Abstract
The dynamic nature of the chromatin fiber provides the structural and functional flexibility required for the accurate transcriptional responses to various stimuli. In living cells, structural proteins such as the linker histone H1 and the High Mobility Group (HMG) proteins continuously modulate the local and global architecture of the chromatin fiber and affect the binding of regulatory factors to their nucleosomal targets. HMGN proteins specifically bind to the nucleosome core particle through a highly conserved “nucleosomal binding domain” (NBD) and reduce chromatin compaction. HMGN5 (NSBP1), a new member of the HMGN protein family, is ubiquitously expressed in mouse and human tissues. Similar to other HMGNs, HMGN5 is a nuclear protein which binds to nucleosomes via NBD, unfolds chromatin and affects transcription. This protein remains mainly uncharacterized and its biological function is unknown. In this review we describe the structure of the HMGN5 gene and the known properties of the HMGN5 protein. We present recent findings related to the expression pattern of the protein during development, the mechanism of HMGN5 action on chromatin, and discuss the possible role of HMGN5 in pathological and physiological processes.
Keywords: HMGN5, NSBP1, chromatin, linker histone H1, transcription, nucleosome
1. Introduction
Essential cellular processes such as transcription, DNA repair, replication and recombination occur in the context of chromatin. Therefore, the architecture of the chromatin fiber plays a key role in the integration of endogenous and exogenous signals for proper adjustment of cellular responses to a constantly changing environment.
A hallmark of chromatin organization is the dynamic nature of the chromatin fiber [1, 2]. Histones in the nucleosome core particle, the basic unit of chromatin, are subjected to numerous post translational modifications, collectively known as the “histone code” that directly affects the structure of chromatin [3]. The pattern of histone modifications is established and maintained through the balancing action of chromatin modifying enzymes that continuously add and remove modifications to histone tails. Histone modifications are recognized by chromatin remodeling complexes thereby providing the molecular platform for chromatin remodeling [4]. The next level of complexity of chromatin fiber is the formation of higher order chromatin structures, such as the 30 nm fiber, tertiary structures of chromatin loops, and entire chromosomes [5]. The dynamic properties of these structures are regulated by numerous chromatin modifying factors including a complex network of nucleosome-binding chromatin architectural proteins, such as linker histone H1 [6] and High Mobility Group (HMG) proteins [7]. The activity of the linker histone H1 is required for the formation of higher order chromatin structures and chromatin compaction whereas HMGNs, such as HMGN1 and HMGN2 compete with histone H1 for the binding to nucleosomes and counteract this function [7–9,14].
High Mobility Group N (HMGN) proteins are a family of ubiquitous nuclear proteins which are expressed in higher eukaryotes and specifically interact with nucleosomes without any known preference for the underlying DNA [10]. This interaction is mediated by the Nucleosome Binding Domain (NBD), a highly conserved sequence motif that is the signature of the HMGN protein family. Additionally, all members of the HMGN family contain negatively charged and highly unstructured C-terminal regulatory domain which increases the affinity of HMGN binding to the nucleosome and is involved in transcriptional activation [11]. However, the molecular function of this domain is not fully understood. Based on these structural properties, six proteins have been identified as members of HMGN protein family: HMGN1 (HMG14), HMGN2 (HMG17), HMGN3a and 3b, HMGN4, and HMGN5, previously known as NBP-45, GARP45 and NSBP1.
Fluorescence Recovery after Photobleaching (FRAP) experiments demonstrate that HMGN proteins are highly dynamic and are constantly moving in the nucleus in a “stop and go” manner [12]. HMGNs destabilize linker histone H1 binding to nucleosomes by competing for chromatin binding sites [9], unfold higher order chromatin structures [13, 14] and modulate transcription [15, 16]. In addition, HMGNs affect several histone modifications thus adjusting the chromatin architecture at the level of the nucleosome [17–19]. Also, it has been recently reported that recombinant HMGN proteins can counteract the ATP-dependent chromatin remodeling activities in vitro in reversible and dynamic manner [20].
Studies of HMGN1 knock out (KO) mice reveal that these mice are hypersensitive to UV and ionizing radiation and have a higher incidence of multiple malignant tumors and metastases [21, 22] The underlying molecular mechanism for the increased tumor burden of HMGN1 KO mice lies in the ability of HMGN1 to optimize the interaction of the DNA repair machinery with chromatin [23]. Aberrant expression of HMGN proteins during mouse development and cellular differentiation is detrimental for early embryonic development, myotube formation and chondrocyte differentiation, indicating that proper regulation of HMGN protein expression is important for these processes [16, 24, 25].
HMGN5 protein was first identified in our laboratory based on its structural similarity to other HMGN proteins [26]. HMGN5 is a typical member of the HMGN family which is localized to the nucleus and contains a functional NBD and negatively charged C-terminus [26]. Unlike other HMGNs, the C-terminus of HMGN5 is unusually long (more than 300 amino acids long in the mouse protein and around 200 amino acids long in the human protein) and affects cellular localization and architectural properties of the protein [27].
HMGN5 remains mainly uncharacterized and its biological function is unknown. In this review we summarize the available data on molecular and biochemical properties of HMGN5 gene and protein, expression pattern during development and its role in the regulation of chromatin architecture and transcription. We discuss the potential association of HMGN5 with physiological and pathological processes, including cancer progression. We note that most of the recent data come from work on the mouse HMGN5 protein and further research is required to elucidate the functional importance of this unusual chromatin architectural protein in other species.
2. The structure of the HMGN5 gene
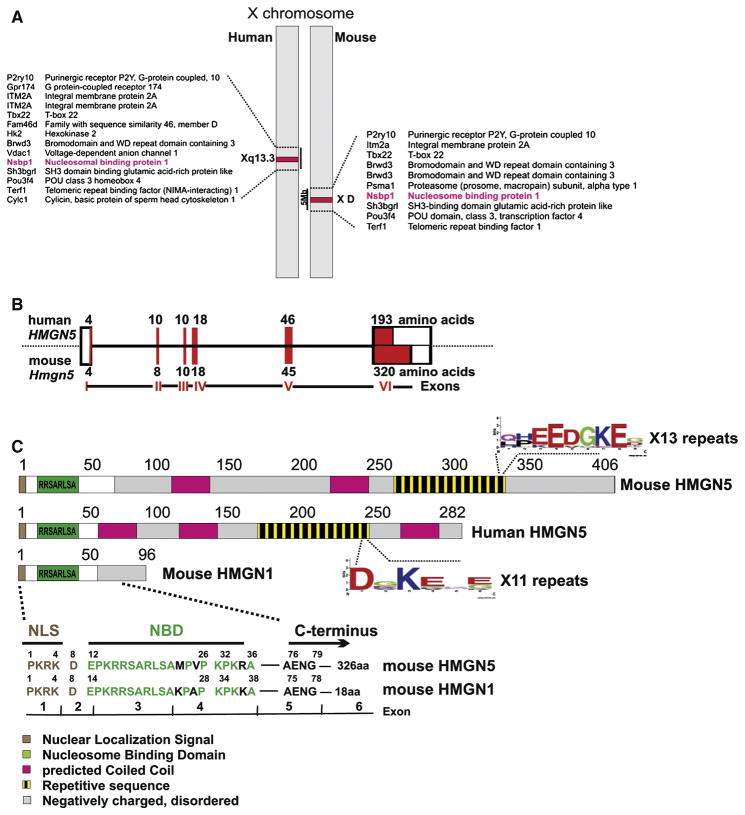
Analysis of the databases of ortholog genes suggest that the Hmgn5 gene appeared late in evolution and is only present in rats, mice, cows, monkeys and human. For other species such as dogs, pigs, chicken and lower eukaryotes no similar gene or protein are predicted. Genes coding for both mouse and human HMGN5 proteins are located at syntenic regions of the X chromosome (Xq13.3 for human gene and X D for mouse gene, Fig 1A) and span approximately 8 Kb regions. In contrast to the Hmgn1, which has multiple copies of non functional retropseudogenes Hmgn5 gene is present in the genome in a single copy [28]. The open reading frame of Hmgn5 is flanked by a short 5′ UTR and a long 3′ UTR which contains several polyadenylation signals. Hmgn5 is composed of 6 exons and 5 introns which encode a 282 amino acids long human protein and 406 amino acids long mouse protein. A significantly shorter exon 6 of the human HMGN5 gene accounts for the length difference between the two proteins (Fig 1B). In humans the protein region encoded by the last exon of HMGN5 contains sequences highly similar to HAL1 and LINE1 retrotransposable elements and sequences corresponding to the HERVH endogenous retrovirus [28].
Figure 1. Comparison of human and mouse HMGN5.
(A) Human and mouse HMGN5 genes are located at syntenic regions of the X-chromosome. (B) Organization of the Hmgn5 gene. Exons are marked by red blocks and untranslated regions are marked by white boxes. The number of amino acids encoded by each exon is indicated. (C) Schematic representation of structural domains of mouse and human HMGN5 and mouse HMGN1 proteins. Localization of each domain is related to the amino acid sequences of the proteins numbered on top of each scheme. Proteins are drawn to scale. Sequence alignment of the N-terminus of mouse HMGN5 and mouse HMGN1 and the corresponding gene organization are shown. Only amino acids from the highly similar regions are indicated. NLS – nuclear localization signal. NBD – nucleosome binding domain.
3. The properties of the HMGN5 protein
Human and mouse HMGN5 proteins show 59% amino acid identity (86% similarity) and are structurally similar (Fig 1C). The N-terminal part of HMGN5 contains a nuclear localization signal and the NBD. RRSARLSA is the highly conserved functional domain of the NBD which defines the ability of HMGN proteins to specifically interact with nucleosomes [29]. Additional features of HMGN5 associated with HMGN origin include the asymmetric charge distribution along the molecule: the N-terminal region containing NBD is positively charged, whereas C-terminal tail is highly acidic [10]. Analysis of the sequence of HMGN5 protein also predicts the existence of several coiled coil structures which may facilitate interaction with yet unidentified protein partners (Fig. 1C).
Specific binding of HMGN proteins to nucleosomes has been detected by mobility shift assay with purified proteins and core particles [30]. At low ionic strength (0.5X TBE) HMGN1 binds to core particles in noncooperative manner and forms two distinct complexes corresponding to the binding of one or two molecules of HMGN1 per core particle. At “physiological” ionic strength (2X TBE) the binding of HMGN1 is cooperative and only complexes with two HMGN1 molecules per core particle are observed [30]. As expected, under “physiological” conditions HMGN5 cooperatively binds to core particles and forms a complex of 2 molecules of HMGN5 per one core particle with a dissociation constant similar to the HMGN1 (0.4×10−7 × m−1) [26]. Interestingly, in contrast to HMGN1, at low ionic strength HMGN5 does not reveal a noncooperative binding [26]. Importantly, mutations in two serine residues in the core sequence of the NBD of HMGN5, as well as other HMGN proteins, abrogate the interaction of HMGNs with the nucleosome core particles [26, 29, 31].
It has been demonstrated in cells that different members of HMGN protein family cluster into distinct chromatin domains [32]. Nucleosomes containing a heterodimer, i.e. two different HMGN proteins on the single core particle have not been detected either in vitro or in vivo [33]. In agreement with this observation, mobility shift assays indicate that HMGN1 and HMGN5 form separate complexes with core particles [26].
The unique structural feature of HMGN5 is the long and highly acidic C-terminus which contains a stretch of amino acid repeats with a strong consensus sequence EEDGKE for the mouse protein and a more degenerative consensus for the human protein (Fig 1C). These repeats are present 13 times in mouse and 11 times in human HMGN5. Though the biological role of these repeats is not known, the long acidic tail of mouse HMGN5 was associated with general transcriptional activation activity and increased HMGN5-nucleosome interaction in the mobility shift assay [26].
As predicted from its protein sequence, HMGN5, especially its negatively charged C-terminal domain, is highly disordered in solution suggesting that the protein does not have a well defined conformation under native conditions [27]. Long, intrinsically disordered regions are found in around 30% of eukaryotic proteins [34]. For example, linker histone H1, histone acetyltransferase p300 and c-AMP response element binding protein (CREB) contain highly disordered sequences which are required for the proper function of these proteins [35, 36]. This structural feature of HMGN5 is important for understanding its genome wide effect on gene expression. Disordered proteins are prone to form promiscuous low affinity complexes with multiple targets [37]. These proteins are especially common in the regulation of transcription, cell cycle and chaperone folding of proteins. Recently it was demonstrated that the increased sensitivity of cells to overexpression of certain genes is highly correlated with the intrinsically disordered structure of the expressed proteins [37]. When overexpressed, such intrinsically disordered proteins were predicted to have a significant phenotypic effect due to promiscuous molecular interactions. In agreement with this interaction promiscuity theory, overexpression of highly disordered HMGN1 protein inhibited myotube formation and chondrocyte differentiation in cultured cells [16, 25], whereas HMGN1 knock out cells behave normally unless subjected to stress [21]. We also observed that elevated expression of HMGN5 leads to formation of hyperfused myotubes following differentiation of C2C12 myoblasts in vitro (Rochman, M., unpublished observations) and significantly changes cellular transcription profile [27].
4. Intracellular localization of HMGN5
As other HMGN proteins, HMGN5 is also localized to the nucleus of the cell [26]. More detailed analysis reveals that mouse HMGN5 protein is specifically localized to less condensed euchromatic areas and is excluded from more condensed constitutive heterochromatin domains [27]. This localization is unusual because another member of the HMGN family, HMGN1, localizes to both eu- and heterochromatic areas of the nucleus [29]. The major structural distinction between these proteins which accounts for the different nuclear localization is the long acidic C-terminal tail of HMGN5. Indeed, by gradual deletions of the C-terminal tail of HMGN5 it was shown that the tail serves as a module that targets HMGN5 to euchromatin [27]. However, this function of the tail is limited to HMGN proteins. The heterochromatic localization of Heterochromatic Protein 1 (HP1) was not affected by the addition of HMGN5 tail, whereas HMGN1 was relocalized to euchromatin [27]. Moreover, the targeting effect of the acidic tail of HMGN5 protein cannot be substituted by the acidic tail of Acidic Repeat Containing (ACRC) protein, suggesting the importance of the sequence of the HMGN5 tail rather than its charge. Interestingly, intranuclear localization of the human HMGN5 protein, which has a significantly shorter acidic tail, is not restricted to euchromatin (Malicet, C., unpublished observations).
5. Expression pattern of HMGN5 during development
Despite the ubiquitous expression of HMGN5, expression levels of the protein vary significantly between tissues and cell lines [26, 28]. A query of the Gene Expression Omnibus (GEO) expression database suggests the highest expression of mouse HMGN5 protein in the pituitary gland, which is supported by the relatively high expression of the protein in the AtT20 mouse pituitary cell line [27].
As indicated by mRNA and protein analysis, both human and mouse HMGN5 proteins are ubiquitously expressed in adult tissues, although the expression varies during embryonic development [38]. At early stages of mouse development, up to blastocyst stage, HMGN5 is ubiquitously expressed; however starting from 7.5 days post coitum (dpc) its expression is detectable predominantly in ectoplacental cone, an extra embryonic precursor of the placenta [38]. In contrast, HMGN1 protein was mainly localized to the embryonic tissues and excluded from the ectoplacental cone. At 9.5–11.5 dpc HMGN5 transcripts are detected in the giant trophoblast, spongiotrophoblast and decidual cells of the placenta, while expression in the developing embryo remains relatively weak [38]. The most prominent expression was detected in trophoblast giant cells. This expression pattern is recapitulated in rat choriocarcinoma Rcho1 cells, an in vitro model of trophoblast giant cell differentiation. Using this model, it was demonstrated that deregulation of HMGN5 expression by overexpression or siRNA-mediated down regulation altered the expression of prominent markers of giant cell differentiation, suggesting the role of HMGN5 in placental differentiation and function [38]. Consistent with this idea, expression array analysis revealed the two fold increase in HMGN5 transcript in the implantation sites of mice [39]. We also observed similar increase of HMGN5 by immunofluorescence studies (Furusawa, T., unpublished observations).
6. HMGN5 unfolds chromatin and modulates transcription
The concept of dynamic chromatin interactions is paramount in understanding how a relatively low amount of protein (sufficient to bind to less than 2% of the nucleosomes) has a global effect on chromatin architecture [40]. As evident from FRAP analysis, HMGN proteins are highly mobile. They are constantly moving through the nucleus, randomly interacting with nucleosomes and forming metastable protein complexes [8, 41]. The mobility properties of HMGNs are defined by the binding of the proteins to nucleosomes, as apparent from the increased mobility rate of the proteins which are mutated in the NBD and do not bind to nucleosomes [29]. Following binding to chromatin, HMGNs reduce the compaction of higher order chromatin structures thereby affecting transcription and DNA repair [13, 23].
From this perspective, HMGN5 is a characteristic member of the HMGN protein family. It moves very fast through the nucleus and may be the fastest moving member of the HMGN protein family described so far [27]. Altered expression levels of mouse HMGN5 affect transcription of specific markers of differentiation in Rcho-1 placental cells [38] and have a global effect on transcription of pituitary AtT20 cells [27]. Importantly, overexpression of HMGN5 at only twice the endogenous protein level in AtT20 cells alters the expression of more than 2,500 genes. This effect correlates with the ability of HMGN5 to unfold chromatin in vivo and counteract linker histone-mediated chromatin compaction in vitro. Transcription changes induced by HMGN5 are linked to the ability of the protein to interact with nucleosomes because a mutated HMGN5 which does not bind to the nucleosome has no effect on transcription [27]. Though HMGN5 affects the expression of many genes, the magnitude of the effect on each gene is moderate. This suggests that HMGN5 modulates transcription fidelity of the cell.
Alterations in chromatin architecture induced by the binding of HMGN5 provide a molecular explanation for the effects of the protein on transcription and other cellular processes. The effect of HMGN5 binding on chromatin architecture was assessed in a well characterized LacO-LacR array system [42] (Fig 2A). In this system, 256 copies of the LacO sequence are stably integrated into the genome of cells. Fusion of the protein of interest to LacR will tether the protein to LacO sequence and can induce alterations in local chromatin architecture of the array which are detected by fluorescence microscopy. Tethering of wild type HMGN5 protein to the LacO array caused dramatic decondensation of an otherwise compact array structure (Fig 2A), which resembles the unlooping of higher order chromatin structures by the viral transcriptional activator VP16 [43]. The decondensation effect of HMGN5 on the chromatin fiber was supported by increased sensitivity of the chromatin to micrococcal nuclease digestion and by visualization of global chromatin reorganization by microscopy. Significantly, the reorganization and unfolding of chromatin fiber required the proper interaction of HMGN5 with the nucleosome [27].
Figure 2. Model for HMGN5-mediated chromatin unfolding.
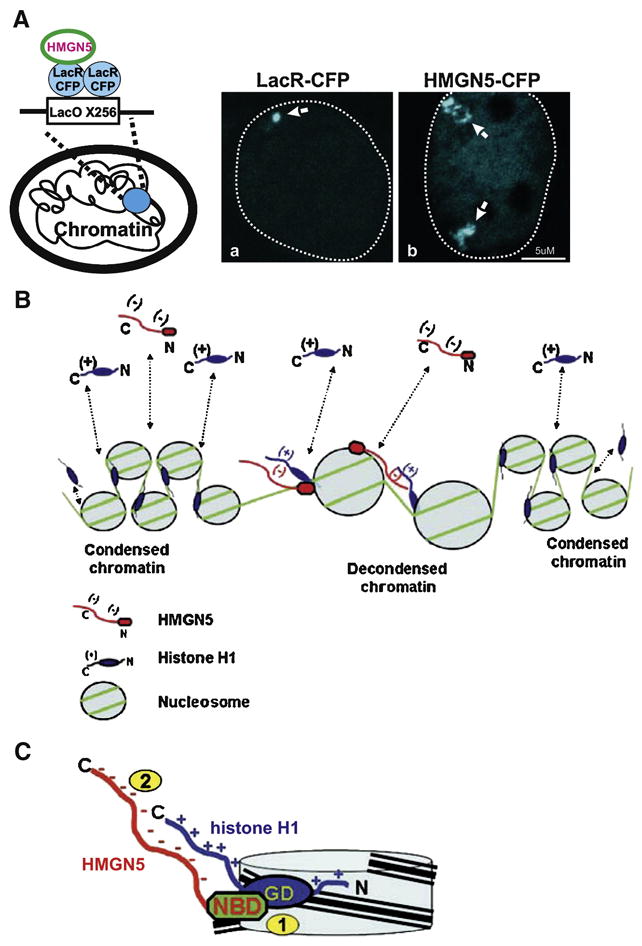
(A) The interaction of HMGN5 with the nucleosomes decondenses a LacO chromatin array. Schematic drawing (left) demonstrates the strategy for tethering HMGN5 to the array by fusion to LacR-CFP. The condensed array, visualized by tethering LacR-CFP (a) appears as a compact dot. Tethering of the wild type HMGN5-CFP (b) to the array induced formation of a highly decondensed array as judged by formation of extended structures with irregular shape (arrows). Nucleus of the cells is marked by a broken line. (B) Effect of HMGN5 on chromatin compaction. In living cells H1 and HMGN5 bind dynamically to chromatin (dotted lines) randomly colliding on nucleosomes and counteracts the H1- induced stabilization of a compact chromatin structure. (C) The binding of HMGN5 to H1-containing nucleosome leads to two molecular events: (1) competition of the NBD of HMGN5 with the Globular Domain (GD) of linker histone for the binding to the nucleosome and (2) juxtaposition and binding of the C-terminal domains of the proteins. Both events apparently contribute to the HMGN5-mediated chromatin unfolding. (B) and (C) are reproduced from [27].
The ability of HMGN5 to unfold chromatin, its overlapping binding site on the nucleosome, and its unusually long acidic tail suggest that direct interaction of HMGN5 with linker histone H1 may be a part of the molecular mechanism of HMGN5-mediated chromatin unfolding. Indeed, a direct interaction between negatively charged HMGN5 tail and positively charged C-terminus of histone H5 (a variant of linker histone H1) was demonstrated both in vitro and in vivo [27]. Significantly, this interaction was not observed for HMGN1, the protein which lacks a long, negatively charged C-terminus uniquely present in HMGN5. In agreement with this data, HMGN5 but not HMGN1 could effectively counteract the compacting effect of linker H5 on a chromatin array in vitro. Similar to HMGN5, it has been recently reported that HMGB1 protein and histone H1 interact in vitro through their acidic and basic tails and that this interaction has the potential to affect global chromatin architecture in the cell [44]. Same kind of acidic-basic interaction was also shown for prothymosin alpha [45], a highly conserved acidic polypeptide localized to the cell nucleus [46].
A major remaining question however is how HMGN5 affects chromatin architecture and transcription if the protein can only bind to a small fraction of the nucleosomes. Our model (Fig 2B) predicts that in the nucleus HMGN5 and linker histones are constantly moving and randomly collide on the nucleosomes. This brief encounter leads to two molecular events: first is a competition of the NBD of HMGN5 with the globular domain of linker histone H1 for the binding site on the nucleosome and a second is a transient interaction of these proteins through their tails (Fig 2C). Subsequently, the efficiency of binding of linker histone H1 to the nucleosome is decreased and chromatin is transiently decondensed. This decondensation can be limited to certain nucleosomes or can have a more global effect if the formation of higher order chromatin structures such as chromatin loops is affected. We believe that HMGN5 can serve as part of multiprotein complexes which determine the preferential association of HMGN5 with defined chromatin regions. Indeed, preferential localization of HMGN1 to the promoters of active genes was suggested [47].
The ability of HMGNs to compete with the binding of linker histones to chromatin is contingent on the correct binding of HMGNs to nucleosomes, because HMGN1 mutants that do not bind to nucleosomes, do not compete with linker histone H1 [8]. However, the contribution of individual structural domains of HMGNs, such as NBD or regulatory domain to this competition is not clear. It has been shown that a C-terminal deletion mutant of HMGN1 is ineffective in competing with linker histone H1 in mixed chromatin domains [8]. On the other hand, the mobility of tailless HMGN5 mutant is significantly faster that that of HMGN1 [27], implying that the sequence differences in the regions adjacent to the core NDB contribute to different functional properties of HMGNs. Thus, HMGNs differ in their localization pattern, nuclear mobility [27], effect on chromatin modifications [48] and chromatin compaction [27]. Furthermore, analysis of HMGN knock out mice revealed that each HMGN protein has different physiological functions [21, 49]. We therefore suggest, that functional integrity of the N-terminal NBD and the C-terminal regulatory domain will define the consequence of the interaction of individual HMGN proteins with the chromatin fiber, including competition with linker histones and transcription regulation. Further deciphering the relationship between the structural domains of individual proteins is required to understand the specificity and/or redundancy between the HMGNs on the level of individual cell and the whole organism.
7. HMGN5 association with physiological and pathological processes
Aberrant expression of HMGN proteins is associated with developmental defects, hypersensitivity to stress and increased tumorigenic potential in mice [50]. Though the molecular role of HMGN5 is not yet understood, there are indications in the literature that suggest the potential involvement of HMGN5 in disease and in normal cellular functions (Table 1 and references therein). For example, microarray analysis and overexpression experiments identified HMGN5 as a factor that induces differentiation in mouse embryonic stem cells [51]. Upregulation of HMGN5 proteins was observed in prostate cancer [52] and squamous cell carcinoma in humans [53], adenocarcinoma in aging mice [54] and in the highly metastatic MDA-MB-435HM breast cancer cell line [55], suggesting a role for HMGN5 in tumorigenesis. The association of increased protein expression with cancer can be also predicted from the structure of the protein which is defined as intrinsically disordered [37].
Table 1.
HMGN5 association with pathological and physiological processes.
| The protein is | Rodent HMGN5 | Human HMGN5 |
|---|---|---|
| Upregulated | • 4.7-fold in Ngn3-induced differentiation of mPACL20 cells into pancreatic islands [58] 1.6-fold in the androgen receptor KO mice [59] 1.6-fold in rat retinal ganglion cells cultured in glia-conditioned medium [60] 2-fold in MRL/lrp but not in NZB/W mouse model of lupus following IFN alpha stimulation of mononuclear cells [61] 2-fold in the embryo implantation site [39] In LPS-stimulated dendritic cells [62] In adult mouse uterus following the early exposure of mice to estrogenic compound diethylstilbestrol [54] |
• By SATB1 in highly metastatic MDA-MB-231 breast cancer cells [63] 16-fold in a primary cutaneous squamous cell [53] 4.8-fold in MDA-MB-435HM (highly metastatic) compared to MDA-MB-435LM (low metastatic) breast cancer cells [55] |
| Down regulated | • 2.32-fold in the amyloid precursor protein TG mouse model of Alzheimer’s disease [64] | |
| Other effects | • Promotes ES cells differentiation when overexpressed [51] Potentially associates with spontaneous ovarian granulose [65] Elevated expression is correlated with uterine adenocarcinoma in aging animals [54] |
• Potentially associates with Martin–Probst deafness mental retardation syndrome, spinal muscular atrophy, coronary heart disease (OMIM database) Potentially associates with premature ovarian cancer [66] Potentially associates with androgen-dependent prostate cancer [52,67] |
The localization of HMGN5 gene on the X chromosome which contains a disproportionally high number of genes associated with mental functions and the late appearance of HMGN5 in evolution imply the possible association of the protein with behavior and cognition [56]. If demonstrated experimentally, this might shed light on the role of chromatin architecture in brain function. Interestingly, human ACRC protein, which has an HMGN5-like, bipartite structure composed of a positively charged C-terminal domain and negatively charged N-terminus containing a long stretch of acidic repeats, is also localized in proximity to the HMGN5 gene on the X chromosome [57].
8. Conclusions and perspectives
Understanding chromatin architecture and its regulation is crucial for deciphering the dynamics of biological processes, such as development and differentiation. It is progressively becoming clear that the regulation of chromatin architecture requires coordinative efforts of multiple proteins and that HMGN protein family is an integral part of this network. The discovery of HMGN5 extends the functional potential of HMGNs in the regulation of the structure of the chromatin fiber and emphasizes the variety of regulatory effects of different members of the same protein family on transcription. It is apparent now that even though HMGN5 binds to nucleosomes in a similar manner as other HMGNs, the outcome of this interaction on the global chromatin architecture is quite different. The unusually long and highly acidic tail which affects the interaction of linker histones with chromatin may confer to HMGN5 an added capacity to unfold higher order chromatin structures. Indeed, specific expression of HMGN5 in the placental giant cells which undergo endoreduplication without cell division suggests that the protein is required for the regulation of transcription from highly compact chromatin. The proposed dual mode of competition between HMGN5 and linker histones offers a new mechanism of the regulation of chromatin dynamics by HMGN proteins. Additional experiments are required to elucidate the molecular mechanism of this competition and its role in chromatin structure and function. We suggest that HMGN5 plays a significant role in maintaining the integrity of the chromatin fiber in cells and its aberrant expression is deleterious for the cell fate. Potential association of HMGN5 with cancer progression and brain activity makes this protein an attractive candidate for further research. However, similar to HMGN1, HMGN5 is probably not a “life or death” decision maker in the organism but rather a protein which affects the cellular phenotype by optimizing chromatin responses to the constantly changing environment.
Acknowledgments
This project was supported by the Intramural Research Program of NIH, National Cancer Institute (NCI). We thank The Fellows Editorial Board for editorial assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wachsmuth M, Caudron-Herger M, Rippe K. Genome organization: balancing stability and plasticity. Biochim Biophys Acta. 2008;1783:2061–2079. doi: 10.1016/j.bbamcr.2008.07.022. [DOI] [PubMed] [Google Scholar]
- 2.Kumaran RI, Thakar R, Spector DL. Chromatin dynamics and gene positioning. Cell. 2008;132:929–934. doi: 10.1016/j.cell.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Y, Fischle W, Cheung W, Jacobs S, Khorasanizadeh S, Allis CD. Beyond the double helix: writing and reading the histone code. Novartis Found Symp. 2004;259:3–17. discussion 17–21, 163–169. [PubMed] [Google Scholar]
- 4.Marmorstein R, Trievel RC. Histone modifying enzymes: structures, mechanisms, and specificities. Biochim Biophys Acta. 2009;1789:58–68. doi: 10.1016/j.bbagrm.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 6.Bustin M, Catez F, Lim JH. The dynamics of histone H1 function in chromatin. Mol Cell. 2005;17:617–620. doi: 10.1016/j.molcel.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 7.Gerlitz G, Hock R, Ueda T, Bustin M. The dynamics of HMG protein-chromatin interactions in living cells. Biochem Cell Biol. 2009;87:127–137. doi: 10.1139/O08-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Catez F, Brown DT, Misteli T, Bustin M. Competition between histone H1 and HMGN proteins for chromatin binding sites. EMBO Rep. 2002;3:760–766. doi: 10.1093/embo-reports/kvf156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Catez F, Yang H, Tracey KJ, Reeves R, Misteli T, Bustin M. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol Cell Biol. 2004;24:4321–4328. doi: 10.1128/MCB.24.10.4321-4328.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bustin M, Reeves R. High-mobility-group chromosomal proteins: architectural components that facilitate chromatin function. Prog Nucleic Acid Res Mol Biol. 1996;54:35–100. doi: 10.1016/s0079-6603(08)60360-8. [DOI] [PubMed] [Google Scholar]
- 11.Bustin M. Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol Cell Biol. 1999;19:5237–5246. doi: 10.1128/mcb.19.8.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Catez F, Lim JH, Hock R, Postnikov YV, Bustin M. HMGN dynamics and chromatin function. Biochem Cell Biol. 2003;81:113–122. doi: 10.1139/o03-040. [DOI] [PubMed] [Google Scholar]
- 13.Trieschmann L, Alfonso PJ, Crippa MP, Wolffe AP, Bustin M. Incorporation of chromosomal proteins HMG-14/HMG-17 into nascent nucleosomes induces an extended chromatin conformation and enhances the utilization of active transcription complexes. EMBO J. 1995;14:1478–1489. doi: 10.1002/j.1460-2075.1995.tb07134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ding HF, Bustin M, Hansen U. Alleviation of histone H1-mediated transcriptional repression and chromatin compaction by the acidic activation region in chromosomal protein HMG-14. Mol Cell Biol. 1997;17:5843–5855. doi: 10.1128/mcb.17.10.5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ueda T, Furusawa T, Kurahashi T, Tessarollo L, Bustin M. The Nucleosome Binding Protein HMGN3 Modulates The Transcription Profile Of Pancreatic {beta}-Cells And Affects Insulin Secretion. Mol Cell Biol. 2009;19:5264–76. doi: 10.1128/MCB.00526-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Furusawa T, Lim JH, Catez F, Birger Y, Mackem S, Bustin M. Down-regulation of nucleosomal binding protein HMGN1 expression during embryogenesis modulates Sox9 expression in chondrocytes. Mol Cell Biol. 2006;26:592–604. doi: 10.1128/MCB.26.2.592-604.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lim JH, Catez F, Birger Y, West KL, Prymakowska-Bosak M, Postnikov YV, Bustin M. Chromosomal protein HMGN1 modulates histone H3 phosphorylation. Mol Cell. 2004;15:573–584. doi: 10.1016/j.molcel.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 18.Postnikov YV, Belova GI, Lim JH, Bustin M. Chromosomal protein HMGN1 modulates the phosphorylation of serine 1 in histone H2A. Biochemistry. 2006;45:15092–15099. doi: 10.1021/bi0613271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lim JH, West KL, Rubinstein Y, Bergel M, Postnikov YV, Bustin M. Chromosomal protein HMGN1 enhances the acetylation of lysine 14 in histone H3. EMBO J. 2005;24:3038–3048. doi: 10.1038/sj.emboj.7600768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rattner BP, Yusufzai T, Kadonaga JT. HMGN proteins act in opposition to ATP-dependent chromatin remodeling factors to restrict nucleosome mobility. Mol Cell. 2009;34:620–626. doi: 10.1016/j.molcel.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Birger Y, Catez F, Furusawa T, Lim JH, Prymakowska-Bosak M, West KL, Postnikov YV, Haines DC, Bustin M. Increased tumorigenicity and sensitivity to ionizing radiation upon loss of chromosomal protein HMGN1. Cancer Res. 2005;65:6711–6718. doi: 10.1158/0008-5472.CAN-05-0310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Birger Y, West KL, Postnikov YV, Lim JH, Furusawa T, Wagner JP, Laufer CS, Kraemer KH, Bustin M. Chromosomal protein HMGN1 enhances the rate of DNA repair in chromatin. EMBO J. 2003;22:1665–1675. doi: 10.1093/emboj/cdg142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim YC, Gerlitz G, Furusawa T, Catez F, Nussenzweig A, Oh KS, Kraemer KH, Shiloh Y, Bustin M. Activation of ATM depends on chromatin interactions occurring before induction of DNA damage. Nat Cell Biol. 2009;11:92–96. doi: 10.1038/ncb1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mohamed OA, Bustin M, Clarke HJ. High-mobility group proteins 14 and 17 maintain the timing of early embryonic development in the mouse. Dev Biol. 2001;229:237–249. doi: 10.1006/dbio.2000.9942. [DOI] [PubMed] [Google Scholar]
- 25.Pash JM, Alfonso PJ, Bustin M. Aberrant expression of high mobility group chromosomal protein 14 affects cellular differentiation. J Biol Chem. 1993;268:13632–13638. [PubMed] [Google Scholar]
- 26.Shirakawa H, Landsman D, Postnikov YV, Bustin M. NBP-45, a novel nucleosomal binding protein with a tissue-specific and developmentally regulated expression. J Biol Chem. 2000;275:6368–6374. doi: 10.1074/jbc.275.9.6368. [DOI] [PubMed] [Google Scholar]
- 27.Rochman M, Postnikov Y, Correll S, Malicet C, Wincovitch S, Karpova T, McNally J, Wu X, Bubunenko N, Grigoryev S, Bustin M. The Interaction Of NSBP1/HMGN5 With Nucleosomes In Euchromatin Counteracts Linker Histone-Mediated Chromatin Compaction And Modulates Transcription. Mol Cell. 2009;35:642–656. doi: 10.1016/j.molcel.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.King LM, Francomano CA. Characterization of a human gene encoding nucleosomal binding protein NSBP1. Genomics. 2001;71:163–173. doi: 10.1006/geno.2000.6443. [DOI] [PubMed] [Google Scholar]
- 29.Ueda T, Catez F, Gerlitz G, Bustin M. Delineation of the protein module that anchors HMGN proteins to nucleosomes in the chromatin of living cells. Mol Cell Biol. 2008;28:2872–2883. doi: 10.1128/MCB.02181-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Postnikov YV, Lehn DA, Robinson RC, Friedman FK, Shiloach J, Bustin M. The cooperative binding of chromosomal protein HMG-14 to nucleosome cores is reduced by single point mutations in the nucleosomal binding domain. Nucleic Acids Res. 1994;22:4520–4526. doi: 10.1093/nar/22.21.4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prymakowska-Bosak M, Misteli T, Herrera JE, Shirakawa H, Birger Y, Garfield S, Bustin M. Mitotic phosphorylation prevents the binding of HMGN proteins to chromatin. Mol Cell Biol. 2001;21:5169–5178. doi: 10.1128/MCB.21.15.5169-5178.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Postnikov YV, Herrera JE, Hock R, Scheer U, Bustin M. Clusters of nucleosomes containing chromosomal protein HMG-17 in chromatin. J Mol Biol. 1997;274:454–465. doi: 10.1006/jmbi.1997.1391. [DOI] [PubMed] [Google Scholar]
- 33.Cherukuri S, Hock R, Ueda T, Catez F, Rochman M, Bustin M. Cell cycle-dependent binding of HMGN proteins to chromatin. Mol Biol Cell. 2008;19:1816–1824. doi: 10.1091/mbc.E07-10-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337:635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 35.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 36.Hansen JC, Lu X, Ross ED, Woody RW. Intrinsic protein disorder, amino acid composition, and histone terminal domains. J Biol Chem. 2006;281:1853–1856. doi: 10.1074/jbc.R500022200. [DOI] [PubMed] [Google Scholar]
- 37.Vavouri T, Semple JI, Garcia-Verdugo R, Lehner B. Intrinsic protein disorder and interaction promiscuity are widely associated with dosage sensitivity. Cell. 2009;138:198–208. doi: 10.1016/j.cell.2009.04.029. [DOI] [PubMed] [Google Scholar]
- 38.Shirakawa H, Rochman M, Furusawa T, Kuehn MR, Horigome S, Haketa K, Sugita Y, Inada T, Komai M, Bustin M. The nucleosomal binding protein NSBP1 is highly expressed in the placenta and modulates the expression of differentiation markers in placental Rcho-1 cells. J Cell Biochem. 2009;106:651–658. doi: 10.1002/jcb.22046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen Y, Ni H, Ma XH, Hu SJ, Luan LM, Ren G, Zhao YC, Li SJ, Diao HL, Xu X, Zhao ZA, Yang ZM. Global analysis of differential luminal epithelial gene expression at mouse implantation sites. J Mol Endocrinol. 2006;37:147–161. doi: 10.1677/jme.1.02009. [DOI] [PubMed] [Google Scholar]
- 40.Phair RD, Scaffidi P, Elbi C, Vecerova J, Dey A, Ozato K, Brown DT, Hager G, Bustin M, Misteli T. Global nature of dynamic protein-chromatin interactions in vivo: three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Mol Cell Biol. 2004;24:6393–6402. doi: 10.1128/MCB.24.14.6393-6402.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lim JH, Bustin M, Ogryzko VV, Postnikov YV. Metastable macromolecular complexes containing high mobility group nucleosome-binding chromosomal proteins in HeLa nuclei. J Biol Chem. 2002;277:20774–20782. doi: 10.1074/jbc.M200404200. [DOI] [PubMed] [Google Scholar]
- 42.Belmont AS, Li G, Sudlow G, Robinett C. Visualization of large-scale chromatin structure and dynamics using the lac operator/lac repressor reporter system. Methods Cell Biol. 1999;58:203–222. doi: 10.1016/s0091-679x(08)61957-3. [DOI] [PubMed] [Google Scholar]
- 43.Tumbar T, Sudlow G, Belmont AS. Large-scale chromatin unfolding and remodeling induced by VP16 acidic activation domain. J Cell Biol. 1999;145:1341–1354. doi: 10.1083/jcb.145.7.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cato L, Stott K, Watson M, Thomas JO. The interaction of HMGB1 and linker histones occurs through their acidic and basic tails. J Mol Biol. 2008;384:1262–1272. doi: 10.1016/j.jmb.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 45.Karetsou Z, Sandaltzopoulos R, Frangou-Lazaridis M, Lai CY, Tsolas O, Becker PB, Papamarcaki T. Prothymosin alpha modulates the interaction of histone H1 with chromatin. Nucleic Acids Res. 1998;26:3111–3118. doi: 10.1093/nar/26.13.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hannappel E, Huff T. The thymosins. Prothymosin alpha, parathymosin, and beta-thymosins: structure and function. Vitam Horm. 2003;66:257–296. doi: 10.1016/s0083-6729(03)01007-0. [DOI] [PubMed] [Google Scholar]
- 47.Hock R, Wilde F, Scheer U, Bustin M. Dynamic relocation of chromosomal protein HMG-17 in the nucleus is dependent on transcriptional activity. EMBO J. 1998;17:6992–7001. doi: 10.1093/emboj/17.23.6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ueda T, Postnikov YV, Bustin M. Distinct domains in high mobility group N variants modulate specific chromatin modifications. J Biol Chem. 2006;281:10182–10187. doi: 10.1074/jbc.M600821200. [DOI] [PubMed] [Google Scholar]
- 49.Ueda T, Furusawa T, Kurahashi T, Tessarollo L, Bustin M. The nucleosome binding protein HMGN3 modulates the transcription profile of pancreatic {beta} cells and affects insulin secretion. Mol Cell Biol. 2009;29:5264–5276. doi: 10.1128/MCB.00526-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hock R, Furusawa T, Ueda T, Bustin M. HMG chromosomal proteins in development and disease. Trends Cell Biol. 2007;17:72–79. doi: 10.1016/j.tcb.2006.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pritsker M, Ford NR, Jenq HT, Lemischka IR. Genomewide gain-of-function genetic screen identifies functionally active genes in mouse embryonic stem cells. Proc Natl Acad Sci U S A. 2006;103:6946–6951. doi: 10.1073/pnas.0509861103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang C, Zhou LQ, Song G. Effect of nucleosomal binding protein 1 in androgen-independent prostatic carcinoma. Zhonghua Yi Xue Za Zhi. 2008;88:657–660. [PubMed] [Google Scholar]
- 53.Green J, Ikram M, Vyas J, Patel N, Proby CM, Ghali L, Leigh IM, O’Toole EA, Storey A. Overexpression of the Axl tyrosine kinase receptor in cutaneous SCC-derived cell lines and tumours. Br J Cancer. 2006;94:1446–1451. doi: 10.1038/sj.bjc.6603135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tang WY, Newbold R, Mardilovich K, Jefferson W, Cheng RY, Medvedovic M, Ho SM. Persistent hypomethylation in the promoter of nucleosomal binding protein 1 (Nsbp1) correlates with overexpression of Nsbp1 in mouse uteri neonatally exposed to diethylstilbestrol or genistein. Endocrinology. 2008;149:5922–5931. doi: 10.1210/en.2008-0682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li DQ, Hou YF, Wu J, Chen Y, Lu JS, Di GH, Ou ZL, Shen ZZ, Ding J, Shao ZM. Gene expression profile analysis of an isogenic tumour metastasis model reveals a functional role for oncogene AF1Q in breast cancer metastasis. Eur J Cancer. 2006;42:3274–3286. doi: 10.1016/j.ejca.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 56.Khil PP, Camerini-Otero RD. Molecular features and functional constraints in the evolution of the mammalian X chromosome. Crit Rev Biochem Mol Biol. 2005;40:313–330. doi: 10.1080/10409230500356703. [DOI] [PubMed] [Google Scholar]
- 57.Nolte D, Ramser J, Niemann S, Lehrach H, Sudbrak R, Muller U. ACRC codes for a novel nuclear protein with unusual acidic repeat tract and maps to DYT3 (dystonia parkinsonism) critical interval in xq13.1. Neurogenetics. 2001;3:207–213. doi: 10.1007/s100480100120. [DOI] [PubMed] [Google Scholar]
- 58.Gasa R, Mrejen C, Leachman N, Otten M, Barnes M, Wang J, Chakrabarti S, Mirmira R, German M. Proendocrine genes coordinate the pancreatic islet differentiation program in vitro. Proc Natl Acad Sci U S A. 2004;101:13245–13250. doi: 10.1073/pnas.0405301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.MacLean HE, Chiu WS, Notini AJ, Axell AM, Davey RA, McManus JF, Ma C, Plant DR, Lynch GS, Zajac JD. Impaired skeletal muscle development and function in male, but not female, genomic androgen receptor knockout mice. FASEB J. 2008;22:2676–2689. doi: 10.1096/fj.08-105726. [DOI] [PubMed] [Google Scholar]
- 60.Goritz C, Thiebaut R, Tessier LH, Nieweg K, Moehle C, Buard I, Dupont JL, Schurgers LJ, Schmitz G, Pfrieger FW. Glia-induced neuronal differentiation by transcriptional regulation. Glia. 2007;55:1108–1122. doi: 10.1002/glia.20531. [DOI] [PubMed] [Google Scholar]
- 61.Lu Q, Shen N, Li XM, Chen SL. Genomic view of IFN-alpha response in pre-autoimmune NZB/W and MRL/lpr mice. Genes Immun. 2007;8:590–603. doi: 10.1038/sj.gene.6364421. [DOI] [PubMed] [Google Scholar]
- 62.Efron PA, Tsujimoto H, Bahjat FR, Ungaro R, Debernardis J, Tannahill C, Baker HV, Edwards CK, Moldawer LL. Differential maturation of murine bone-marrow derived dendritic cells with lipopolysaccharide and tumor necrosis factor-alpha. J Endotoxin Res. 2005;11:145–160. doi: 10.1179/096805105X46583. [DOI] [PubMed] [Google Scholar]
- 63.Han HJ, Russo J, Kohwi Y, Kohwi-Shigematsu T. SATB1 reprogrammes gene expression to promote breast tumour growth and metastasis. Nature. 2008;452:187–193. doi: 10.1038/nature06781. [DOI] [PubMed] [Google Scholar]
- 64.Jee SW, Cho JS, Kim CK, Hwang DY, Shim SB, Lee SH, Sin JS, Park JH, Kim YS, Choi SY, Kim YK. Oligonucleotide-based analysis of differentially expressed genes in hippocampus of transgenic mice expressing NSE-controlled APPsw. Neurochem Res. 2006;31:1035–1044. doi: 10.1007/s11064-006-9117-8. [DOI] [PubMed] [Google Scholar]
- 65.Beamer WG, Shultz KL, Tennent BJ, Nadeau JH, Churchill GA, Eicher EM. Multigenic and imprinting control of ovarian granulosa cell tumorigenesis in mice. Cancer Res. 1998;58:3694–3699. [PubMed] [Google Scholar]
- 66.Rizzolio F, Bione S, Sala C, Goegan M, Gentile M, Gregato G, Rossi E, Pramparo T, Zuffardi O, Toniolo D. Chromosomal rearrangements in Xq and premature ovarian failure: mapping of 25 new cases and review of the literature. Hum Reprod. 2006;21:1477–1483. doi: 10.1093/humrep/dei495. [DOI] [PubMed] [Google Scholar]
- 67.Zhou LQ, Song G, He ZS, Hao JR, Na YQ. Effects of inhibiting nucleosomal binding protein 1 on proliferation of human prostate cancer cells. Zhonghua Yi Xue Za Zhi. 2007;87:404–408. [PubMed] [Google Scholar]