High mobility group nucleosomal proteins (HMGNs) are small non-histone proteins associated with chromatin. HMGNs have the unique ability to bind to nucleosomes with higher affinity than to naked DNA [1]. They have been studied extensively for their ability to modulate transcription. Although initially viewed as general transcriptional activators on chromatin templates, it is now appreciated that they are instead highly specific modulators of gene expression. We review the mechanisms for targeting HMGNs to specific genes and for how they subsequently regulate transcription.
HMGN and transcriptionally active genes – early studies
A body of early research in the field implicated HMGNs in preferentially associating with DNase I-sensitive, or “active” genes in the nuclei. In a highly intriguing publication, Weisbrod and Weintraub first demonstrated that addition of 10% TCA-soluble extracts containing HMGN1/2 could reconstitute the DNase I sensitivity of the β-globin gene in chicken erythrocyte chromatin stripped of non-histone proteins [2]. Although somewhat controversial at the time (as described below), this initial result suggested that HMGNs are not only present on actively transcribed genes, but are also essential for maintaining an active chromatin structure. A number of subsequent studies on model genes using similar in vitro reconstitution assays with more purified proteins, as well as analyses of the DNA sequences contained in chromatin precipitated by HMGN1/2 antibodies, further reinforced the notion that HMGNs preferentially associate with active genes [3–10]. However, others reported poor correlation between HMGN1/2 binding and gene activity [6, 11–13]. Finally, investigations into the characteristics of HMGN-bound versus -unbound chromatin yielded conflicting results.
Some studies suggested that hypomethylated DNA, hyperacetylated histone, or longer linker DNA distinguished chromatin preferentially bound by HMGN proteins [5, 14–17], however other studies failed to detect any differences in these parameters [7, 17, 18]. Further compounding the controversy were reports demonstrating rearrangement of HMGNs at low ionic strength, a condition typically used to solubilize chromatin in these studies [19, 20]. Because of this potential rearrangement of proteins during the isolation process, the DNA ultimately associated with HMGN may not represent its true binding sites in vivo, a potential cause for the conflicting data that were reported. One study of mononucleosomes from the chicken β-globin cluster suggested that HMGN2 binding, although tissue specific, did not correlate directly with either DNase I sensitivity or active gene transcription [21]. On the other hand, experiments in which protein and DNA were first crosslinked before fractionation to prevent the rearrangement of HMGNs showed a modest enrichment of HMGN on the transcribed chromatin of chicken embryonic β-globin gene [9]. Finally, immunofluorescence studies using antibody against HMGN1 showed preferential staining of transcriptionally active regions in the polytene chromosomes of Chironomus pallidvittatus [22].
Recent studies examining HMGN association with chromatin using the chromatin immunoprecipitation (ChIP) technique have definitively shown enrichment of HMGN on specific target genes in vivo. Enrichment was observed on HMGN3 target genes Glyt1, Glut2 and on the HMGN1 target gene Sox9 [23–25]. Moreover, dynamic association of HMGN1 with promoters of estrogen-induced as well as immediate early genes has been reported [26, 27]. With the development of techniques for genome-wide analysis for association of protein with chromatin in vivo, the field is now poised to finally answer the old question of whether HMGN1 associates preferentially with transcribed genes in vivo (see more detailed discussion in conclusion and future prospectives).
Although the association of HMGN with DNase I-sensitive genes or “active genes” remains to be conclusively addressed, other studies indicated that HMGN does play a functional role in transcriptional regulation. In one initial study in this regard, microinjection of antibodies against HMGN2 into human fibroblasts inhibited RNA synthesis [28]. In a further characterization it was shown that HMGN2 specifically affected RNA polymerase II (pol II)-dependent transcription in cells [29]. In particular, HMGN2 colocalized with nascent transcript and microinjection of a peptide corresponding to the nucleosomal binding domain of HMGN2 blocked pol II-dependent transcription. Taken together, these studies provided evidence for the functional involvement of HMGNs in overall transcription in vivo.
Dissecting HMGN functionality as a general chromatin-dependent transcriptional activator by in vitro transcription assays
The association of HMGNs with transcriptionally active genes implied that HMGNs might activate transcription, thereby raising the question of their potential mechanism of action. When tested in a LexA fusion system, HMGN did not function as a conventional transcription activator on a synthetic reporter construct containing LexA binding sites [30]. Insights into a general role for HMGNs in transcription were obtained instead by in vitro transcription studies using chromatin templates. In particular, these studies utilized either SV40 minichromosomes isolated from CV-1 cells or minichromosomes assembled in Drosophila embryo or Xenopus egg extracts [31–36]. In each system, HMGNs stimulated transcription only from chromatin but not from naked DNA templates, demonstrating that HMGN1/2 are chromatin-specific transcriptional activators. The mechanism of stimulation by HMGN1/2 on transcription varied depending on the system, however: HMGNs enhanced the overall rate of transcription on chromatin assembled in Xenopus egg extract [32, 33, 35, 37], enhanced the rate of transcription initiation on chromatin assembled in Drosophila embryo extract [34, 38], or enhanced transcription elongation on SV40 minichromosomes isolated from CV-1 cells [31, 36]. In both reconstitution systems, HMGN1/2 had to be added during chromatin assembly in order to stimulate transcription. In contrast, adding HMGN1 to preassembled SV40 minichromosomes was sufficient to enhance transcription. This difference between SV40 minichromosomes and chromatin reconstituted in vitro presumably reflects the fact that isolated SV40 minichromosomes represent transcription-competent cellular chromatin [39]. This distinction is likely also to explain why the initiation of transcription was enhanced only in the reconstituted chromatin systems; on preassembled SV40 minichromosomes, the promoter was already in an active configuration. In summary, despite the observed differences based on experimental design, these studies all corroborated that HMGNs function as chromatin-specific transcriptional activators.
The in vitro transcription studies were also able to elucidate how HMGNs altered chromatin in order to activate transcription. The structure of minichromosomes assembled in vitro, in the absence and presence of HMGNs, has been characterized by restriction enzyme accessibility, micrococcal nuclease digestion, and sucrose gradient centrifugation. Such analyses demonstrated that HMGN1/2 enhances transcription in the Xenopus egg chromatin assembly system by inducing an extended chromatin conformation, with no effect on nucleosomal repeat length. Moreover, analysis of HMGN truncation mutants showed that the C-terminal acidic domain is responsible for enhancing transcription [32, 33, 37]. Similar structural results were obtained in the SV40 minichromosome system (see below). In contrast, a more recent analysis of linear chromatin arrays failed to observe any effect of HMGNs on MgCl2-induced chromatin compaction [40]. One potential explanation for this difference may be that Mg2+ induces a higher degree/order of chromatin compaction than of the chromatin used in previous studies. Consistent with this interpretation, one report showed that HMGN2 potentiates transcription from minichromosomes assembled in the absence of Mg2+, but not in its presence [37].
Using the SV40 minichromosome in vitro system, structural and functional antagonism between HMGN1/2 and histone H1 was also first identified. On these chromatin templates, histone H1 inhibited pol II-mediated transcription, as anticipated, correlating with compaction of higher order chromatin structure assayed by sedimentation analysis. HMGN1 alleviated both the histone H1-mediated compaction of chromatin and the histone H1-mediated transcriptional repression. The connection between compaction and repression was solidified by showing that a mutant HMGN1 lacking the C-terminal acidic domain was unable to induce relaxation of the chromatin, and also unable to stimulate transcription [31, 36]. This interplay between HMGN and histone H1 was subsequently corroborated and further characterized in vivo by immunofluorescence studies. Microinjection of HMGN1/2 into cells stably expressing histone H1-GFP decreased the residence time of nuclear H1-GFP, demonstrating that HMGN1/2 competes for histone H1 binding sites on chromatin [41]. In contrast, HMGNs do not compete with HMGB or HMGA proteins for binding to chromatin. However, HMGNs do synergize with HMGB and HMGA to weaken binding of histone H1 [41, 42]. Together, these results showed that one mechanism by which HMGN enhances transcription is by competing for binding of histone H1 to chromatin, both in vitro and in vivo.
HMGN: from transcription activator to transcription modulator
The studies discussed above, combined with the fact that HMGNs do not exhibit any intrinsic preference for binding particular DNA sequences [43] suggested that HMGNs act as general transcriptional activators. This view implies that HMGNs would enhance expression from all transcriptionally competent genes. However, this viewpoint was challenged by gene expression profiling analysis. Comparison of mRNA profiles from Hmgn1−/− and Hmgn1+/+ MEFs indicated that loss of HMGN1 alters levels from only 3% of the expressed genes [44]. Similarly, stable overexpression of HMGN3 affected only 0.8% of expressed genes [45]. Therefore in vivo, HMGNs only modulate expression from a subset of genes and appear not to be general transcriptional activators. Moreover, a number of studies uncovered instances in which HMGNs also inhibit gene expression, including inhibition by HMGN1 of anisomycin-induced immediate-early (IE) genes in MEFs [26, 27], inhibition by HMGN1 of estrogen-induced gene expression in MCF7 cells [26] and inhibitory of gene expression by HMGN3 in Hepa-1 cells [45] and MIN6 cells [25]. In principle, such inhibition could be a secondary effect due to consequences of primary targets of HMGNs, given the length of time during which HMGNs levels were altered, in the range of days, weeks or even months. However, in the cases of rapid induction of IE genes by anisomycin treatment and of estrogen-responsive genes, ChIP experiments demonstrated that HMGNs were either selectively depleted from the fosB promoter or specifically recruited to the TFF1 promoter in response to signaling. These findings strongly suggest that these promoters are direct targets of HMGNs, and therefore support the notion that HMGNs can not only stimulate, but also directly suppress gene expression. The groundwork for understanding how HMGNs might suppress transcription was laid by the Bustin laboratory, when they demonstrated that HMGNs affect the levels of certain histone modifications (for detailed analysis see review by Dr. Yuri Postnikov in this issue). In particular, the loss of HMGN1 elevated total cellular levels of histone H3 Ser10 phosphorylation and H3 Lys9 acetylation, both of which correlate with gene activation. At the gene level, HMGN1 specifically reduced the level of these modifications on the fosB promoter in MEFs and the TFF1 promoter in MCF7 cells. This alteration in histone modifications at the gene regulatory regions provides an explanation for the HMGN1-mediated inhibition of gene expression, reinforcing the conclusion that HMGN1 can act as a transcriptional repressor.
A critical question regarding how HMGNs regulate transcription is how they are targeted to specific promoters. The importance of this issue is highlighted by the ability of HMGNs to regulate only a subset of genes. Association of HMGNs with specific regulatory regions cannot be targeted by specific DNA sequences, since there is no underlying sequence preference upon binding of HMGNs to nucleosomes [43]. An alternative possibility would be that certain features of the local chromatin, such as histone modification patterns, facilitate binding of HMGNs. This hypothesis was tested early on in vitro, without consistent results [5, 7, 14, 17, 18]. Furthermore, since the loss of HMGN1 alters histone modifications, it is more likely that HMGN1 binding causes changes in histone modifications rather than the opposite [27, 46, 47]. A third possibility is that HMGNs are targeted to specific regions by virtue of being present in larger protein complexes [48]. Shirakawa et al showed that HMGNs exist in complexes inside cells [43, 49]. However the major components of the identified complex were snRNPs and annexin II [49], which cannot readily account for gene regulatory binding specificity of HMGNs. On the other hand, associations between HMGNs and specific transcription factors have been identified, which can target HMGNs to specific promoters.
The first demonstration of a specific interaction between HMGNs and transcription factors was that between HMGN3 and thyroid hormone receptor (TR). In fact, HMGN3 was initially identified in a yeast two hybrid screen as thyroid hormone-dependent TR interacting protein [50]. A subsequent study demonstrated that HMGN3 interacts with TR/RAR in vitro, forms a complex with TR on a TR response element (TRE) in vitro and stimulates TR/RAR-mediated transcription on a TRE-driven reporter construct upon injection into Xenopus oocytes [51]. The biological relevance of interaction of HMGNs with specific transcription factors in mediating effects of HMGNs on transcription was first demonstrated in a study of estrogen-mediated transcription activation. In MCF7 cells, the interaction of HMGN1 with ERα and SRF directed HMGN recruitment to the TFF1 and FOS regulatory regions, respectively, and reduced estrogen-mediated transcriptional activation of these two genes. A mutant of HMGN1, HMGN1 S20E/S24E with diminished nucleosome binding activity, unexpectedly differentially impacted these two HMGN-transcription factor interactions. Whereas this mutant still interacts with ERα, retaining its ability to inhibit TFF1 gene activation, it no longer interacts with SRF, and has lost the ability to regulate induction of FOS. This result demonstrates that HMGN1 can be recruited to specific promoters through association with particular transcription factors and that this interaction is required for HMGN1 to regulate transcription of these genes [26]. In another study, Amen et al described a complex between HMGN2, the homeodomain transcription factor PITX2 and β-catenin. The HMGN2-PITX2 interaction decreased PITX2 binding to target sequences in vitro and inhibited the ability of PITX2 to activate reporter genes in a transient transfection assay. Interestingly, activation of the Wnt/β-catenin pathway by several mechanisms relieves HMGN2-mediated repression of PITX2 transcriptional activation [52]. The mechanism by which β-catenin relieves the repression is not known and effects on endogenous genes need to be demonstrated. Nonetheless, this study provides another example of interaction of HMGNs with specific transcription factors to influence gene expression. The mechanism by which HMGNs affect gene expression is very different between HMGN1/ERα and HMGN2/PTIX complexes. Whereas HMGN1 does not affect binding of estrogen receptor to the estrogen response element, either in vitro or in vivo [26], HMGN2 decreased PITX2 DNA-binding, at least in vitro. Another example of HMGN being recruited to a specific promoter involves a complex between NFIX, CCGBP1 and HMGN1, required to regulate HSF1 transcription. At normal temperature, this complex represses HSF1 transcription while at elevated temperature, the complex is heat sensitive and thus repression is lost [53]. In this case, CCGBP1 directly binds the DNA, apparently recruiting HMGN1, which is required for association of NFIX to the promoter. The specific role that HMGN1 plays in the recruitment of NFIX is not clear. Finally, in MIN6 cells, HMGN3 increases binding of the transcription factor PDX1 to the Glut2 promoter, consistent with its ability to enhance Glut2 transcription [25]. No complex between HMGN3 and PDX1 was detectable in this instance. Instead, a stochastic interaction between HMGN3 and PDX1 was proposed to mediate the effect. These findings in multiple specific instances suggest that by forming a stable complex or even potentially a transient interaction with specific transcription factors, HMGNs can modulate the expression of specific genes.
In summary, our understanding of HMGNs transcriptional effects has evolved from a role as a general transcription activator via potential binding to all nucleosomes to that of a specific transcription modulator (see also [48] for a recent review). To date, all evidence indicates that HMGNs are targeted to specific promoters through protein-protein interactions with specific transcription factors. It has also become clear that HMGNs can affect gene transcription through multiple effects on local or global chromatin composition and structure, including the following:
HMGNs can compete with binding of histone H1 to chromatin. This mechanism was first elucidated by in vitro transcription experiments, and later reinforced by in vivo immunofluorescence studies. Although this model was originally used to explain how HMGNs enhanced transcription, it could potentially also explain transcriptional inhibition by HMGNs. Based on gene expression profiling in histone H1 knockout mice, it is now appreciated that histone H1 may enhance, as well as inhibit, transcription [54].
HMGN1 can modulate gene transcription by locally affecting histone modifications. The ability to alter histone modification was first demonstrated elegantly in HMGN1 knockout MEFs, and later validated on specific gene regulatory regions. Again, depending on which histone modifications are affected, the end result can be either inhibitory or activating on transcription of specific genes. The question remains as to the molecular mechanism by which HMGN1 exerts this effect.
A very recent study on the effect of HMGNs on chromatin remodeling demonstrated that HMGNs can inhibit ATP-dependent chromatin activity in vitro [55]. Chromatin remodeling can either facilitate or repress transcription. By inhibiting chromatin remodeling, HMGNs could potentially alter gene expression in either direction, depending on the local chromatin context. However, in another study no effect of HMGN was observed on chromatin remodeling activity [40], and the in vivo relevance of HMGNs altering chromatin remodeling activity remains to be demonstrated.
HMGN1 modifications and effects on gene regulation
HMGNs are subjected to a number of modifications including acetylation, methylation and phosphorylation. The best studied of these modifications is the phosphorylation of HMGN1 during IE gene induction in response to stress, mitogenic and pharmacological stimuli. Stimulation of mouse fibroblasts with epidermal growth factor (EGF), anisomycin, or tetradecanoyl phorbol acetate (TPA) results in transient phosphorylation specifically of HMGN1, but not of HMGN2, on Ser6 and Ser20 [27, 56–58], as well as Ser24 [27], and phosphorylation of histone H3 on Ser10 [27, 56–58]. Phosphorylation of HMGN1 and histone H3 occurs concomitantly with IE gene induction. The role of such phosphorylation was tested by inhibiting MSK1/2, a MAPK downstream effector kinase responsible for the phosphorylation of both H3 and HMGN; inhibition of the kinase impaired IE gene induction. Mechanistically, phosphorylation of HMGN1 on Ser20 and Ser24 decreases its binding to chromatin [59–61], relieving the inhibition by HMGN1 of histone H3 Ser10 phosphorylation, thus leading to gene activation. The role of Ser6 phosphorylation in gene activation, however, was not investigated in this system.
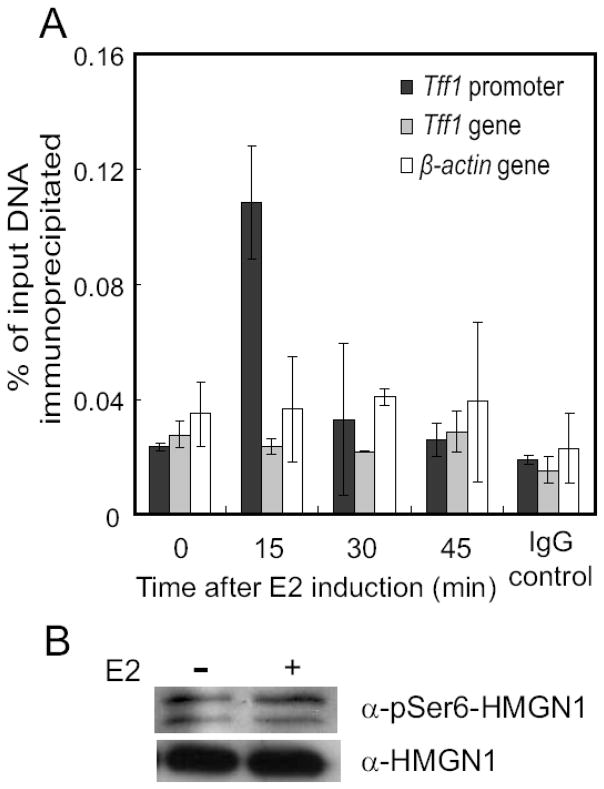
Isolation of HMGN1 from MCF7 cells propagated in constant levels of estrogen and analysis by mass spectrometry also identified multiple sites of phosphorylation on HMGN1, including in this case Ser6, Ser85, Ser88 and Ser98 [62]. This raised the possibility that Ser6 phosphorylation plays a role in regulation of gene expression by estrogen. We used ChIP analysis to examine whether estrogen stimulation altered the level of HMGN1 phosphorylation, specifically at Ser6, associated with estrogen-regulated gene regulatory regions. In these experiments, a dramatic but transient induction of HMGN1pSer6 was observed in response to estrogen stimulation on the TFF1 promoter (Fig. 1A), consistent with rapid phosphorylation correlating with IE gene induction [58]. However, estrogen stimulation did not result in any detectable change in the overall phosphorylation level of total cellular HMGN1 (Fig. 1B). This transient increase of HMGN1pSer6, whose association is limited only to the promoter region of this estrogen-inducible gene (Fig. 1A), is likely due to recruitment of MSK1 to the promoter, rather than specific recruitment of this phosphorylated form of HMGN1. In support of this model, a modest increase of MSK1 was observed on TFF1 promoter in response to estrogen induction in another study [63]. Further study indicated, however, that phosphorylation of HMGN1 at Ser6 did not affect binding of HMGN1 to chromatin in vivo, its binding to ERα in vitro, or its ability to inhibit TFF1 gene activation in vivo [64]. The lack of functionality of phosphorylation at Ser6 in these particular assays, related to stimulation by estrogen, may either suggest redundancy with the function of other phosphorylation sites on the same protein, or that such other modifications are functionally more important than Ser6 phosphorylation, despite the dramatic regulation observed. Obviously, there is still much to be learned of the biological consequences of specific modifications of HMGN.
Figure 1. Rapid and transient association of pSer6HMGN1 with the TFF1 promoter upon estrogen induction.
MCF-7 cells were cultured under estrogen-deprived conditions and then treated with 100 nM 17-β-estradiol (E2) for the indicated times. (A) ChIP using antibodies against pSer6HMGN1. Input DNA and immunoprecipitated DNA were quantified by real time PCR using the primers described previously [26]. The amount of binding of protein to a specific promoter or gene region is expressed as percentage of DNA immunoprecipitated by either the specific antibody or IgG (right most set of bars), as compared to the total chromosomal DNA before immunoprecipitation. Error bars represent standard deviation from two independent experiments. (B) Immunoblotting from a representative experiment, showing the levels of total cellular pSer6HMGN1 and HMGN1 with and without estrogen treatment.
Conclusions and perspectives
Recent studies on HMGNs have discovered exciting and expanding frontiers as to the transcriptional regulatory functions of this family of proteins. Long viewed as architectural chromatin proteins that enhance transcription, we now appreciate that HMGNs are specific transcription modulators that can both enhance as well as inhibit transcription. Many questions still remain to fully elucidate the function of HMGNs in transcription, however. One critical question is what the genome-wide binding pattern of HMGNs to chromatin is. Are HMGNs present only on regions of the genome that are functional (active in transcription, replication, recombination or repair) or are they present on chromatin in general with enrichment on regions of chromatin where their functions are needed? The lack of sequence preference in binding to nucleosomes combined with the unique characteristic of higher affinity to nucleosomes over DNA suggest a promiscuous binding property, in the absence of other considerations. The observed, specific enrichment or depletion of HMGNs on regulatory regions in a number of different cell types is only modest, being in the range of 1.5- to 5-fold, unlike the much more prominent enrichment of transcription factors on their target promoters. Furthermore, HMGN1 moves rapidly throughout the nucleus [65]. All these data suggest that HMGNs may generally associate with all chromatin, and only exhibit enrichment or depletion in local regions when exerting their biological function. With the development of techniques such as ChIP-chip and ChIP-seq, it is now possible to map genome wide association of HMGNs in order to answer the question.
All the described mechanisms whereby HMGNs regulate transcription involve their interplay with proteins or protein complexes that modify chromatin; these include modifying higher order structure by histone H1, altering levels of histone modifications, and modulating chromatin remodeling. This raises the question of whether HMGNs employ only one of these players at a time, or if it is possible that multiple such mechanisms operate concomitantly. Another unresolved question relates to the specificity within the family of HMGN proteins, whether they regulate transcription of distinct genes and in distinct circumstances, or whether there is functional redundancy between family members. The HMGN proteins share a high degree of similarity and are indistinguishable in in vitro biochemical studies [32, 33]. However, the phenotypes of mouse knockouts of the different genes are quite distinct [25, 66]. Differential post-translational modification in response to specific signals [58] may impart distinct biological roles. In addition, the tissue specificity of HMGN expression may result in different physiological functions. For example, HMGN3 is expressed at higher levels in pancreas and pituitary glands and consistent with this expression pattern, HMGN3 ablation affects pancreatic gene expression [25, 67]. Nonetheless, some redundancy has been observed between family members, such as on the Sox9 promoter where an increase of HMGN2 was seen in HMGN1 knockout cells [24]. Finally, the target gene specificity of HMGNs has been demonstrated to involve interacting with and recruitment by specific transcription factors. The question of whether different HMGN family members interact with the same or different sets of transcription factors also must be addressed in order to understand the distinct versus redundant roles. Notably, whether or not such interacting transcription factors are present may also vary, depending on cell type. In order to understand the network of regulation of target genes, regulatory regions of genes whose expression is affected by HMGN depletion can be analyzed for enrichment of transcription factor binding sites.
In summary, it is now appreciated that HMGNs can be very specific, rather than general, transcriptional regulators, and that such specificity of gene regulation is generated in part through interactions with only a subset of transcription factors. It has also been elucidated that once associated with chromatin at specific regions, HMGNs can regulate transcription through a variety of mechanisms, by affecting multiple aspects of chromatin function. However, the basis for differential regulation of different genes remains unclear, and will require investigations of the HMGN network using global interaction assays, both with proteins and with DNA, full analysis of states of HMGN modifications, and computational analyses of the gene regulatory regions with which HMGNs are more highly associated.
Acknowledgments
This work was supported by National Institutes of Health grant R01 GM54808 and a Boston University SPRInG award to U.H.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bustin M, Reeves R. High-mobility-group chromosomal proteins: architectural components that facilitate chromatin function. Prog Nucleic Acid Res Mol Biol. 1996;54:35–100. doi: 10.1016/s0079-6603(08)60360-8. [DOI] [PubMed] [Google Scholar]
- 2.Weisbrod S, Weintraub H. Isolation of a subclass of nuclear proteins responsible for conferring a DNase I-sensitive structure on globin chromatin. Proc Natl Acad Sci USA. 1979;76:630–4. doi: 10.1073/pnas.76.2.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gazit B, Panet A, Cedar H. Reconstitution of a deoxyribonuclease I-sensitive structure on active genes. Proc Natl Acad Sci USA. 1980;77:1787–90. doi: 10.1073/pnas.77.4.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kootstra A. Isolation of high mobility group-containing mononucleosomes from avian erythrocyte nuclei and their sensitivity to DNase I. J Biol Chem. 1982;257:13088–94. [PubMed] [Google Scholar]
- 5.Weisbrod ST. Properties of active nucleosomes as revealed by HMG 14 and 17 chromatography. Nucleic Acids Res. 1982;10:2017–42. doi: 10.1093/nar/10.6.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nicolas RH, Wright CA, Cockerill PN, Wyke JA, Goodwin GH. The nuclease sensitivity of active genes. Nucleic Acids Res. 1983;11:753–72. doi: 10.1093/nar/11.3.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brotherton TW, Ginder GD. Preferential in vitro binding of high mobility group proteins 14 and 17 to nucleosomes containing active and DNase I sensitive single-copy genes. Biochemistry. 1986;25:3447–54. doi: 10.1021/bi00359a053. [DOI] [PubMed] [Google Scholar]
- 8.Dorbic T, Wittig B. Chromatin from transcribed genes contains HMG17 only downstream from the starting point of transcription. EMBO J. 1987;6:2393–9. doi: 10.1002/j.1460-2075.1987.tb02517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Postnikov YV, Shick VV, Belyavsky AV, Khrapko KR, Brodolin KL, Nikolskaya TA, Mirzabekov AD. Distribution of high mobility group proteins 1/2, E and 14/17 and linker histones H1 and H5 on transcribed and non-transcribed regions of chicken erythrocyte chromatin. Nucleic Acids Res. 1991;19:717–25. doi: 10.1093/nar/19.4.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goodwin GH, Mathew CG, Wright CA, Venkov CD, Johns EW. Analysis of the high mobility group proteins associated with salt-soluble nucleosomes. Nucleic Acids Res. 1979;7:1815–35. doi: 10.1093/nar/7.7.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barsoum J, Levinger L, Varshavsky A. On the chromatin structure of the amplified, transcriptionally active gene for dihydrofolate reductase in mouse cells. J Biol Chem. 1982;257:5274–82. [PubMed] [Google Scholar]
- 12.Levinger L, Barsoum J, Varshavsky A. Two-dimensional hybridization mapping of nucleosomes. comparison of DNA and protein patterns. J Mol Biol. 1981;146:287–304. doi: 10.1016/0022-2836(81)90389-2. [DOI] [PubMed] [Google Scholar]
- 13.Goodwin GH, Nicolas RH, Cockerill PN, Zavou S, Wright CA. The effect of salt extraction on the structure of transcriptionally active genes; evidence for a DNAseI-sensitive structure which could be dependent on chromatin structure at levels higher than the 30 nm fibre. Nucleic Acids Res. 1985;13:3561–79. doi: 10.1093/nar/13.10.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malik N, Smulson M, Bustin M. Enrichment of acetylated histones in polynucleosomes containing high mobility group protein 17 revealed by immunoaffinity chromatography. J Biol Chem. 1984;259:699–702. [PubMed] [Google Scholar]
- 15.Swerdlow PS, Varshavsky A. Affinity of HMG17 for a mononucleosome is not influenced by the presence of ubiquitin-H2A semihistone but strongly depends on DNA fragment size. Nucleic Acids Res. 1983;11:387–401. doi: 10.1093/nar/11.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stein A, Townsend T. HMG 14/17 binding affinities and DNAase I sensitivities of nucleoprotein particles. Nucleic Acids Res. 1983;11:6803–19. doi: 10.1093/nar/11.19.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weisbrod S, Groudine M, Weintraub H. Interaction of HMG 14 and 17 with actively transcribed genes. Cell. 1980;19:289–301. doi: 10.1016/0092-8674(80)90410-9. [DOI] [PubMed] [Google Scholar]
- 18.Weisbrod S, Weintraub H. Isolation of actively transcribed nucleosomes using immobilized HMG 14 and 17 and an analysis of alpha-globin chromatin. Cell. 1981;23:391–400. doi: 10.1016/0092-8674(81)90134-3. [DOI] [PubMed] [Google Scholar]
- 19.Albright SC, Wiseman JM, Lange RA, Garrard WT. Subunit structures of different electrophoretic forms of nucleosomes. J Biol Chem. 1980;255:3673–84. [PubMed] [Google Scholar]
- 20.Landsman D, Mendelson E, Druckmann S, Bustin M. Exchange of proteins during immunofractionation of chromatin. Exp Cell Res. 1986;163:95–102. doi: 10.1016/0014-4827(86)90561-6. [DOI] [PubMed] [Google Scholar]
- 21.Brotherton TW, Reneker J, Ginder GD. Binding of HMG 17 to mononucleosomes of the avian beta-globin gene cluster in erythroid and non-erythroid cells. Nucleic Acids Res. 1990;18:2011–6. doi: 10.1093/nar/18.8.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Westermann R, Grossbach U. Localization of nuclear proteins related to high mobility group protein 14 (HMG 14) in polytene chromosomes. Chromosoma. 1984;90:355–65. doi: 10.1007/BF00294162. [DOI] [PubMed] [Google Scholar]
- 23.West KL. HMGN proteins play roles in DNA repair and gene expression in mammalian cells. Biochem Soc Trans. 2004;32:918–9. doi: 10.1042/BST0320918. [DOI] [PubMed] [Google Scholar]
- 24.Furusawa T, Lim JH, Catez F, Birger Y, Mackem S, Bustin M. Down-regulation of nucleosomal binding protein HMGN1 expression during embryogenesis modulates Sox9 expression in chondrocytes. Mol Cell Biol. 2006;26:592–604. doi: 10.1128/MCB.26.2.592-604.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ueda T, Furusawa T, Kurahashi T, Tessarollo L, Bustin M. The nucleosome binding protein HMGN3 modulates the transcription profile of pancreatic beta-cells and affects insulin secretion. Mol Cell Biol. 2009;29:5264–76. doi: 10.1128/MCB.00526-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu N, Hansen U. HMGN1 modulates estrogen-mediated transcriptional activation through interactions with specific DNA-binding transcription factors. Mol Cell Biol. 2007;27:8859–73. doi: 10.1128/MCB.01724-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lim JH, Catez F, Birger Y, West KL, Prymakowska-Bosak M, Postnikov YV, Bustin M. Chromosomal protein HMGN1 modulates histone H3 phosphorylation. Mol Cell. 2004;15:573–84. doi: 10.1016/j.molcel.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 28.Einck L, Bustin M. Inhibition of transcription in somatic cells by microinjection of antibodies to chromosomal proteins. Proc Natl Acad Sci USA. 1983;80:6735–9. doi: 10.1073/pnas.80.22.6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hock R, Wilde F, Scheer U, Bustin M. Dynamic relocation of chromosomal protein HMG-17 in the nucleus is dependent on transcriptional activity. EMBO J. 1998;17:6992–7001. doi: 10.1093/emboj/17.23.6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Landsman D, Bustin M. Assessment of the transcriptional activation potential of the HMG chromosomal proteins. Mol Cell Biol. 1991;11:4483–9. doi: 10.1128/mcb.11.9.4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ding HF, Rimsky S, Batson SC, Bustin M, Hansen U. Stimulation of RNA polymerase II elongation by chromosomal protein HMG-14. Science. 1994;265:796–9. doi: 10.1126/science.8047885. [DOI] [PubMed] [Google Scholar]
- 32.Trieschmann L, Alfonso PJ, Crippa MP, Wolffe AP, Bustin M. Incorporation of chromosomal proteins HMG-14/HMG-17 into nascent nucleosomes induces an extended chromatin conformation and enhances the utilization of active transcription complexes. EMBO J. 1995;14:1478–89. doi: 10.1002/j.1460-2075.1995.tb07134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trieschmann L, Postnikov YV, Rickers A, Bustin M. Modular structure of chromosomal proteins HMG-14 and HMG-17: definition of a transcriptional enhancement domain distinct from the nucleosomal binding domain. Mol Cell Biol. 1995;15:6663–9. doi: 10.1128/mcb.15.12.6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tremethick DJ, Hyman L. High mobility group protein 14 and 17 can prevent the close packing of nucleosomes by increasing the strength of protein contacts in the linker DNA. J Biol Chem. 1996;271:12009–16. doi: 10.1074/jbc.271.20.12009. [DOI] [PubMed] [Google Scholar]
- 35.Weigmann N, Trieschmann L, Bustin M. Enhancement of the transcription potential of nascent chromatin by chromosomal proteins HMG-14/-17 is coupled to nucleosome assembly and not DNA synthesis. DNA Cell Biol. 1997;16:1207–16. doi: 10.1089/dna.1997.16.1207. [DOI] [PubMed] [Google Scholar]
- 36.Ding HF, Bustin M, Hansen U. Alleviation of histone H1-mediated transcriptional repression and chromatin compaction by the acidic activation region in chromosomal protein HMG-14. Mol Cell Biol. 1997;17:5843–55. doi: 10.1128/mcb.17.10.5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crippa MP, Trieschmann L, Alfonso PJ, Wolffe AP, Bustin M. Deposition of chromosomal protein HMG-17 during replication affects the nucleosomal ladder and transcriptional potential of nascent chromatin. EMBO J. 1993;12:3855–64. doi: 10.1002/j.1460-2075.1993.tb06064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paranjape SM, Krumm A, Kadonaga JT. HMG17 is a chromatin-specific transcriptional coactivator that increases the efficiency of transcription initiation. Genes Dev. 1995;9:1978–91. doi: 10.1101/gad.9.16.1978. [DOI] [PubMed] [Google Scholar]
- 39.Hansen U. Transcriptional and structural analyses of isolated SV40 chromatin. Methods Mol Biol. 1999;119:261–90. doi: 10.1385/1-59259-681-9:261. [DOI] [PubMed] [Google Scholar]
- 40.Hill DA, Peterson CL, Imbalzano AN. Effects of HMGN1 on Chromatin Structure and SWI/SNF-mediated Chromatin Remodeling. J Biol Chem. 2005;280:41777–83. doi: 10.1074/jbc.M509637200. [DOI] [PubMed] [Google Scholar]
- 41.Catez F, Brown DT, Misteli T, Bustin M. Competition between histone H1 and HMGN proteins for chromatin binding sites. EMBO Rep. 2002;3:760–6. doi: 10.1093/embo-reports/kvf156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Catez F, Yang H, Tracey KJ, Reeves R, Misteli T, Bustin M. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol Cell Biol. 2004;24:4321–8. doi: 10.1128/MCB.24.10.4321-4328.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shirakawa H, Herrera JE, Bustin M, Postnikov Y. Targeting of high mobility group-14/-17 proteins in chromatin is independent of DNA sequence. J Biol Chem. 2000;275:37937–44. doi: 10.1074/jbc.M000989200. [DOI] [PubMed] [Google Scholar]
- 44.Rubinstein YR, Furusawa T, Lim JH, Postnikov YV, West KL, Birger Y, Lee S, Nguyen P, Trepel JB, Bustin M. Chromosomal protein HMGN1 modulates the expression of N-cadherin. FEBS J. 2005;272:5853–63. doi: 10.1111/j.1742-4658.2005.04980.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.West KL, Castellini MA, Duncan MK, Bustin M. Chromosomal proteins HMGN3a and HMGN3b regulate the expression of glycine transporter 1. Mol Cell Biol. 2004;24:3747–56. doi: 10.1128/MCB.24.9.3747-3756.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Postnikov YV, Belova GI, Lim JH, Bustin M. Chromosomal Protein HMGN1 Modulates the Phosphorylation of Serine 1 in Histone H2A. Biochemistry. 2006;45:15092–15099. doi: 10.1021/bi0613271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lim JH, West KL, Rubinstein Y, Bergel M, Postnikov YV, Bustin M. Chromosomal protein HMGN1 enhances the acetylation of lysine 14 in histone H3. EMBO J. 2005;24:3038–48. doi: 10.1038/sj.emboj.7600768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hock R, Furusawa T, Ueda T, Bustin M. HMG chromosomal proteins in development and disease. Trends Cell Biol. 2007;17:72–9. doi: 10.1016/j.tcb.2006.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lim JH, Bustin M, Ogryzko VV, Postnikov YV. Metastable macromolecular complexes containing high mobility group nucleosome-binding chromosomal proteins in HeLa nuclei. J Biol Chem. 2002;277:20774–82. doi: 10.1074/jbc.M200404200. [DOI] [PubMed] [Google Scholar]
- 50.Lee JW, Choi HS, Gyuris J, Brent R, Moore DD. Two classes of proteins dependent on either the presence or absence of thyroid hormone for interaction with the thyroid hormone receptor. Mol Endocrinol. 1995;9:243–54. doi: 10.1210/mend.9.2.7776974. [DOI] [PubMed] [Google Scholar]
- 51.Amano T, Leu K, Yoshizato K, Shi YB. Thyroid hormone regulation of a transcriptional coactivator in Xenopus laevis: implication for a role in postembryonic tissue remodeling. Dev Dyn. 2002;223:526–35. doi: 10.1002/dvdy.10075. [DOI] [PubMed] [Google Scholar]
- 52.Amen M, Espinoza HM, Cox C, Liang X, Wang J, Link TM, Brennan RG, Martin JF, Amendt BA. Chromatin-associated HMG-17 is a major regulator of homeodomain transcription factor activity modulated by Wnt/beta-catenin signaling. Nucleic Acids Res. 2008;36:462–76. doi: 10.1093/nar/gkm1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Singh U, Bongcam-Rudloff E, Westermark B. A DNA sequence directed mutual transcription regulation of HSF1 and NFIX involves novel heat sensitive protein interactions. PLoS One. 2009;4:e5050. doi: 10.1371/journal.pone.0005050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Happel N, Doenecke D. Histone H1 and its isoforms: contribution to chromatin structure and function. Gene. 2009;431:1–12. doi: 10.1016/j.gene.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 55.Rattner BP, Yusufzai T, Kadonaga JT. HMGN proteins act in opposition to ATP-dependent chromatin remodeling factors to restrict nucleosome mobility. Mol Cell. 2009;34:620–6. doi: 10.1016/j.molcel.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Soloaga A, Thomson S, Wiggin GR, Rampersaud N, Dyson MH, Hazzalin CA, Mahadevan LC, Arthur JS. MSK2 and MSK1 mediate the mitogen- and stress-induced phosphorylation of histone H3 and HMG-14. EMBO J. 2003;22:2788–97. doi: 10.1093/emboj/cdg273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thomson S, Clayton AL, Hazzalin CA, Rose S, Barratt MJ, Mahadevan LC. The nucleosomal response associated with immediate-early gene induction is mediated via alternative MAP kinase cascades: MSK1 as a potential histone H3/HMG-14 kinase. EMBO J. 1999;18:4779–93. doi: 10.1093/emboj/18.17.4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barratt MJ, Hazzalin CA, Zhelev N, Mahadevan LC. A mitogen- and anisomycin-stimulated kinase phosphorylates HMG-14 in its basic amino-terminal domain in vivo and on isolated mononucleosomes. EMBO J. 1994;13:4524–35. doi: 10.1002/j.1460-2075.1994.tb06774.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cherukuri S, Hock R, Ueda T, Catez F, Rochman M, Bustin M. Cell cycle-dependent binding of HMGN proteins to chromatin. Mol Biol Cell. 2008;19:1816–24. doi: 10.1091/mbc.E07-10-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Prymakowska-Bosak M, Hock R, Catez F, Lim JH, Birger Y, Shirakawa H, Lee K, Bustin M. Mitotic phosphorylation of chromosomal protein HMGN1 inhibits nuclear import and promotes interaction with 14.3.3 proteins. Mol Cell Biol. 2002;22:6809–19. doi: 10.1128/MCB.22.19.6809-6819.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hock R, Scheer U, Bustin M. Chromosomal proteins HMG-14 and HMG-17 are released from mitotic chromosomes and imported into the nucleus by active transport. J Cell Biol. 1998;143:1427–36. doi: 10.1083/jcb.143.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zou Y, Jiang X, Wang Y. Identification of novel in vivo phosphorylation sites in high mobility group N1 protein from the MCF-7 human breast cancer cells. Biochemistry. 2004;43:6322–9. doi: 10.1021/bi0362828. [DOI] [PubMed] [Google Scholar]
- 63.Espino PS, Li L, He S, Yu J, Davie JR. Chromatin modification of the trefoil factor 1 gene in human breast cancer cells by the Ras/mitogen-activated protein kinase pathway. Cancer Res. 2006;66:4610–6. doi: 10.1158/0008-5472.CAN-05-4251. [DOI] [PubMed] [Google Scholar]
- 64.Zhu N. Ph.D. dissertation. Boston University; 2007. Role of high mobility group nucleosomal binding protein HMGN1 in estrogen-mediated transcription activation; pp. 105–114. [Google Scholar]
- 65.Phair RD, Misteli T. High mobility of proteins in the mammalian cell nucleus. Nature. 2000;404:604–9. doi: 10.1038/35007077. [DOI] [PubMed] [Google Scholar]
- 66.Birger Y, West KL, Postnikov YV, Lim JH, Furusawa T, Wagner JP, Laufer CS, Kraemer KH, Bustin M. Chromosomal protein HMGN1 enhances the rate of DNA repair in chromatin. EMBO J. 2003;22:1665–75. doi: 10.1093/emboj/cdg142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.West KL, Ito Y, Birger Y, Postnikov Y, Shirakawa H, Bustin M. HMGN3a and HMGN3b, two protein isoforms with a tissue-specific expression pattern, expand the cellular repertoire of nucleosome-binding proteins. J Biol Chem. 2001;276:25959–69. doi: 10.1074/jbc.M101692200. [DOI] [PubMed] [Google Scholar]