate spine morphology may contribute to cognitive impairment.
Actin is the major cytoskeleton in spines, and actin as well as a number of its binding proteins have been implicated in synaptic function and plasticity (Carlisle and Kennedy, 2005; Lippman and Dunaevsky, 2005; Schubert and Dotti, 2007; Tada and Sheng, 2006). For example, SPAR (Spine-Associated Rap GTPase-activating protein (GAP)) is a large, multidomain protein that inhibits Rap small GTPases and also binds to the postsynaptic density (PSD) scaffold protein PSD-95 (Fig. 1A) together with NMDA receptors in brain (Pak et al., 2001). Additionally, SPAR induces profound reorganization of F-actin filaments and promotes dendritic spine head growth (Pak et al., 2001). SPAR protein levels are tightly regulated at synapses by phosphorylation-dependent degradation mediated by the activity-inducible polo kinase Plk2/SNK (Pak and Sheng, 2003).
Figure 1. Identification of PDLIM5 as a SPAR interacting protein.
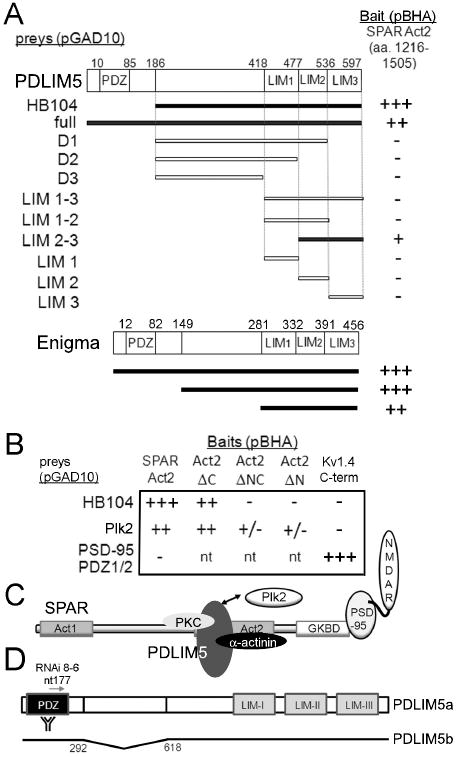
(A) Yeast 2-hybrid screen (Y2H) was performed using as bait SPAR Act2 domain in vector pBHA. Interaction strength for all Y2H assays was scored in terms of β-gal activity (+++, 0-15 min; ++, 15-30 min; +, 30 min to 2 hrs; +/-, 2 hrs to overnight; -, not detected; nt, not tested). Region of PDLIM5 encoded by positive clone HB104 and deletion analysis of PDLIM5 are shown below schematic diagram of PDLIM5 primary structure. Bottom, regions of Enigma tested in Y2H assay are shown below schematic diagram of Enigma primary structure. Numbers shown are amino acid residues; interacting clones are highlighted in black. (B) Deletion analysis of SPAR Act2. Domain constructs lacking the N terminal portion of Act2 (ΔC), the C terminal portion (ΔN), or both (ΔNC) were assayed by Y2H for interaction with PDLIM5 clone HB104. Plk2 in vector pGAD10 is used as a positive control for interaction with Act2, and PSD-95 PDZ1/2 domain in pGAD10 is used as negative control. Kv1.4 C-terminus is used as a positive control for PSD-95 PDZ1/2 interaction. (C) Schematic of SPAR interactions. SPAR domains shown are Act1 and 2, actin-associated domains 1 and 2; GKBD, guanylate kinase binding domain that interacts with the guanylate kinase-like domain of PSD-95. NMDAR, NMDA receptors. Double arrow denotes PDLIM5 and Plk2 interaction with the same region of SPAR. (D) Schematic of PDLIM5 reagents used in the study. PDLIM5a (full-length) and PDLIM5b (deletion of nt 292-618), position of RNAi 8-6, and PDZ region used as immunogen for PDLIM5 rabbit antibodies are shown.
Here we show that SPAR interacts with a PDZ-LIM domain family protein called PDZ and LIM domain 5 (PDLIM5), formerly known as Enigma Homolog (ENH). PDZ-LIM proteins are scaffolds consisting of a single PDZ domain and one to three LIM domains, each acting as protein-protein interaction modules. In addition to PDLIM5, this family includes the founding member Enigma (LMP-1/PDLIM7), as well as ALP, RIL, Elfin (CLP36), Mystique, ZASP (Cypher / Oracle), LMO7 and LIM domain kinases LIMK1 and LIMK2 (te Velthuis and Bagowski, 2007). Of major interest, single nucleotide polymorphisms in PDLIM5 have been associated with schizophrenia, depression, and bipolar disorder (Kato et al., 2005; Li et al., 2008; Liu et al., 2008), although the physiological functions of PDLIM5 are not well understood. The PDZ domain of PDLIM5 has been shown to interact with α-actinin in cardiomyocytes (Nakagawa et al., 2000), and its LIM domains have been identified as interaction sites for various PKC isoforms (Chen et al., 2006; Kuroda et al., 1996; Maeno-Hikichi et al., 2003). Presynaptically, PDLIM5 has been reported to physically tether PKCε with N-type calcium channels, thereby promoting phosphorylation-dependent modulation of channel properties (Kuroda et al., 1996; Maeno-Hikichi et al., 2003), although this point is now controversial (Gardezi et al., 2009). Thus, the precise molecular roles of PDLIM5 in severe psychiatric illnesses remain unclear.
We now report that PDLIM5 is expressed postsynaptically in excitatory pyramidal neurons of the hippocampus and associates with SPAR protein in the brain. In hippocampal neurons, PDLIM5 promotes decreased dendritic spine size, opposite to the effect of SPAR overexpression that causes spine head enlargement, and PDLIM5 spine localization is regulated by PKC. These results expand the repertoire of morphogens operating in dendritic spines and shed new light on the potential functions of PDLIM5 in plasticity and disease.
Results and Discussion
To better understand the mechanism by which SPAR regulates dendritic spine morphology, we performed a yeast two-hybrid (Y2H) screen to identify SPAR-associated proteins (Fig. 1A). The Act2 domain has been shown to be sufficient for association with F-actin filaments in hippocampal neurons as well as heterologous cells, and is required for SPAR to cause dendritic spine enlargement (Pak et al., 2001). Through a Y2H screen using SPAR Act2 as bait, we identified a positive interacting clone (HB104) representing a portion of the rat PDLIM5 coding region (Fig. 1A). Full-length PDLIM5a (see below for isoform nomenclature) also interacted, albeit not as strongly as HB104, with SPAR Act2. Deletion analysis using Y2H assays showed that the LIM2/3 domains of PDLIM5 were sufficient for the interaction with SPAR (Fig. 1A), although additional regions of PDLIM5 may also contribute to strength of interaction. A construct containing all 3 LIM domains did not interact with SPAR, suggesting either that LIM1 inhibits the interaction, or that the LIM1-3 fusion protein was not properly folded or expressed. Conversely, to narrow down the portion of SPAR that interacted with PDLIM5, we found that clone HB104 bound preferentially to the N-terminal part of Act2 domain, similar to Plk2 (Fig. 1B; see Fig. 1C for schematic model of proposed interactions).
To ask whether other PDZ-LIM proteins besides PDLIM5 also interact with SPAR, we tested the related family member Enigma by Y2H assay against the SPAR Act2 domain. We found that, like PDLIM5, full-length Enigma could also bind to Act2, and that the Enigma LIM domains were sufficient for this association (Fig. 1A). Thus, LIM domains of at least two PDZ-LIM domain family proteins are capable of recognizing SPAR Act2.
PDLIM5 isoforms are in the postsynaptic density
Several PDLIM5 isoform sequences have been deposited in GenBank. We used PCR to recover PDLIM5 from a rat brain cDNA library for further analysis, using primers against the extreme amino and carboxyl termini of the longest form of rat PDLIM5 (accession #NM_053326). In addition to a full-length isoform PDLIM5a (equivalent to human accession # NM_006457), encoding a protein product of ∼80 kD, we also recovered an additional isoform, termed PDLIM5b (orthologous to human accession # NP_001011513), that contains an in-frame deletion of nt 292-618 and encodes a smaller protein of ∼55 kD (Fig. 1D).
In neurons, PDLIM5 has previously been found to colocalize with synaptophysin (Maeno-Hikichi et al., 2003), a presynaptic marker, whereas SPAR is primarily found postsynaptically (Pak et al., 2001). To determine if PDLIM5 isoforms and SPAR could plausibly form a physiological complex, we raised rabbit antibodies against the N-terminal PDZ domain of rat PDLIM5 that should recognize all isoforms of PDLIM5 (Fig. 1D). Using these antibodies we analyzed PDLIM5 subcellular distribution in biochemical fractions of adult rat brain lysates. By immunoblotting, we observed only two prominent bands for PDLIM5, and these bands comigrated with recombinant PDLIM5a and PDLIM5b (Fig. 2A). Consistent with previous findings (Maeno-Hikichi et al., 2003), PDLIM5 co-distributed with synaptophysin in the synaptic vesicle-enriched fraction (LP2). Importantly, however, a subset of PDLIM5 also co-fractionated with PSD-95 in the PSD, where synaptophysin was absent, although the degree of enrichment for PDLIM5 in this fraction was not as great as for PSD-95 (Fig. 2A). Thus, PDLIM5 is widely found throughout various subcellular domains including both pre- and postsynaptic compartments, but is not a ‘core’ component of PSDs.
Figure 2. Biochemical association of PDLIM5 with SPAR.
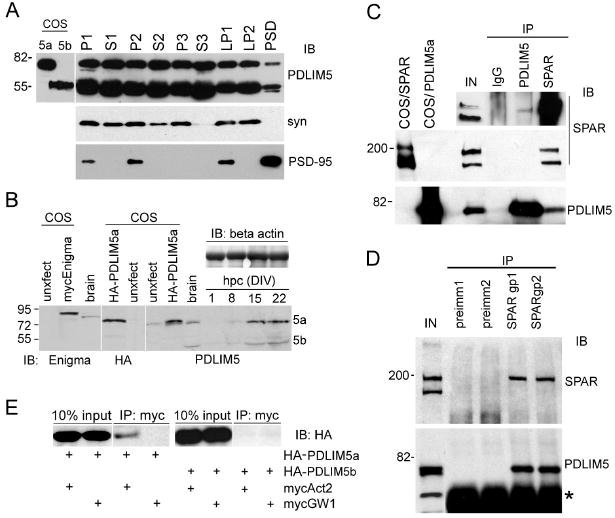
(A) Adult rat brains were homogenized, subjected to biochemical fractionation, and immunoblotted for PDLIM5 (GU8 rabbit antibodies), synaptophysin (syn), or PSD-95 as indicated. P1, crude pellet; P2, crude synaptosomes; P3, light membrane; S3, cytosolic; LP1, synaptic membrane; LP2, synaptic vesicle; PSD, postsynaptic density. PDLIM5 co-fractionated with both pre- and postsynaptic markers. COS-7 lysates expressing recombinant PDLIM5a or PDLIM5b are included (left lanes) to show the expected sizes of isoforms. (B) Cultured hippocampal neuron lysates (hpc) were harvested at various days in vitro (DIV) and immunoblotted for PDLIM5 (a and b isoforms are indicated). Beta actin levels are shown as loading control. To show the expected size of PDLIM5 and the specificity of antibodies, whole brain lysates or COS-7 transfected cell lysates of myc-Enigma or HA-PDLIM5a are included (left lanes as indicated) and immunoblotted with antibodies shown at bottom of blots. Note recombinant myc-Enigma and HA-PDLIM5a migrate slightly higher than endogenous counterparts due to the epitope tag. Unxfect, untransfected. (C) Adult rat brain lysates were immunoprecipitated with antibodies against PDLIM5 (GU8 rabbit) or SPAR rabbit, or nonimmune rabbit IgG, and immunoblotted for endogenous SPAR and PDLIM5, as indicated. COS-7 lysates expressing recombinant SPAR or PDLIM5 are included (left lanes) to show the expected size of full length proteins. Inset at top shows a longer exposure of a SPAR western from a duplicate experiment showing weak but detectable SPAR coimmunoprecipitated with PDLIM5 antibodies. IN, 10% lysate used per IP (1% lysate for overexposed inset). (D) Rat brain lysates immunoprecipitated with two independent anti-SPAR guinea pig sera (gp1 and gp2), or preimmune sera from the same animals (preimm1 and preimm2). Precipitates were immunoblotted for SPAR or PDLIM5, as indicated. Asterisk represents IgG heavy chain. IN, 10% of input used per immunoprecipitation reaction. (E) COS-7 cells were transfected with myc-tagged Act2 domain, HA-tagged full-length PDLIM5a or PDLIM5b, or empty mycGW1 vector as indicated. Lysates were immunoprecipitated with myc agarose beads and immunoblotted with HA epitope antibodies. Input is shown demonstrating equal expression of PDLIM5 between conditions. All molecular weights shown in kilodaltons.
In cultured hippocampal neurons, the expression levels of both PDLIM5a and b isoforms increased with in vitro development (Fig. 2B), with prominent expression at 15DIV coinciding with the peak of synaptogenesis and persisting through mature stages at 22DIV. To compare PDLIM5 distribution to that of Enigma, we found that Enigma was also expressed in adult rat brain lysates as well as in hippocampal neurons, and increased in abundance with developmental maturation (Supplemental Fig. S1A). Unlike PDLIM5, which was present at similar levels in membrane and cytosolic fractions, Enigma was found primarily in the cytosolic compartment. However, a minority of Enigma could be detected in various other biochemical fractions including in the PSD (Supplemental Fig. S1B). The antibodies against PDLIM5 and Enigma were specific and did not cross react, which could be shown unambiguously due to the slightly higher migration of Enigma relative to PDLIM5a (Fig. 2B). These results suggest that PDLIM5 and Enigma have overlapping subcellular distributions and thus may perform shared as well as distinct functions in neurons.
Biochemical interaction of PDLIM5 and SPAR
Based on their common presence in the PSD, we next tested for interaction between native PDLIM5 and SPAR in brain. Lysates from adult rat whole brain were detergent extracted using 1% deoxycholate and subjected to immunoprecipitation. We observed robust immunoprecipitation of PDLIM5a with our affinity purified PDLIM5 antibodies, and these antibodies also specifically coimmunoprecipitated SPAR, although the coprecipitation was weak (Fig. 2C). Interestingly, PDLIM5 was specifically associated with the higher molecular weight SPAR band at ∼200 kD, the species found primarily in the PSD fraction (Pak et al., 2001)(Supplemental Fig. S1B). In the “reverse” direction using SPAR rabbit antibodies, we recovered both bands of SPAR along with a strong recovery of PDLIM5a (Fig. 2C), suggesting that a higher proportion of SPAR is complexed with PDLIM5 than vice versa. This conclusion is consistent with the biochemical fractionation profiles indicating that PDLIM5 is more widely distributed than SPAR (Fig. 2A). PDLIM5b migrated in SDS-PAGE with IgG heavy chain and could not be visualized in this experiment. Control nonimmune rabbit IgG failed to precipitate either SPAR or PDLIM5. As additional confirmation of these interactions, we performed another set of coimmunoprecipitations using two independent guinea pig SPAR antisera (gp1 and gp2). Both antisera immunoprecipitated SPAR (preferentially recognizing the higher molecular weight species) in addition to robustly recovering PDLIM5a, whereas their respective negative control preimmune sera did not recover either protein (Fig. 2D). Furthermore, SPAR could also coimmunoprecipitate with Enigma from rat brain, and vice versa (Supplemental Fig. S1C), lending support to the idea that SPAR can form functional interactions with PDZ-LIM proteins.
We next cotransfected COS-7 cells with HA-epitope tagged PDLIM5a or PDLIM5b and myc-epitope tagged Act2 domain constructs to test whether Act2 of SPAR was sufficient to interact with PDLIM5 in a mammalian cellular environment. Immunoprecipitation of mycAct2 led to a clear but modest coprecipitation of HA-PDLIM5a (Fig. 2E). In contrast, HA-PDLIM5b was not precipitated by Act2 domain, suggesting that SPAR is preferentially associated with the larger PDLIM5 isoform. Thus, the linker sequence (nt 292-618) between the PDZ and LIM domains, which is lacking in PDLIM5b (Fig. 1D), may contribute to PDLIM5 interaction with SPAR.
SPAR recruits PDLIM5 to clusters in COS-7 cells
One possible explanation for low level of coimmunoprecipitation between SPAR and PDLIM5 could be that SPAR is known to be tightly associated with actin cytoskeleton and is far more difficult to solubilize than PDLIM5 (Fig. 2A and Supplemental Fig. S1B, compare relative abundance in P1 nonsolubilized fraction). Conditions that extract SPAR away from actin could therefore perturb other protein-protein interactions as well. To circumvent this issue and study the SPAR/PDLIM5 complex using an independent and less disruptive method, we employed immunocytochemistry of heterologous cells. As observed previously, recombinant SPAR overexpressed in COS-7 cells formed self-aggregating clusters (Fig. 3A) that potently recruit and reorganize F-actin (Pak et al., 2001). PDLIM5a expressed in COS-7 cells by itself was diffusely distributed throughout the cytoplasm and was occasionally enriched at apparent lamellopodial ruffles (Fig. 3B, arrows). In contrast, PDLIM5a became highly recruited to SPAR clusters when both were coexpressed (Fig. 3D, arrowheads). On the other hand, SPAR was not redistributed to membrane ruffles, where a fraction of PDLIM5a remained (Fig. 3D, arrow), suggesting that SPAR serves to recruit PDLIM5a, rather than the reverse. Similarly, SPAR and Enigma colocalized very strongly when coexpressed in COS-7 cells (Supplemental Fig. S2).
Figure 3. Colocalization of SPAR and PDLIM5 in COS-7 cells.

(A-C) COS-7 cells were singly transfected for SPAR, PDLIM5a, or SPAR ΔAct2 as indicated and immunostained for the exogenous protein. Arrows in (B) indicate examples of PDLIM5a enrichment at apparent membrane ruffles. (D) COS-7 cells doubly transfected with SPAR and PDLIM5a, and immunostained for SPAR (D1) or PDLIM5a (D2), with merged image shown in D3. Arrowheads, SPAR clusters that recruit PDLIM5. Arrow, example of PDLIM5 enrichment at membrane ruffle that does not recruit SPAR. (E) COS-7 cells doubly transfected with PDLIM5a and SPAR ΔAct2 and immunostained for SPAR (E1) or PDLIM5a (E2), with merged image shown in E3. (F) COS-7 cells doubly transfected with PDLIM5a and Act2 domain and immunostained for Act2 (F1) or PDLIM5a (F2), with merged image in F3. (G) COS-7 cells doubly transfected with PDLIM5b and SPAR and immunostained for SPAR (G1) or PDLIM5b (G2), with merged image in G3. Scale bar in (A), 5 μm.
The colocalization of PDLIM5a and SPAR required the Act2 domain of SPAR, since a SPAR deletion construct lacking this domain (ΔAct2) did not recruit PDLIM5a, which remained diffuse in its presence (Fig. 3E2). The ΔAct2 mutant was also unable to form clusters, either expressed alone (Fig. 3C) or with PDLIM5a (Fig. 3E1), and instead displayed a thin, fibrous distribution. Finally, the isolated Act2 domain fragment by itself was capable of forming clusters which colocalized nearly perfectly with PDLIM5a (Fig. 3F). Thus, the Act2 region is necessary and sufficient for PDLIM5a interaction with SPAR. Because the Act2 domain colocalized more strongly with PDLIM5a than the full length SPAR did, SPAR may contain additional regulatory elements that modulate its interaction with PDLIM5. In contrast, and consistent with the negative coimmunoprecipitation results, PDLIM5b was not recruited to SPAR clusters and the majority of PDLIM5b remained diffusely distributed in the presence of wildtype SPAR (Fig. 3G).
Distribution of PDLIM5 at excitatory synapses in hippocampal neurons
To determine the localization of endogenous PDLIM5, we employed immunocytochemistry of mature cultured hippocampal neurons at 19 days in vitro (DIV), a stage of development that expressed abundant PDLIM5 (Fig. 2B). We observed PDLIM5 colocalized with synaptophysin (Fig. 4G-I) as reported previously (Maeno-Hikichi et al., 2003), although the overlap was partial: many synaptophysin puncta did not coexist with detectable PDLIM5, and many clusters of PDLIM5 did not overlap with synaptophysin (Fig. 4M for quantification and 4N for linescan analysis). Similarly, PDLIM5 showed partial overlap with postsynaptic proteins SPAR and PSD-95 (Fig. 4A-C and D-F, respectively; quantified in 4M and linescans in 4N). However, little colocalization was observed between PDLIM5 and the inhibitory synaptic marker glutamic acid decarboxylase (GAD), particularly for larger puncta (Fig. 4J-L; quantified in 4M and linescan in 4N). These data are consistent with the biochemical fractionation experiments indicating both pre- and postsynaptic localization of PDLIM5 and demonstrate further that PDLIM5 is associated with a subset of excitatory synapses, but not with inhibitory synapses.
Figure 4. Colocalization of PDLIM5 with pre- and postsynaptic elements of excitatory synapses.
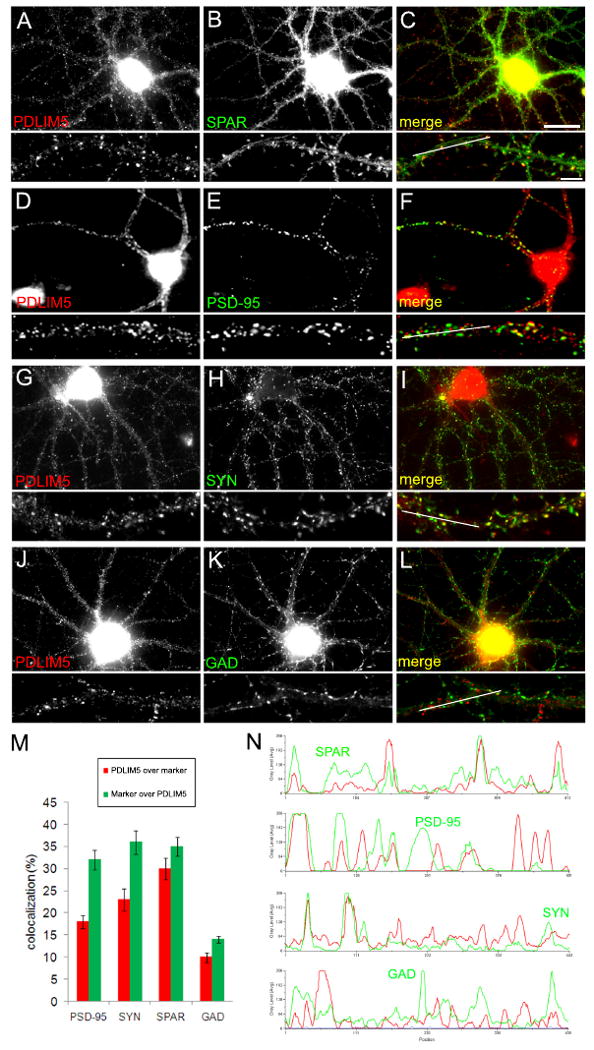
(A-L) Cultured hippocampal neurons at 19DIV were immunostained for endogenous PDLIM5 (GU8 rabbit antibodies, left panels) along with various synaptic markers (middle panels) as indicated, with merged images shown in right panels. Below each panel is a higher magnification view of a representative dendritic segment. White lines in merged dendrite images indicate regions used for linescan analysis in (N). Scale bar, 20 μm in (C), 5 μm for magnified dendrites. (M) Quantification of colocalization between PDLIM5 and synaptic markers PSD-95, synaptophysin (SYN), SPAR, or GAD. Data are means +/- SEM and are shown as percent colocalization of PDLIM5 with marker (red bars) or percent colocalization of marker with PDLIM5 (green bars). (N) Linescans showing representative overlap of PDLIM5 distribution (red lines) with SPAR, PSD-95, SYN, or GAD as indicated (green lines).
PDLIM5a promotes excitatory synapse contraction
To further understand the physiological postsynaptic roles of PDLIM5 in neurons, we generated several different PDLIM5 shRNA constructs to knock down endogenous protein levels by RNA interference (RNAi). These shRNAs were expressed from the vector pSuper, and varied in effectiveness as assessed in COS-7 cells co-transfected with HA-PDLIM5a (Fig. 5A and data not shown; quantified in Fig. 5B). The RNAi construct 8-6 was the most effective with ∼90% knockdown and was used in subsequent experiments. As RNAi 8-6 is targeted against a region of PDLIM5 shared by both PDLIM5a and PDLIM5b (see Fig. 1D), this construct also knocked down expression of HA-PDLIM5b to below detection in COS-7 cells (data not shown). Transfection of cultured hippocampal neurons with RNAi 8-6 and GFP for 3 days caused a marked loss (∼95%) in endogenous dendritic PDLIM5 expression levels (Fig. 5D) compared to GFP plus empty pSuper vector (Fig. 5C, quantified in Fig. 5B). In addition to its expression in neurons, there was highly abundant PDLIM5 protein observed in glial cells which was also knocked down to a similar degree (∼97%) by RNAi 8-6 (data not shown; quantified in Fig. 5B).
Figure 5. Generation of PDLIM5 knockdown and rescue constructs.
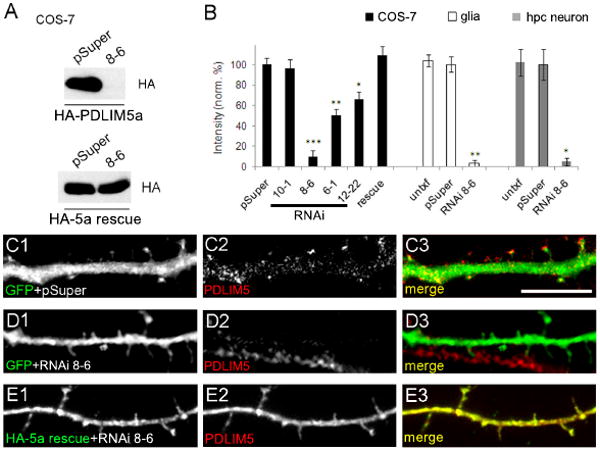
(A) COS-7 cells were cotransfected with HA-tagged PDLIM5a (upper blot) or HA-5a rescue construct (lower blot) and either pSuper vector or RNAi 8-6 as indicated. Total protein lysates were immunostained with HA antibodies. Results shown are representative of three independent experiments. (B) Quantification of PDLIM5 RNAi results against exogenous PDLIM5a transfected in COS-7 cells (black bars, n=3 western blots) or against endogenous PDLIM5 in glia (white bars, n=12 cells) and in neurons (gray bars, n=12 cells) as indicated. Untxf = untransfected neighboring cells used as internal negative control. Data are normalized to pSuper control and shown as mean +/- SEM. ***p<0.001, **p<0.01, one-way ANOVA with Tukey's post hoc test (COS-7 data) or Student's t-test (glial and hippocampal data). (C-E) Hippocampal neurons co-transfected with pEGFP and pSuper control (C), with pEGFP and PDLIM5 RNAi 8-6 (D), or with HA-5a rescue and RNAi 8-6 (E) were immunostained for GFP or HA antibodies (left panels) to identify transfected cells and for PDLIM5 in middle panels (C2, D2, E2). Merged images are shown in right panels. Scale bar, 5 μm.
Knockdown of PDLIM5 protein in neurons was accompanied by increased staining intensity of endogenous SPAR in neuronal dendrites as well as in spines (Fig. 6B). Knockdown of PDLIM5 also increased PSD-95 intensity (Fig. 6F) but not to the same extent as for SPAR (quantification in Fig. 6L, M). However, PDLIM5 knockdown caused no significant effect on total levels of synaptophysin (Fig. 6J; quantified in 6N). No effect was observed on any examined proteins by empty pSuper vector alone (Fig. 6A, E, I).
Figure 6. Effect of PDLIM5 knockdown on synaptic protein expression.
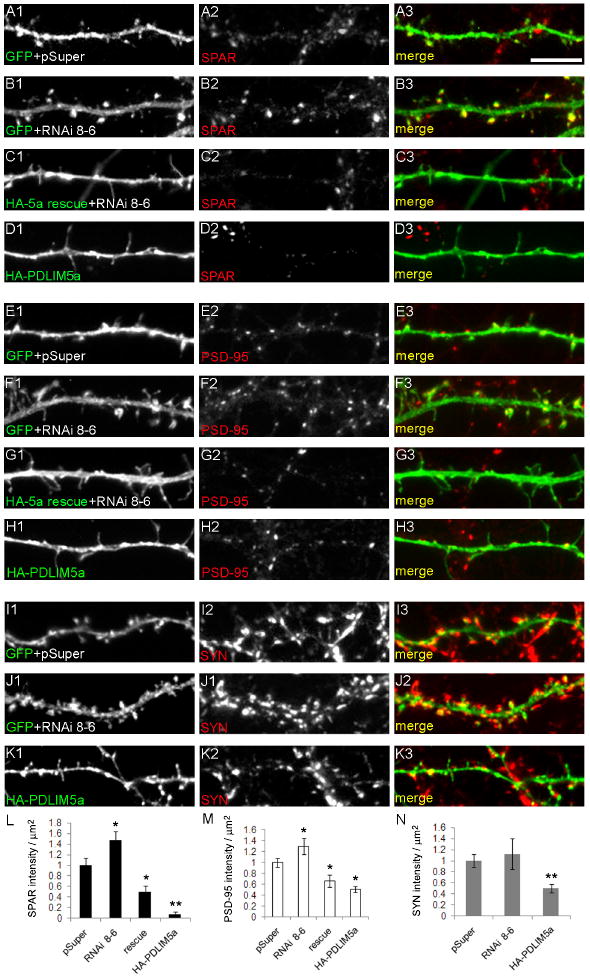
(A-K) Hippocampal neurons co-transfected with pEGFP and pSuper control (A, E, I), with pEGFP and PDLIM5 RNAi 8-6 (B, F, J), with HA-5a rescue and RNAi 8-6 (C, G), or with HA-PDLIM5a (D, H, K) were immunostained for GFP or HA as indicated (left panels) to identify transfected cells and for SPAR (A2-D2), PSD-95 (E2-H2), or synaptophysin (SYN; I2-K2) in middle panels. Merged images are shown in right panels. Scale bar, 5 μm. (L, M, N) Quantification of data in A-K. Data are shown as mean +/- SEM. **p<0.01, *p<0.05, one-way ANOVA with Tukey's post hoc test.
To verify the specificity of the PDLIM5 knockdown, we generated an HA-tagged PDLIM5a rescue construct (HA-5a) containing silent point mutations within the shRNA complementarity region conferring resistance to RNAi 8-6 in COS-7 cells (Fig. 5A) and in neurons (Fig. 5E). Detection of the exogenous rescue protein with PDLIM5 antibodies also confirms the efficacy of the immunostaining of PDLIM5 in neurons (Fig. 5E2). By coexpression of RNAi 8-6 with the HA-5a rescue construct, the increase in SPAR/PSD-95 staining intensity could be fully reversed, and was actually greatly reduced to a level significantly below control values (Fig. 6C and G, quantified in L, M).
The above data suggested that the rescue construct was not merely preventing the action of the RNAi 8-6, but was exerting a gain-of-function overexpression phenotype. To test this possibility, we overexpressed wild-type PDLIM5a in the absence of RNAi and observed more pronounced reductions in both SPAR (Fig. 6D; quantified in 6L) and PSD-95 (Fig. 6H; quantified in 6M). Furthermore, PDLIM5a overexpression also caused significant reductions in synaptophysin (Fig. 6K; quantified in 6N). Thus, native PDLIM5 expression is required to restrain postsynaptic growth of excitatory synapses, as judged by immunodetection of synaptic markers, and PDLIM5a actively promotes synaptic reduction.
PDLIM5a causes dendritic spine shrinkage
To determine whether the observed effects on synaptic puncta size reflected changes in dendritic spines, we cotransfected 15-19DIV cultured hippocampal neurons with HA epitope-tagged PDLIM5a and pEGFP After 3 days of expression, the recombinant PDLIM5a protein became distributed throughout the neuron, including dendritic protrusions, and we frequently observed long, thin filopodia-like structures and fewer mature mushroom shaped heads (Fig. 7B) compared to GFP control (Fig. 7A). Quantification indicated that spines were markedly longer in PDLIM5a expressing neurons, leading to an increased length-to-width ratio (Fig. 7H). To test whether PDLIM5a binding to SPAR was important for its effects on spines, we examined two constructs that were unable to interact with SPAR: PDLIM5b, and PDLIM5aΔLIM lacking LIM domains 2 and 3. Overexpression of either of these constructs caused a significant increase in dendritic spine head diameter (Fig. 7C, D), similar to the effect of SPAR overexpression (Pak et al., 2001). Thus, one postsynaptic function of PDLIM5a may be to interact with and thereby inhibit SPAR's ability to cause spine head growth. In contrast to these effects on spine size, there was no significant increase in the number of dendritic protrusions under any of our conditions compared to neurons transfected with GFP alone (Fig. 7I).
Figure 7. Role of PDLIM5 in spine morphogenesis.
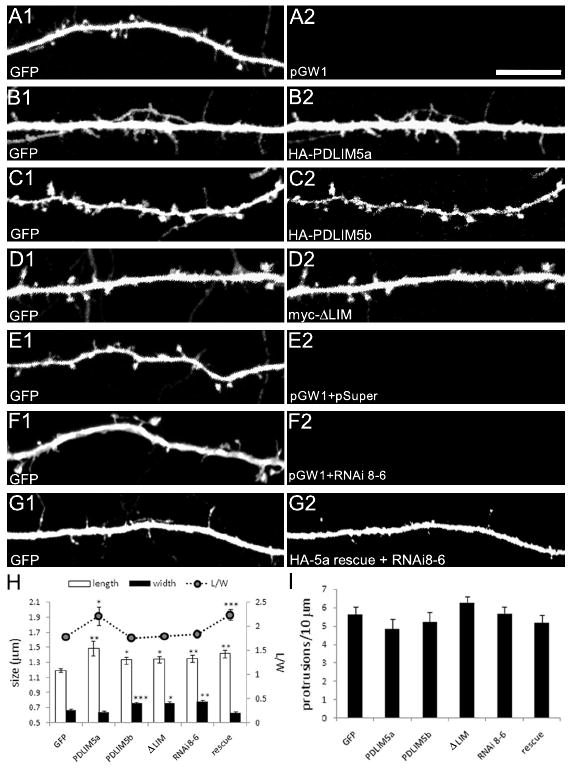
(A-G) Cultured hippocampal neurons were transfected with constructs as indicated and analyzed by confocal microscopy. In all cases, GFP (left panels) was included to visualize neuronal morphology and was used for quantification of spines. All cotransfected proteins were immunostained to verify expression (right panels). Scale bar, 10 μm. (H) Quantification of spine length and width (left axis) and ratio of length to width (L/W, right axis) for indicated conditions. ***p<0.001, **p<0.01, *p<0.05; one-way ANOVA with Tukey's post hoc test. (I) Quantification of linear density of dendritic protrusions for conditions as indicated.
Next we examined the effect of PDLIM5 knockdown on spine morphology. Quantification of spine dimensions and comparison to control GFP neurons cotransfected with empty pSuper and GW1 (Fig. 7E) revealed that loss of PDLIM5 protein resulted in both increased width and increased length of protrusions (Fig. 7F), similar to the effect of overexpression of PDLIM5b or ΔLIM (Fig. 7C, D, H). Thus, it is likely that the PDLIM5b and ΔLIM constructs act in a dominant negative manner to inhibit endogenous PDLIM5a. Both length and width of spines were increased to a similar degree, leading to an unchanged length-to-width ratio, suggesting that spine heads were enlarged uniformly in three dimensions (Fig. 7H). This enlargement of spines could be completely reversed by the HA-5a rescue construct coexpressed with RNAi 8-6, resulting in excessively thin spines resembling those in neurons overexpressing PDLIM5a alone (Fig. 7G, compare to 7B; quantified in H).
Finally, because PDLIM5 is known to bind PKC, we hypothesized that PDLIM5 function or localization may be regulated by PKC activity. Thus, we incubated cultured hippocampal neurons (27DIV) with phorbol 12-myristate 13-acetate (PMA), a potent and specific activator of PKC. In control neurons treated with vehicle (DMSO), SPAR and PDLIM5 immunostaining were observed in dendritic spines as well as dendritic shaft (Fig. 8A), as shown above (Fig. 4). However, neurons treated with PMA (1 μM, 10 min) showed a prominent enrichment of PDLIM5 in dendritic spine heads (Fig. 8B1). Analysis of integrated intensity within dendritic spines and in adjacent dendritic shaft (Fig. 8C2, D2) showed that the overall level of PDLIM5 in spines increased with PMA application with a slight but nonsignificant decrease in the dendrite (Fig. 8F). Together, these shifts produced a highly significant increase in the ratio of PDLIM5 in spines vs. dendrite (Fig. 8H). In contrast, SPAR levels were slightly increased in spines and decreased in dendrites by PMA, but neither to a significant degree (Fig 8C1, D1, quantified in 8E). However, the ratio of SPAR in spines vs. dendrites was significantly but modestly increased (Fig. 8G). These results suggest a redistribution of PDLIM5 (and SPAR to a lesser extent) from shaft to spine with PKC activation. The lack of significant decreases in dendrite staining may reflect the much larger volume in dendrite vs. spine, such that movement of PDLIM5 into the spines may not reduce the available dendritic pool to a measurable degree.
Figure 8. PKC activation drives PDLIM5 to dendritic spines.
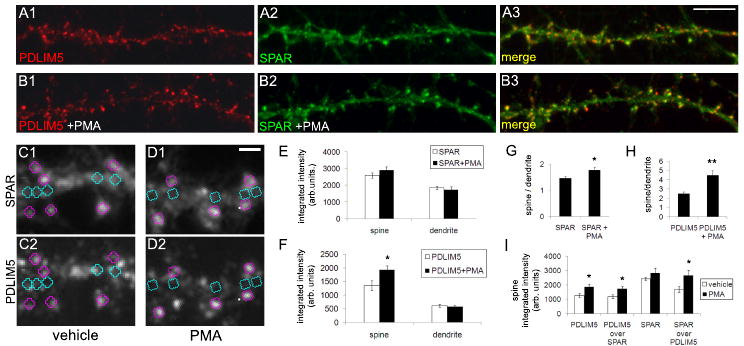
Cultured hippocampal neurons (27DIV) were incubated with (A) vehicle (DMSO) or (B) phorbol 12-myristate 13-acetate (PMA, 1 μM) for 10 min, then fixed and immunostained for PDLIM5 (A1, B1) or SPAR (A2, B2), with merged image shown at right (A3, B3). Scale bar in A3, 5 μm. (C-D) Integrated intensities of immunofluorescence signals for vehicle (C) and PMA treated neurons (D) was measured in 0.5 μm diameter circular regions in dendritic spines (pink) and adjacent dendrite (teal) of SPAR images (C1, D1) and regions of interest then transferred to corresponding PDLIM5 images (C2, D2) for intensity and colocalization measurement. Scale bar in D1, 1 μm. (E-F) Quantification of integrated intensity of (E) SPAR and (F) PDLIM5 in dendritic spines and dendritic shaft from data in (A, B). (G-H) Quantification of ratio of integrated intensity in dendritic spines vs. dendritic shaft for (G) SPAR and (H) PDLIM5 from data in (A, B). (I) Quantification of integrated intensity of PDLIM5 and SPAR in dendritic spines under conditions as indicated. PDLIM5 over SPAR and SPAR over PDLIM5 represent integrated intensity of colocalized pixels. *p<0.05, **p<0.01.
The increase in PDLIM5 in spines with PMA was accompanied by a corresponding increase in the integrated intensity of pixels colocalized with SPAR (Fig. 8I). However, the percent colocalization of PDLIM5 with SPAR was unchanged, as most of the PDLIM5 specifically within spines was already highly colocalized with SPAR (93.3 +/- 1.7% PDLIM5 colocalized with SPAR in spines with vehicle, 94.2 +/- 1.0% with PMA). The integrated pixel intensity of SPAR colocalized with PDLIM5 also increased with PMA (Fig. 8I), and notably the percent colocalization of SPAR with PDLIM5 increased considerably as well (69.0 +/- 6.14% SPAR colocalized with PDLIM5 in spines with vehicle, 92.1 +/- 2.8% with PMA; p=0.004 vs. vehicle), suggesting recruitment of PDLIM5 by SPAR. Thus, these data are consistent with the idea that SPAR may act as a targeting factor for PKC-regulated PDLIM5 spine delivery.
Our data have identified a novel protein complex involving SPAR and PDLIM5 in neurons that regulates dendritic spine morphogenesis. PDLIM5 is found at both pre- and postsynaptic sites in hippocampal neurons, and exclusively at excitatory synapses. The interaction with SPAR required the LIM2 and LIM3 domains of PDLIM5, regions previously implicated in binding to PKC (Chen et al., 2006; Kuroda et al., 1996; Maeno-Hikichi et al., 2003). However, the naturally occurring PDLIM5b isoform, which lacks only a linker region between the PDZ and LIM domains, was unable to bind to SPAR, suggesting that the linker and LIM2/3 may cooperate to interact with SPAR within the context of the full-length PDLIM5a protein. In addition to PDLIM5, the related protein Enigma also formed interactions with SPAR in vitro and in vivo, implicating Enigma in potential regulation of postsynaptic functions as well.
The interaction with PDLIM5 occurred through the Act2 domain of SPAR, a region required for SPAR to cause spine enlargement (Pak et al., 2001). Thus, one function for PDLIM5 could be to link SPAR to the F-actin cytoskeleton and thereby regulate morphological rearrangement; in this regard, the reported binding of PDLIM5 to the F-actin crosslinking protein α-actinin (Nakagawa et al., 2000) could facilitate such regulation. We recently found that SPAR Act2 domain also binds to α-actinin (Hoe et al., 2009), suggesting that PDLIM5 and α-actinin may form a triple complex with SPAR. Alternatively, these proteins may compete for SPAR binding. Such competition could explain the partial colocalization of PDLIM5a and SPAR in heterologous cells and neurons. In addition, the polo kinase Plk2 binds to the same region of SPAR Act2 as PDLIM5. Because Plk2 promotes SPAR degradation, PDLIM5 may assist Plk2 binding and thereby enhance SPAR destruction. Such a role could explain the spine enlargement and shrinkage due to decreased and increased PDLIM5 expression, respectively (see below). Clearly, the SPAR Act2 domain coordinates a complex assembly of several proteins (Fig. 1C) and additional studies will be needed to dissect the functional interrelationships among these various factors.
What is the postsynaptic function of PDLIM5? Our data implicate PDLIM5 in the regulation of excitatory synapse size. Overexpression of PDLIM5a caused dramatic loss of SPAR, as well as reductions in both PSD-95 and apposed synaptophysin. Thus, it is likely that the synapse as a whole is downregulated. With such a model, it is possible that loss of SPAR is merely secondary to shrinkage of synapses. However, the observation that SPAR levels are affected by PDLIM5 knockdown and overexpression to a greater degree than PSD-95 suggests that SPAR expression is directly controlled by PDLIM5, rather than indirectly via expansion or contraction of the PSD.
Consistent with this function for PDLIM5 in the inhibition of excitatory synaptic growth, knockdown of PDLIM5 increased the immunofluorescence intensity of SPAR and PSD-95. However, PDLIM5 knockdown did not cause a significant increase in synaptophysin staining. This observation may simply reflect a more modest effect on presynaptic terminals which could be masked due to the extremely strong synaptophysin signal normally present in these neurons (Fig. 6I). Alternatively, the growth of presynaptic terminals may lag temporally behind the postsynaptic effect. A final possibility is that PDLIM5 may actually be required to couple postsynaptic and presynaptic size changes in addition to restraining postsynaptic growth.
Interestingly, the localization of PDLIM5 in dendritic spines was stimulated by PKC, leading to greater colocalization with SPAR protein. Such regulated delivery of PDLIM5 to spines and SPAR could explain the relatively modest coprecipitation and partial colocalization of these proteins in brain, neuronal cultures, and heterologous systems, as well as the partial cofractionation of PDLIM5 with the PSD. It is not currently known which PKC isoforms are involved postsynaptically, although several PKC isoforms are known to be important for various forms of synaptic plasticity (Abeliovich et al., 1993; Sacktor, 2008; Wang and Feng, 1992) and can phosphorylate NMDA receptors (Tingley et al., 1993) as well as AMPA receptors (Blackstone et al., 1994; Roche et al., 1996; Tan et al., 1994). Another study reported the involvement of a related PDZ-LIM domain protein, RIL, in AMPA receptor trafficking (Schulz et al., 2004). Further work is required to determine whether PDLIM5 is capable of tethering PKC to channels or receptors in the PSD.
Regulation of dendritic spine morphology by PDLIM5 isoforms
With respect to dendritic spine morphology, SPAR and PDLIM5 function in opposing ways. Whereas SPAR promotes spine enlargement, PDLIM5a expression favors spine thinning and elongation. Consistent with this effect, knocking down PDLIM5 caused spine enlargement. It should be noted that the RNAi 8-6 construct knocks down both the spine-thinning PDLIM5a isoform and the spine-promoting PDLIM5b, but causes overall spine enlargement; thus the default pathway in the absence of PDLIM5 isoforms may be in favor of spine enlargement. Alternatively, the function of PDLIM5b may be simply to antagonize PDLIM5a. Similar to PDLIM5 depletion, overexpression of PDLIM5ΔLIM or PDLIM5b also increased spine size. Since these constructs did not associate with SPAR by immunoprecipitation or by colocalization, these results suggest SPAR binding could be important for PDLIM5 effects on spines. We hypothesize that PDLIM5b and ΔLIM sequester other as yet unidentified PDLIM5-associated factors from properly targeting to and inhibiting SPAR function.
The spine morphogenetic effects described here are all the more intriguing in view of accumulating evidence implicating PDLIM5 in brain disorders. PDLIM5 is located on human chromosome 4q22.3, a region implicated in schizophrenia by several independent linkage studies (Arinami, 2003; Gurling et al., 2001; Mowry et al., 2000). Furthermore, single nucleotide polymorphisms (SNPs) in PDLIM5 have been linked to schizophrenia (Horiuchi et al., 2006; Li et al., 2008), major depressive disorder (Liu et al., 2008), and bipolar disorder (Kato et al., 2005), suggesting that PDLIM5 may be a general susceptibility gene for psychiatric illnesses. Certain SNPs located within the 5′ untranslated region of PDLIM5 are associated with increased levels of PDLIM5 expression (Horiuchi et al., 2006), and mRNA levels of PDLIM5 have been found to be elevated in post-mortem brains of patients with schizophrenia, bipolar disorder, and major depression (Iwamoto et al., 2004a; Iwamoto et al., 2004b; Kato et al., 2005). Mimicking this upregulation by overexpressing PDLIM5 in cultured hippocampal neurons resulted in excessively thin dendritic spines. Such aberrant spine phenotypes resemble the thin spines or filopodia characteristic of a variety of mental retardations (Blanpied and Ehlers, 2004), leading to the hypothesis that impairment of SPAR or PDLIM5 spine regulation may be a contributing factor to major psychiatric illnesses as well.
PDLIM5 is also a homolog of an Alzheimer's Disease-associated Neuronal Thread Protein (AD7c-NTP) that is upregulated during the early stages of the disease (Wu et al., 2004), further emphasizing connections between PDZ-LIM proteins and clinically relevant processes. Thus, elucidating the link between PDLIM5 and dendritic spine morphogenesis may help to understand the cognitive deficits that accompany a variety of pathological conditions.
Experimental Methods
Constructs
The following constructs have been described: PSD-95 PDZ1/2 domains in pGAD10 vector and Kv1.4c in pBHA vector (Kim et al., 1995); SPAR-Act2 in pBHA and Plk2 in pGAD10 (Pak and Sheng, 2003); and myc-SPAR GW1, mycAct2 GW1, and mycSPARΔAct2 GW1 (Pak et al., 2001). PDLIM5a, PDLIM5b, and Enigma cDNAs were obtained by PCR from a rat brain Marathon cDNA library using amino- and carboxy-terminus primers and cloned into vector HA-GW1 or myc-GW1. Deletion constructs were generated by PCR with appropriate primers. For PDLIM5 and Enigma deletions in pGAD10, regions are denoted in Fig. 1A. HA-PDLIM5aΔLIM in GW1 lacks aa477-497. Deletions for SPAR-Act2 domain cloned in pBHA were ΔN (aa 1313-1505), ΔNC (aa 1313-1415) and ΔC (1216-1415). To generate PDLIM5 siRNA constructs, we used pSuper vector, which expresses a short interfering RNA (siRNA) under the control of the H1 promoter. PDLIM5 siRNA target sequences were as follows: 6-1, GCAGAGAGCTGAGCACATT; 8-6, GGGAATGACTCATCTCGAA (underlined nucleotides mutated in PDLIM5 rescue contruct, see below); 10-1, CCTTCTACTGGGAGAATTT; and 12-22, GCTGTATCCAAAGTCACTTCC. BLAST search was used to verify specificity of target sequences. PDLIM5a-rescue construct was generated by QuikChange (Stratagene) site directed mutagenesis system with the following silent mutations: ACGCACCTA. All constructs were verified by DNA sequencing.
Yeast two hybrid
Two-hybrid screen and assays were performed using the yeast strain L40 harboring β-gal and HIS3 reporters, as described (Niethammer and Sheng, 1998). 1 × 106 clones of a rat brain cDNA library in pGAD10 (Clontech) were screened using as bait the Act2 domain of SPAR (aa 1216-1505) cloned in pBHA.
Antibodies
The rabbit-anti-SPAR antibodies used for western blotting and immunoprecipitation have been described (Pak et al., 2001). SPAR rabbit antibodies GU4 (for immunocytochemistry) and guinea pig anti-SPAR gp1 and gp2 antibodies (for immunoprecipitation) were produced by immunization of rabbits or guinea pigs with a fusion protein pMal-SPAR GKBD region (Cocalico Biologicals). PDLIM5 antibodies GU8 were produced by immunization of rabbits with pMal-PDLIM5 PDZ domain (Cocalico Biologicals). The following antibodies were purchased from commercial sources: myc 9E10, myc agarose conjugate, and HA rabbit polyclonal (Santa Cruz Biotechnology); GFP monoclonal 3E6 (MP Biologicals); GAD65 clone GAD-6 (BD Biosciences); synaptophysin SVP38 monoclonal, and nonimmune goat and rabbit IgG (Sigma); K28/43 (PSD-95 specific) monoclonal antibodies (NeuroMabs); and Enigma/PDLIM7 monoclonal (Abnova) and rabbit polyclonal (Imgenex).
Western blotting
Proteins were transferred to a nitrocellulose membrane at 80V for 2 hours at 4°C. Blots were stained with Ponceau S for 15 min to verify protein levels and then blocked in TBST with 5% non-fat dried milk. Blots were incubated with primary antibodies at appropriate concentration for 2 hours at room temperature or overnight at 4°C. Blots were then rinsed 3 × 15 min in TBST and incubated with HRP-conjugated secondary antibody (1:2500) for two hours at room temperature in TBST with 5% non-fat dried milk. Blots were again rinsed 3 × 15 min in TBST and developed with Western Lightning (Perkin Elmer) or SuperSignal West Femto (Pierce) enhanced chemiluminesence reagent.
COS-7 transfections, immunostaining, and immunoprecipitation
COS-7 were maintained in DMEM with 10% fetal bovine serum and 0.1% gentamicin (Invitrogen). COS-7 cells were transiently transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol, and immunostained as described (Kim et al., 1995). For immunoprecipitation, cells were harvested in RIPA buffer and lysates centrifuged at 16,000 g for 15 minutes. Antibodies, or non-immune rabbit and mouse IgG/protein A or protein G sepharose conjugates were mixed with supernatants for 2 hr at 4°C. After washing 5× in RIPA buffer, immunoprecipitates were analyzed by immunoblotting.
Brain immunoprecipitation and fractionation
Immunoprecipitations from rat brain homogenates were performed as described (Dunah et al., 1998) except that whole brain lysates were extracted by 1% sodium deoxycholate for 1 hr at 4°C. For each immunoprecipitation, clarified extract (200 μg protein) was incubated with 10 μg of desired antibodies (or nonimmune IgG or preimmune serum) for 2 hr at 4°C. Precipitates were washed in 50 mM Tris pH 7.5, 150 mM NaCl, 0.1% Triton X-100. PSD purification (Cho et al., 1992) and subcellular biochemical fractionation (Huttner et al., 1979) of rat brain were performed essentially as described.
Neuronal culture and transfection
For spine morphology studies, hippocampal primary neuronal cultures prepared from embryonic day (E) 18-19 rat embryos were plated at high density (∼750 cells mm-2) (Sala et al., 2000). Rats were treated in accordance with NIH and Georgetown guidelines on animal care and use. For colocalization studies, medium density cultures (∼150 cells mm-2) were plated on coverslips coated with poly-D-lysine (30 μg/ml) and laminin (2 μg/ml). Cultures were grown in Neurobasal medium (Invitrogen) supplemented with B27 (Invitrogen), 0.5 mM glutamine and 12.5 μM glutamate. Neurons were transfected using calcium phosphate (Xia et al., 1996) with modifications (Jiang and Chen, 2006).
Immunocytochemistry
Coverslips, either neurons or COS-7 cells, were fixed in PBS containing 4% paraformaldehyde and 4% sucrose for 10 min at room temperature, or in cold methanol for 10 min at -20°C. For PDLIM5 immunocytochemistry neurons were fixed in 1% paraformaldehyde / 4% sucrose / PBS for 5 min at room temperature, followed by methanol fixation for 10 min at -20°C. Cells were rinsed 3 × 5 minutes in PBS, and then incubated in primary antibody either overnight at 4°C or 3 hrs at room temperature in 1× GDB antibody dilution buffer (Sala et al., 2000). Coverslips were then rinsed 3 × 15 min in PBS or neuron wash buffer (20 mM NaPO4, pH 7.4, 400 mM NaCl), and then incubated with the appropriate secondary antibody for two hrs at room temperature in GDB. Secondary antibodies used were donkey-anti-rabbit or anti-mouse AlexaFluor 488 (1:100) or AlexaFluor 555 (1:300); and donkey-anti-goat AlexaFluor 647, 1:50 (Invitrogen). Cover slips were washed 3 × 15 min in PBS or neuron wash buffer, rinsed with distilled water, and mounted on glass slides with VectaShield (Vector) mounting media. For some experiments neurons were treated with bath application of 1 μM phorbol 12-myristate 13-acetate for 10 min (Calbiochem).
Quantitation and image analysis
Images were acquired using a Fluoview confocal laser scanning microscope (Olympus) or Axiovert 200M epifluorescence inverted microscope (Zeiss) using consistent laser intensity or camera exposure levels, respectively, for each fluorescent marker in each experiment. Confocal z-series image stacks encompassing entire dendrite segments were analyzed using MetaMorph software (Universal Imaging Corporation, Downingtown, PA, USA). For quantitation of spines and staining intensity, 3 dendritic segments of 50 μm were collected from at least 12 neurons. For each condition, individual spine/intensity measurements were first grouped and averaged per neuron; means from several neurons were then averaged to obtain a population mean. For morphological classification, filopodia were defined as dendritic protrusions of greater than 2 μm in length, a length-to-width ratio of greater than 3. All other protrusions less than 2 μm in length were classified as spines. Filopodia classification was validated by visual inspection on a random sampling of protrusions to verify lack of discernible spine head.
Statistical analyses
All data are expressed as mean ± standard error of the mean. Data were analyzed for multiple groups using one-way ANOVA with Tukey's post hoc test or using Student's t-test for two group comparisons. Significance was determined at p<0.05.
Supplementary Material
(A) Dissociated hippocampal neuron cultures were homogenized at the indicated day in vitro (DIV) and immunoblotted with Enigma rabbit antibodies. Adult Br, total homogenate of adult rat brain. rEnigma, recombinant Engima expressed in COS-7 cells. NT, nontransfected COS-7 cells. (B) Adult rat brains were homogenized, subjected to biochemical fractionation, and immunoblotted with rabbit antibodies for Enigma or SPAR as indicated. H, total homogenate; P1, crude pellet; P2, crude synaptosomes; S3, cytosolic; PSD, postsynaptic density. (C) Adult rat brain lysates were immunoprecipitated (IP) with nonimmune mouse IgG (mIgG), Enigma monoclonal antibodies, nonimmune rabbit IgG (rIgG) or with anti-SPAR rabbit antibodies as indicated. Pellets were immunoblotted (IB) with rabbit antibodies for endogenous SPAR or Enigma as shown. IN, 5% of input used per immunoprecipitation reaction. Molecular weights shown in kilodaltons.
(A) COS-7 cells were transfected with Enigma and immunostained for the exogenous protein. (B-D) COS-7 cells doubly transfected with SPAR and Enigma, and immunostained for Enigma (B) or SPAR (C), with merged image shown in D. Scale bar, 5 μm.
Acknowledgments
SH, DME, JYL, NU and DTSP performed the experiments; DTSP designed the experiments and wrote the manuscript. This work was supported by NIH/NINDS grant NS048085 (DTSP). We thank members of the Pak laboratory for critical reading of this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abeliovich A, Chen C, Goda Y, Silva AJ, Stevens CF, Tonegawa S. Modified hippocampal long-term potentiation in PKC gamma-mutant mice. Cell. 1993;75:1253–1262. doi: 10.1016/0092-8674(93)90613-u. [DOI] [PubMed] [Google Scholar]
- Arinami T. Initial genome-wide scan for linkage with schizophrenia in the Japanese Schizophrenia Sib-Pair Linkage Group (JSSLG) families. Am J Med Genet B Neuropsychiatr Genet. 2003;120B:22–28. doi: 10.1002/ajmg.b.20022. [DOI] [PubMed] [Google Scholar]
- Blackstone C, Murphy TH, Moss SJ, Baraban JM, Huganir RL. Cyclic AMP and synaptic activity-dependent phosphorylation of AMPA-preferring glutamate receptors. J Neurosci. 1994;14:7585–7593. doi: 10.1523/JNEUROSCI.14-12-07585.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpied TA, Ehlers MD. Microanatomy of dendritic spines: emerging principles of synaptic pathology in psychiatric and neurological disease. Biol Psychiatry. 2004;55:1121–1127. doi: 10.1016/j.biopsych.2003.10.006. [DOI] [PubMed] [Google Scholar]
- Bourne J, Harris KM. Do thin spines learn to be mushroom spines that remember? Curr Opin Neurobiol. 2007;17:381–386. doi: 10.1016/j.conb.2007.04.009. [DOI] [PubMed] [Google Scholar]
- Bourne JN, Harris KM. Balancing structure and function at hippocampal dendritic spines. Annu Rev Neurosci. 2008;31:47–67. doi: 10.1146/annurev.neuro.31.060407.125646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlisle HJ, Kennedy MB. Spine architecture and synaptic plasticity. Trends Neurosci. 2005;28:182–187. doi: 10.1016/j.tins.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Chen Y, Lai M, Maeno-Hikichi Y, Zhang JF. Essential role of the LIM domain in the formation of the PKCepsilon-ENH-N-type Ca2+ channel complex. Cell Signal. 2006;18:215–224. doi: 10.1016/j.cellsig.2005.04.007. [DOI] [PubMed] [Google Scholar]
- Cho KO, Hunt CA, Kennedy MB. The rat brain postsynaptic density fraction contains a homolog of the Drosophila discs-large tumor suppressor protein. Neuron. 1992;9:929–942. doi: 10.1016/0896-6273(92)90245-9. [DOI] [PubMed] [Google Scholar]
- De Roo M, Klauser P, Garcia PM, Poglia L, Muller D. Spine dynamics and synapse remodeling during LTP and memory processes. Prog Brain Res. 2008;169:199–207. doi: 10.1016/S0079-6123(07)00011-8. [DOI] [PubMed] [Google Scholar]
- Dunah AW, Luo J, Wang YH, Yasuda RP, Wolfe BB. Subunit composition of N-methyl-D-aspartate receptors in the central nervous system that contain the NR2D subunit. Mol Pharmacol. 1998;53:429–437. doi: 10.1124/mol.53.3.429. [DOI] [PubMed] [Google Scholar]
- Ethell IM, Pasquale EB. Molecular mechanisms of dendritic spine development and remodeling. Prog Neurobiol. 2005;75:161–205. doi: 10.1016/j.pneurobio.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Gardezi SR, Weber AM, Li Q, Wong FK, Stanley EF. PDLIM5 is not a neuronal CaV2.2 adaptor protein. Nat Neurosci. 2009;12:957–958. doi: 10.1038/nn0809-957a. author reply 958. [DOI] [PubMed] [Google Scholar]
- Gurling HM, Kalsi G, Brynjolfson J, Sigmundsson T, Sherrington R, Mankoo BS, Read T, Murphy P, Blaveri E, McQuillin A, Petursson H, Curtis D. Genomewide genetic linkage analysis confirms the presence of susceptibility loci for schizophrenia, on chromosomes 1q32.2, 5q33.2, and 8p21-22 and provides support for linkage to schizophrenia, on chromosomes 11q23.3-24 and 20q12.1-11.23. Am J Hum Genet. 2001;68:661–673. doi: 10.1086/318788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoe HS, Lee JY, Pak DT. Combinatorial morphogenesis of dendritic spines and filopodia by SPAR and alpha-actinin2. Biochem Biophys Res Commun. 2009;384:55–60. doi: 10.1016/j.bbrc.2009.04.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi Y, Arai M, Niizato K, Iritani S, Noguchi E, Ohtsuki T, Koga M, Kato T, Itokawa M, Arinami T. A polymorphism in the PDLIM5 gene associated with gene expression and schizophrenia. Biol Psychiatry. 2006;59:434–439. doi: 10.1016/j.biopsych.2005.07.041. [DOI] [PubMed] [Google Scholar]
- Huttner WB, Meyermann R, Neuhoff V, Althaus HH. Neurochemical and morphological studies of bulk isolated rat brain cells. II. Preparation of viable cerebral neurons which retain synaptic complexes. Brain Res. 1979;171:225–237. doi: 10.1016/0006-8993(79)90329-9. [DOI] [PubMed] [Google Scholar]
- Iwamoto K, Bundo M, Washizuka S, Kakiuchi C, Kato T. Expression of HSPF1 and LIM in the lymphoblastoid cells derived from patients with bipolar disorder and schizophrenia. J Hum Genet. 2004a;49:227–231. doi: 10.1007/s10038-004-0136-5. [DOI] [PubMed] [Google Scholar]
- Iwamoto K, Kakiuchi C, Bundo M, Ikeda K, Kato T. Molecular characterization of bipolar disorder by comparing gene expression profiles of postmortem brains of major mental disorders. Mol Psychiatry. 2004b;9:406–416. doi: 10.1038/sj.mp.4001437. [DOI] [PubMed] [Google Scholar]
- Jiang M, Chen G. High Ca2+-phosphate transfection efficiency in low-density neuronal cultures. Nat Protoc. 2006;1:695–700. doi: 10.1038/nprot.2006.86. [DOI] [PubMed] [Google Scholar]
- Kato T, Iwayama Y, Kakiuchi C, Iwamoto K, Yamada K, Minabe Y, Nakamura K, Mori N, Fujii K, Nanko S, Yoshikawa T. Gene expression and association analyses of LIM (PDLIM5) in bipolar disorder and schizophrenia. Mol Psychiatry. 2005;10:1045–1055. doi: 10.1038/sj.mp.4001719. [DOI] [PubMed] [Google Scholar]
- Kim E, Niethammer M, Rothschild A, Jan YN, Sheng M. Clustering of Shaker-type K+ channels by interaction with a family of membrane-associated guanylate kinases. Nature. 1995;378:85–88. doi: 10.1038/378085a0. [DOI] [PubMed] [Google Scholar]
- Kuroda S, Tokunaga C, Kiyohara Y, Higuchi O, Konishi H, Mizuno K, Gill GN, Kikkawa U. Protein-protein interaction of zinc finger LIM domains with protein kinase C. J Biol Chem. 1996;271:31029–31032. doi: 10.1074/jbc.271.49.31029. [DOI] [PubMed] [Google Scholar]
- Li C, Tao R, Qin W, Zheng Y, He G, Shi Y, Li X, Guo Z, Chen H, Feng G, He L. Positive association between PDLIM5 and schizophrenia in the Chinese Han population. Int J Neuropsychopharmacol. 2008;11:27–34. doi: 10.1017/S1461145707007687. [DOI] [PubMed] [Google Scholar]
- Lippman J, Dunaevsky A. Dendritic spine morphogenesis and plasticity. J Neurobiol. 2005;64:47–57. doi: 10.1002/neu.20149. [DOI] [PubMed] [Google Scholar]
- Liu Z, Liu W, Xiao Z, Wang G, Yin S, Zhu F, Wang H, Cheng J, Wang X, He X, Li W. A major single nucleotide polymorphism of the PDLIM5 gene associated with recurrent major depressive disorder. J Psychiatry Neurosci. 2008;33:43–46. [PMC free article] [PubMed] [Google Scholar]
- Maeno-Hikichi Y, Chang S, Matsumura K, Lai M, Lin H, Nakagawa N, Kuroda S, Zhang JF. A PKC epsilon-ENH-channel complex specifically modulates N-type Ca2+ channels. Nat Neurosci. 2003;6:468–475. doi: 10.1038/nn1041. [DOI] [PubMed] [Google Scholar]
- Mowry BJ, Ewen KR, Nancarrow DJ, Lennon DP, Nertney DA, Jones HL, O'Brien MS, Thornley CE, Walters MK, Crowe RR, Silverman JM, Endicott J, Sharpe L, Hayward NK, Gladis MM, Foote SJ, Levinson DF. Second stage of a genome scan of schizophrenia: study of five positive regions in an expanded sample. Am J Med Genet. 2000;96:864–869. [PubMed] [Google Scholar]
- Nakagawa N, Hoshijima M, Oyasu M, Saito N, Tanizawa K, Kuroda S. ENH, containing PDZ and LIM domains, heart/skeletal muscle-specific protein, associates with cytoskeletal proteins through the PDZ domain. Biochem Biophys Res Commun. 2000;272:505–512. doi: 10.1006/bbrc.2000.2787. [DOI] [PubMed] [Google Scholar]
- Newey SE, Velamoor V, Govek EE, Van Aelst L. Rho GTPases, dendritic structure, and mental retardation. J Neurobiol. 2005;64:58–74. doi: 10.1002/neu.20153. [DOI] [PubMed] [Google Scholar]
- Niethammer M, Sheng M. Identification of ion channel-associated proteins using the yeast two-hybrid system. Methods Enzymol. 1998;293:104–122. doi: 10.1016/s0076-6879(98)93010-5. [DOI] [PubMed] [Google Scholar]
- Pak DT, Sheng M. Targeted protein degradation and synapse remodeling by an inducible protein kinase. Science. 2003;302:1368–1373. doi: 10.1126/science.1082475. [DOI] [PubMed] [Google Scholar]
- Pak DT, Yang S, Rudolph-Correia S, Kim E, Sheng M. Regulation of dendritic spine morphology by SPAR, a PSD-95-associated RapGAP. Neuron. 2001;31:289–303. doi: 10.1016/s0896-6273(01)00355-5. [DOI] [PubMed] [Google Scholar]
- Purpura DP. Dendritic spine “dysgenesis” and mental retardation. Science. 1974;186:1126–1128. doi: 10.1126/science.186.4169.1126. [DOI] [PubMed] [Google Scholar]
- Roche KW, O'Brien RJ, Mammen AL, Bernhardt J, Huganir RL. Characterization of multiple phosphorylation sites on the AMPA receptor GluR1 subunit. Neuron. 1996;16:1179–1188. doi: 10.1016/s0896-6273(00)80144-0. [DOI] [PubMed] [Google Scholar]
- Sacktor TC. PKMzeta, LTP maintenance, and the dynamic molecular biology of memory storage. Prog Brain Res. 2008;169:27–40. doi: 10.1016/S0079-6123(07)00002-7. [DOI] [PubMed] [Google Scholar]
- Sala C, Rudolph-Correia S, Sheng M. Developmentally regulated NMDA receptor-dependent dephosphorylation of cAMP response element-binding protein (CREB) in hippocampal neurons. J Neurosci. 2000;20:3529–3536. doi: 10.1523/JNEUROSCI.20-10-03529.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert V, Dotti CG. Transmitting on actin: synaptic control of dendritic architecture. J Cell Sci. 2007;120:205–212. doi: 10.1242/jcs.03337. [DOI] [PubMed] [Google Scholar]
- Schulz TW, Nakagawa T, Licznerski P, Pawlak V, Kolleker A, Rozov A, Kim J, Dittgen T, Kohr G, Sheng M, Seeburg PH, Osten P. Actin/alpha-actinin-dependent transport of AMPA receptors in dendritic spines: role of the PDZ-LIM protein RIL. J Neurosci. 2004;24:8584–8594. doi: 10.1523/JNEUROSCI.2100-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada T, Sheng M. Molecular mechanisms of dendritic spine morphogenesis. Curr Opin Neurobiol. 2006;16:95–101. doi: 10.1016/j.conb.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Tan SE, Wenthold RJ, Soderling TR. Phosphorylation of AMPA-type glutamate receptors by calcium/calmodulin-dependent protein kinase II and protein kinase C in cultured hippocampal neurons. J Neurosci. 1994;14:1123–1129. doi: 10.1523/JNEUROSCI.14-03-01123.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Velthuis AJ, Bagowski CP. PDZ and LIM domain-encoding genes: molecular interactions and their role in development. ScientificWorldJournal. 2007;7:1470–1492. doi: 10.1100/tsw.2007.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tingley WG, Roche KW, Thompson AK, Huganir RL. Regulation of NMDA receptor phosphorylation by alternative splicing of the C-terminal domain. Nature. 1993;364:70–73. doi: 10.1038/364070a0. [DOI] [PubMed] [Google Scholar]
- Wang JH, Feng DP. Postsynaptic protein kinase C essential to induction and maintenance of long-term potentiation in the hippocampal CA1 region. Proc Natl Acad Sci U S A. 1992;89:2576–2580. doi: 10.1073/pnas.89.7.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Li Y, Ji C, Xu J, Zheng H, Zou X, Gu S, Lou Y, Xie Y, Mao Y. Cloning and identification of a novel human gene PDLIM5, a homolog of AD-associated neuronal thread protein (AD7c-NTP) DNA Seq. 2004;15:144–147. doi: 10.1080/10425170310001656756. [DOI] [PubMed] [Google Scholar]
- Xia Z, Dudek H, Miranti CK, Greenberg ME. Calcium influx via the NMDA receptor induces immediate early gene transcription by a MAP kinase/ERK-dependent mechanism. J Neurosci. 1996;16:5425–5436. doi: 10.1523/JNEUROSCI.16-17-05425.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Dissociated hippocampal neuron cultures were homogenized at the indicated day in vitro (DIV) and immunoblotted with Enigma rabbit antibodies. Adult Br, total homogenate of adult rat brain. rEnigma, recombinant Engima expressed in COS-7 cells. NT, nontransfected COS-7 cells. (B) Adult rat brains were homogenized, subjected to biochemical fractionation, and immunoblotted with rabbit antibodies for Enigma or SPAR as indicated. H, total homogenate; P1, crude pellet; P2, crude synaptosomes; S3, cytosolic; PSD, postsynaptic density. (C) Adult rat brain lysates were immunoprecipitated (IP) with nonimmune mouse IgG (mIgG), Enigma monoclonal antibodies, nonimmune rabbit IgG (rIgG) or with anti-SPAR rabbit antibodies as indicated. Pellets were immunoblotted (IB) with rabbit antibodies for endogenous SPAR or Enigma as shown. IN, 5% of input used per immunoprecipitation reaction. Molecular weights shown in kilodaltons.
(A) COS-7 cells were transfected with Enigma and immunostained for the exogenous protein. (B-D) COS-7 cells doubly transfected with SPAR and Enigma, and immunostained for Enigma (B) or SPAR (C), with merged image shown in D. Scale bar, 5 μm.


