Abstract
Hydroxyurea (HU) is an effective drug for the treatment of sickle cell disease (SCD). The main clinical benefit of HU is thought to derive from its capacity to increase fetal hemoglobin (HbF) production. However, other effects leading to clinical benefit, such as improved blood rheology, have been suggested. In order to understand HU-induced changes at the proteomic level, we profiled sickle RBC membranes from of HU-treated and untreated patients. Our previous in vitro profiling studies on sickle RBC membranes identified a significant increase in predominantly anti-oxidant enzymes, protein repair and degradation components and a few RBC cytoskeletal proteins. In the present study, using 2D-DIGE (Two-Dimensional Difference In-Gel Electrophoresis) and tandem mass spectrometry, we detected 32 different proteins that significantly changed in abundance in the HU treatment group. The proteins that significantly increased in abundance were mostly membrane skeletal components involved in the regulation of RBC shape and flexibility, and those showing a significant decrease were components of the protein repair and degradation machinery. RBC palmitoylated membrane protein 55 (p55) is significantly increased in abundance at low (in vitro) and high (in vivo) concentrations of HU. Palmitoylated p55 may be an important target of HU-dependent regulation of the sickle RBC membrane, consistent with our earlier in vitro studies.
Keywords: Sickle Cell Disease, Hydroxyurea, Red Blood Cell Membrane, 2D- DIGE, Tandem Mass Spectrometry, Palmitoylated protein 55 (p55)
1. Introduction
Sickle cell disease (SCD) is a recessive genetic disorder caused by a point mutation in the 6th codon of the β globin gene. At the amino acid level, this results in the substitution of glutamic acid by valine in the β-globin subunit of hemoglobin [1]. In the deoxygenated state, the mutant sickle hemoglobin (HbS) forms rigid and insoluble polymers that distort the shape of the RBCs giving them a characteristic sickle shape. The polymerization of HbS leads to the complex pathophysiology associated with SCD, which includes vaso-occlusion, chronic hemolysis and irreversible tissue damage [2].
HU is thought to be an effective drug for the management of SCD due to its capacity to increase HbF levels. Increased HbF levels inhibit the polymerization of HbS and reduce sickling [2]. However, the Multicenter Study of Hydroxyurea in Sickle Cell Anemia revealed that many patients showed clinical improvement before a significant rise in HbF levels [3]. Various reports have demonstrated that increase in HbF is not the only benefit of HU. Some of the factors involved in ameliorating the pathology of sickle cell disease after HU treatment are increased MCV of sickle cell RBCs [4], reduced adhesion of sickle cell RBCs to the endothelium [5] and increased deformability of sickle cell RBCs [6]. These findings point towards HU-induced alterations of additional cellular mechanisms that are yet to be identified and that may mediate the clinical benefits of HU. The understanding of these pathways and the drug mechanism warrants the need to identify additional sickle RBC membrane proteins whose expression is regulated by HU.
A previous in vitro protein profiling study performed in our laboratory identified significant increases in RBC anti-oxidant enzymes and protein repair and degradation components after exposure of sickle RBC membranes to low concentrations of HU (50 and 100 μM). Through this in vitro study, we further demonstrated that 50 μM HU exposed sickle RBC membranes showed a 2-fold increase in tyrosine phosphorylation of catalase as compared to counterparts not exposed to HU [7]. The in vitro protein profiling system allowed us to look at the same sickle RBC membrane sample from individual SS patients with and without HU exposure to identify dose-dependent proteomic changes in vitro, which is difficult to achieve in an in vivo clinical setting. However, the in vitro system utilizes mature enucleated RBCs that lack the capacity to synthesize new proteins and the in vitro proteomic changes identified mainly reflect post-translational modifications. Moreover, HU acts on late erythroid precursors in the bone marrow and influences the erythropoietic pathway [8]. Hence, in the present study, we have undertaken an in vivo proteomic analysis of sickle RBC membranes with the following aims: 1) Identify common HU-induced proteomic changes in vitro and in vivo, 2) Identify HU-induced changes at concentrations that are actually administered to SS patients in a clinical setting and 3) Identify changes in protein expression as well as protein modification.
Though some roles of HU in pathways other than HbF production have been reported, the protein targets altered in these pathways as a result of HU treatment are not known. With HU being the only FDA-approved drug to date, studies to investigate HU-dependent protein alterations are important to understand the drug’s mechanism of action as well as its toxic and beneficial effects. With the goal of identifying RBC membrane protein alterations in homozygous sickle cell anemia (SS) patients on HU therapy, we performed 2D-DIGE followed by tandem mass spectrometry. A global protein profiling approach eliminates the need to study drug-induced response of individual cellular pathways and provides a common platform for the simultaneous fluorescent detection of thousands of drug-related changes in proteins. In this proteomic study, we report a significant increase in two major classes of proteins after in vivo HU therapy: RBC membrane skeletal components and glycolytic enzymes. A combination of 2D-DIGE and tandem mass spectrometry led to the identification of 32 different sickle RBC membrane proteins of interest showing a significant change in content as a response to an average dose of 35 mg/kg (400μM) administered in vivo. Thirty of these showed a significant increase and belonged to the group of RBC membrane skeletal proteins and glycolytic enzymes. The two proteins that showed a significant decrease represented components of the protein repair and degradation machinery. These results demonstrate some important HU-mediated proteomic changes, besides HbF production, likely to have implications in regulating sickle RBC rheology, metabolism and oxidative damage.
The significant increase in palmitoylated membrane protein 1 (p55) at low HU concentrations in vitro and high HU concentrations in vivo makes this protein an important response marker of HU therapy.
2. Materials and Methods
2.1. Subjects
After informed consent, human blood samples (10ml) were collected from homozygous SS patients by venipuncture using lithium heparin as an anti-coagulant at the Southwestern Comprehensive Sickle Cell Center. The treatment group included three adults and two children with SS who had received HU for at least 4 months. We selected these patients because they had an excellent clinical response to HU, based on a marked reduction in the frequency of hospitalizations for vaso-occlusive (painful) crises and acute chest syndrome as well as patient self-report (improved quality of life and decreased painful episodes at home). Untreated SS controls had not been treated at the time of phlebotomy or previously with HU. No patient from the treated or untreated groups had received a blood transfusion within the preceding 4 months.
2.2. Isolation of reticulocyte-free red blood cells
Reticulocyte free red blood cells were isolated by a density based method developed and assessed in our laboratory by Kakhniashvili et al. [9]. The collected blood samples were used within four hours. All the procedures were performed at room temperature. Five ml of blood was centrifuged at 550g for 10 minutes to remove plasma. The pelleted cells were washed four times with 10 volumes of PBS (11.9mM phosphate, pH 7.4, 137mM NaCl, 2.7mM KCl) by centrifuging at 550 g for 10 minutes. The washed blood cells were finally resuspended in PBS at 50% hematocrit and were loaded onto a single layer of 75% percoll of equal volume and centrifuged at 1000xg for 15 minutes in 15 ml centrifugal tubes in a bucket rotor. 75% percoll solution (GE Healthcare, p = 1.130±0.005g/ml) was prepared by dilution with 10X PBS and water. The bottom layer of the density gradient represents pure population of RBCs which after 3 washes of PBS, was used for membrane preparation.
2.3. Preparation of erythrocyte membranes
Erythrocyte membranes were prepared as described [10]. The RBCs were sedimented at 1000 g for 10 minutes at 4°C and resuspended in PBS (10mM NaPO4, pH 7.6, 150mM NaCl). This step was repeated four times. The RBCs were then resuspended in 10 volumes of PBS and sedimented at 2000 g for 10 minutes. The washed RBCs were lysed in six volumes of lysis buffer (5mM NaPO4, 1mM EDTA, pH 7.6) and were sedimented at 31,000 g for 30 minutes. This step was repeated until the pellet became white or light pink. Membrane protein concentration was measured by Protein assay reagent (Bio-Rad). The membranes containing ~ 4–6 mg of protein/ml were vacuum dried and solubilized in lysis buffer (30mM Tris-HCl, pH 8.5, 7M urea, 2M thiourea, and 2% (w/v) nonionic detergent ASB 14).
2.4. Minimal labeling of SS membrane proteins
SS membrane proteins corresponding to 100 μg each of (−)HU and (+)HU samples solubilized in lysis buffer were minimally labeled with Cy3 and Cy5 fluorophores respectively according to the manufacturer’s protocol (Amersham Biosciences).
2.5. Separation of proteins in first dimension (IEF)
100 μg each of the control and drug treated samples were mixed together and rehydration buffer (1% Pharmalyte 3–10 NL, 7 M urea, 2 M thiourea, 2 % ASB-14, 2 mg/ml DTT) was added to the final protein mixture. The sample, included in the rehydration solution was loaded on the IPG (Immobilized pH Gradient) strip holder. Immobiline DryStrip gel (Amersham Biosciences, pH gradient 3–10 Non-Linear, length of strip 13 cm) was placed over the sample and finally IPG cover fluid was added to minimize evaporation and urea crystallization. Rehydration of the IPG strip proceeded on an Ettan IPGphor Isoelectric Focusing System (Amersham Biosciences) for 12 hours at 20°C. Isoelectric focusing was performed in three steps: at 500 V for 1 hour, at 1000 V for 1hour and at 8000 V for 33,000 Volt-hours.
2.6. Reduction and alkylation of the IEF separated proteins
Prior to running the second dimension, the Immobiline strip with separated proteins were equilibrated and reduced in a solution containing 50 mM Tris-HCl buffer, pH 8.6, 2% SDS, 30% glycerol and 5mg/ml DTT at 90° C for one minute. This step was followed by equilibration and alkylation at room temperature in a solution containing 50 mM Tris-HCl buffer, pH 8.6, 6M urea, 2% SDS, 30% glycerol, and 20mg/ml iodoacetamide for 10 minutes.
2.7. Separation of proteins in the second dimension (SDS-PAGE)
After IEF separation, reduction and alkylation steps, the proteins were further separated on a 10% SDS- polyacrylamide gel. The IPG strip containing isoelectrically focused proteins was directly loaded on the top of the separating gel. Electrophoresis was carried out in the dark (to reduce exposure of fluorophores to light) for 18 hours at a constant voltage of 40 volts.
2.8. Gel Imaging and Analysis
The separated proteins labeled with Cy3 and Cy5 fluorophores were detected using 2920 2D-Master Imager (Amersham Biosciences). The Cy3 (primary) and Cy5 (secondary) gel images were then processed using the DeCyder DIA (Differential In-Gel Analysis) software (Amersham Biosciences). This software detects protein spots (spots migrating to the same position on the SDS gel) and quantifies the spot volumes for each image and expresses these volumes as ratios. This ratio can then be used for protein abundance comparisons between the primary Cy3 labeled (−)HU and Cy5 (+)HU SS RBC membrane samples. DeCyder BVA (Biological Variation Analysis) software was used for matching multiple images from different gels for identification of protein spots that are consistently changing in abundance. Statistical analysis was performed on all the protein spots that exhibited ≥ ± 2.5-fold change in protein content at least in one DIGE experiment. Protein spots with statistically significant change in protein content were chosen for identification by mass spectrometry.
2.9. Statistical Analysis
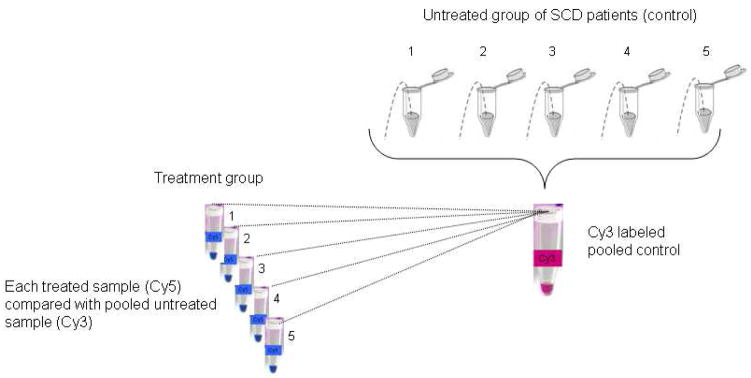
In this study, a pooled membrane sample of 5 SS patients who did not receive hydroxyurea (Cy3 labeled) served as the control group. The treatment group consisted of 5 sickle cell membrane samples from SS patients who received HU (Cy5 labeled). Each individual sickle cell sample from the treatment group was compared with a portion of the pooled control. The data consisted of fold-changes (HU treated/untreated or Cy5/Cy3) in volumes of protein spots in n = 5 pairs of samples. Not every protein spot was observed in every patient. So we only analyzed those spots that were observed in at least three pairs of samples. This resulted in data on 89 spots.
The goal of statistical analysis of these data was to discover protein spots whose mean ratios (HU treated/untreated) differed significantly from one. The fold-changes were converted into ratios and then natural log of the ratios was taken to normalize the data. Since the normality assumption was appropriate for these data, we used the two-sided t-test to compute the p-value for testing the null hypothesis that the distribution of log-ratios on each spot had a mean of zero. The p-value for each individual spot was adjusted to account for multiple testing using the false discovery rate (FDR) method [11]. A cutoff of 0.11 was used for the FDR to assess significance of results. This means that 11% of the spots declared to be significant are expected to be false positives. The statistical analysis was performed using the statistical software R [12].
2.10. Protein identification by tandem mass spectrometry and database search
The selected protein spots were excised from Sypro Ruby- stained gels using an Ettan Spot Picker (Amersham Biosciences). The spots were digested using an in-gel trypsin digestion kit (Pierce). The identity of the tryptic peptides generated out of each excised protein spot was determined using LC/MS/MS (LC-Liquid Chromatography and MS-Mass Spectrometry) and database search. The LC/MS/MS was performed on a HPLC system connected to a LCQ XP ion trap mass spectrometer with a nanospray ionization source (ThermoFinnigan). The parameters for the HPLC system were as described by Kakhniashvili et al. [10]. The database used for tryptic peptide search was NCBI non- redundant protein sequence database (human.nr.fasta, 2007) using the Sequest algorithm. The criteria used to confirm the identity of a certain protein was as followed in our laboratory: 1) Xcorr (X Correlation) of at least 1.5, 2.0 and 2.5 for singly, doubly and triply charged peptides respectively and 2) dCn (delta Correlation) of at least 0.1 regardless of charge state [10].
2.11. Immunoblot analysis of protein p55
Immunoblot analysis was performed to assess the levels of protein p55 in SS patients undergoing or not undergoing HU therapy. Following 2D-electrophoresis, proteins were electroblotted onto nitrocellulose membrane in transfer buffer (25 mM Tris, 192 mM glycine, 0.005% (w/v) SDS, 20% (v/v) methanol) overnight at 4° C and 25 V. Membranes were blocked for 1 hour with 5% milk blocking agent dissolved in 1X PBST (10mM NaPO4, pH 7.4, 150mM NaCl, 0.05 % Tween 20) and incubated in anti-p55 primary antibody (Santa Cruz Biotechnology, Inc.) overnight at 4 °C followed by four washes of PBST for 5 minutes each. Immnoreactive spots were detected by incubating the membranes for 1 hour in horseradish peroxidase conjugated secondary antibody (Amersham Biosciences). After four PBST washes of 5 minutes each, membranes were developed using ECL plus as the chemiluminiscent system (Amersham Biosciences). Densitometric analysis was performed on the spots using an AlphaImager densitometer (Alpha Innotech).
3. Results
Clinical parameters of SS subjects on HU therapy
Table 1 represents the clinical data of SS patients on HU therapy. All five SS patients in the treated group received high doses of HU ranging from 30 to 38 mg/kg/day for duration of 4 months to 5 years. Based on studies performed by Charache et al. [13], an average HU dose of 35mg/kg/day corresponds to a plasma HU concentration of 400μM. All the SS patients on HU therapy showed a marked decrease in vaso-occlusive episodes and acute chest syndrome with the HU doses received at the time when blood was collected. The HU-treated patients had overall event rates of 0/year while on HU.
Table 1.
Clinical data of SS patients on HU therapy
| Patient ID | Age | Gender | Hgb | MCV | Retic | HbF | Pain events/year* | ACS events/year* | HU dose mg/kg/day | Duration |
|---|---|---|---|---|---|---|---|---|---|---|
| 1179 (A) | 44 | F | 6.0 | 117.9 | 7.6 | 15.1 | 3 | 0 | 38 | 7 months |
| 1180 (C) | 16 | F | 9.3 | 102.6 | 9.2 | 9.5 | 1 | 2 | 30 | 4 years |
| 1181 (A) | 23 | M | 11.7 | 102.5 | 12.2 | 15.8 | 1 | 0 | 38 | 7 months |
| 1182 (A) | 38 | M | 11.6 | 122.4 | 13.1 | 34.2 | 5 | 0 | 38 | 4 years |
| 1185 (C) | 13 | M | 9.0 | 75.2 | 2.0 | 6.0 | 4 | 0 | 30 | 4 years, 8 months |
Abbreviations: A-adult, C-child, Hgb-total hemoglobin, MCV-Mean Corpuscular Volume, Retic- Reticulocyte, ACS-Acute Chest Syndrome
Event rates for pain and ACS are lifetime event rates
Table 2 represents clinical data from five patients in the control group who never received HU, their ages ranged from 10–50 years, and they suffered at least one pain episode in the preceding year. Patients from both the groups did not receive blood transfusion in the preceding four months. It should be noted that though patients 1175 and 1176 in the control group showed high HbF levels (19.4% and 21.5% respectively), their corresponding MCVs, frequency of pain events and ACS do not indicate any added advantage due to the high HbF levels. The overall MCV values in the treatment group are higher than those in the control group indicative of improved hydration status of sickle RBCs after HU therapy. Similarly the overall frequency of pain events as well as acute chest syndrome is lower in the treatment group as compared to the control group suggesting clinical improvement after HU therapy. There was no information available on HbF levels from patients 1178 and 1179 probably because HbF levels are not frequently monitored in SS patients not on HU therapy.
Table 2.
Clinical data of SS patients in control group (not on HU therapy)
| Patient ID | Age | Hgb (g/dl) | MCV (fl) | Reticulocyte (%) | HbF (%) | Pain events/year | ACS events/year |
|---|---|---|---|---|---|---|---|
| 1175 | 10 | 8.6 | 67.6 | 4.1 | 21.4 | 2 | 0 |
| 1176 | 43 | 8.4 | 76.2 | 4.2 | 19.2 | 4 | 0 |
| 1177 | 18 | 9.8 | 92.0 | 7.6 | 4.1 | 6 | 4 |
| 1178 | 52 | 9.6 | 84.7 | 10.5 | * | 3 | 5 |
| 1190 | 11 | 7.1 | 82.3 | 20.3 | * | 1 | 2 |
HbF levels of patients 1178 and 1190 were not available at the time of blood collection
Statistically significant proteomic changes in SS patients on HU therapy
Our experimental design (Fig. 1) for in vivo proteomic analysis consisted of a Cy 3 labeled pooled sickle RBC membrane reference control sample obtained from five SS patients not on HU therapy. This was compared against individual Cy 5 labeled sickle RBC membrane samples obtained from five SS patients on HU therapy. Using a False Discovery Rate (FDR) of 11%, a total of 37 spots exhibited significant change after in vivo HU administration. The mean ratio was less than one in only two of these spots, and in the rest, it was more than one. The identity of 32 spots was established using tandem mass spectrometry. Fig. 2 represents a Sypro-Ruby stained 2D gel with the positions of the protein spots of interest marked. Table 3 lists the proteins that demonstrated significant alterations in response to HU therapy.
Fig. 1.
Experimental design for in vivo proteomic analysis of −/+ HU sickle RBC membranes-RBC membrane samples (100 μg each) obtained from reticulocyte free, mature RBCs of five SS subjects not on HU were pooled and labeled with Cy3 fluorophore. This pooled control was compared against individual Cy 5 labeled sickle RBC membrane samples (100μg) obtained from reticulocyte free, mature RBCs of five SS patients on HU therapy.
Fig. 2.
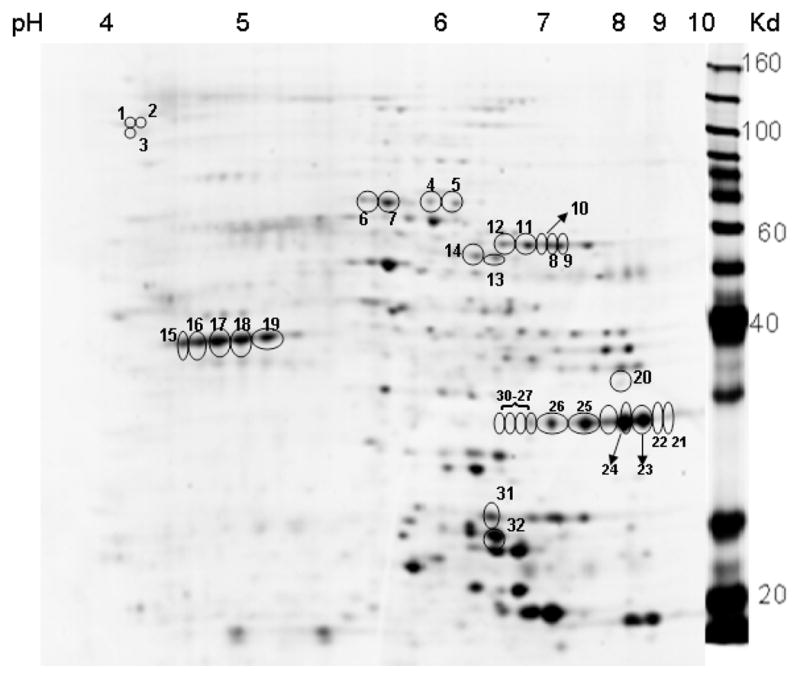
A Sypro-Ruby stained representative 2D gel showing positions of SS RBC membrane proteins that showed a statistically significant change in patients undergoing HU treatment (p< 0.1). The marked protein spots were selected for protein identification by tandem mass spectrometry. The molecular weight marker lane is indicated on the right and the pH gradient used for isoelectric focusing is indicated on the top, determining the 2D position of any given protein spot based on its pI and molecular weight.
Table 3.
Sickle RBC membrane proteins altered in vivo in patients on HU therapy
| Spot no. | Identified protein | Accession no. | Average volume ratio (+HU/−HU) | Adjusted p value (α < 0.1) | Theoretical Mr/pI on 2D gel |
|---|---|---|---|---|---|
| 1 | Solute camel family 4, anion exchanger member 1 (band 3) | 4507021 | 2.45 | 0.09 | 101.8/5.10 |
| 2 | Solute carrier family 4, anion exchanger member 1 (band 3) | 4507021 | 2.45 | 0.095 | 52.30/7.22 |
| Valosin-containing protein | 6005942 | 89.32/5.19 | |||
| 3 | N-acylaminoacyl - peptide hydrolase | 23510451 | 2.85 | 0.095 | 81.22/5.29 |
| 4 | Ankyrin 1 isoform 1 | 105337 | 2.97 | 0.002 | 92.20/6.15 |
| 5 | Ankyrin 1 isoform 1 | 105337 | 3.01 | 0.065 | 92.20/6.15 |
| 6 | Protein 4.1 | 14916944 | 2.59 | 0.085 | 97.59/4.25 |
| 7 | Protein 4.1 | 14916944 | 2.18 | 0.095 | 97.59/4.25 |
| 8 | Palmitoylated membrane protein (p55) | 62898353 | 2.78 | 0.065 | 52.30/7.22 |
| 9 | Palmitoylated membrane protein (p55) | 62898353 | 2.48 | 0.067 | 52.30/7.22 |
| 10 | Palmitoylated membrane protein (p55) | 62898353 | 2.20 | 0.095 | 52.30/7.22 |
| 11 | Palmitoylated membrane protein (p55) | 62898353 | 2.42 | 0.095 | 52.30/7.22 |
| 12 | Palmitoylated membrane protein (p55) | 62898353 | 2.37 | 0.095 | 52.30/7.22 |
| 13 | Chaperonin containing TCP1, subunit 2 | 5453603 | −2.45 | 0.095 | 57.83/6.00 |
| 14 | β Actin | 1419444 | 3.53 | 0.065 | 41.6/5.45 |
| 15 | Actin | 1703156 | 2.70 | 0.09 | 41.77/5.22 |
| 16 | β Actin | 1419444 | 2.33 | 0.09 | 41.6/5.45 |
| 17 | Tropomodulin | 135922 | 2.80 | 0.077 | 40.57/5.03 |
| 18 | Tropornodulin | 135922 | 2.55 | 0.095 | 40.57/5.03 |
| 19 | Fructose - bisphosphate aldolase | 13606 | 2.03 | 0.077 | 39.20/8.01 |
| 20 | Glyceraldehyde-3-Phosphate dehydrogenase | 7669492 | 4.0 | 0.042 | 36.05/8.58 |
| 21 | Glyceraldehyde-3-Phosphate dehydrogenase | 7669492 | 2.77 | 0.042 | 36.05/8.58 |
| 22 | Glyceraldehyde-3-Phosphate dehydrogenase | 7669492 | 3.17 | 0.042 | 36.05/8.58 |
| 23 | Glyceraldehyde-3-Phosphate dehydrogenase | 7669492 | 3.46 | 0.043 | 36.05/8.58 |
| 24 | Glyceraldehyde-3-Phosphate dehydrogenase | 7669492 | 3.49 | 0.043 | 36.05/8.58 |
| 25 | Glyceraldehyde-3-Phosphate dehydrogenase | 7669492 | 3.82 | 0.065 | 36.05/8.58 |
| 26 | Glyceraldehyde-3-Phosphate dehydrogenase | 7669492 | 3.61 | 0.065 | 36.05/8.58 |
| 27 | Glyceraldehyde-3-Phosphate dehydrogenase | 7669492 | 3.17 | 0.067 | 36.05/8.58 |
| 28 | Glyceraldehyde-3-Phosphate dehydrogenase | 7669492 | 2.61 | 0.076 | 36.05/8.58 |
| 29 | Glyceraldehyde-3-Phosphate dehydrogenase | 7669492 | 3.53 | 0.077 | 36.05/8.58 |
| 30 | Glyceraldehyde-3-Phosphate dehydrogenase | 7669492 | 1.95 | 0.095 | 36.05/8.58 |
| 31 | Stomatin isform a | 38016911 | 2.49 | 0.105 | 31.73/7.90 |
| 32 | Proteasome subunit alpha type 4 | 4506185 | −4.13 | 0.105 | 29.48/7.58 |
The altered proteins belonged to four major groups: RBC membrane skeletal proteins, glycolytic enzymes, protein repair participants and protein degradation components. Proteins identified as RBC membrane skeletal components and showing increased expression were protein band 3, ankyrin, protein 4.1, palmitoylated membrane protein (p55), actin, tropomodulin and stomatin. Enzymes participating in glycolysis: glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and fructose bisphosphate aldolase were also identified as significantly increasing after HU treatment. As seen in Table 3, it was of common occurrence for a protein to appear in multiple spots, each spot showing the same direction of change. For example, glyceraldehyde-3-phosphate dehydrogenase was identified in spots 20 through 30 with all spots demonstrating a significant increase suggesting HU-induced increase in expression of the protein during erythropoiesis. Of these proteins, p55 was the only common HU target identified in our present in vivo and previous in vitro analyses of HU- induced changes [7]. The only significant decrease was observed in chaperonin containing TCP1 subunit 2, a component of a large multiprotein TCP1 ring complex involved in the proper folding of actin and tubulin in an ATP-dependent way [14]. A marked decrease in this protein folding/repair component may suggest the failure of the anti-oxidant system to protect against HU-induced oxidative stress at high in vivo HU doses of 400μM or above.
Immunoblot analysis of palmitoylated membrane protein 55 (p55)
Our 2D-DIGE and mass spectrometry based proteomic approach identified a significant increase in palmitoylated membrane protein 55 (p55) in patients on HU therapy. In order to confirm these findings, we assessed the levels of p55 from SS RBC membranes prepared from representative blood samples from the control and treatment group. The results obtained from western blot analysis confirmed increased levels of p55 after in vivo administration of HU (Fig. 3). Patient 1180 (represented as +(HU)1 in lane 2) received 30mg/kg/day of HU for four years and showed a 5-fold increase in p55 levels as compared to the −(HU) sample from the untreated group (lane 1). Patient 1185 (represented as + (HU)2 in lane 3) received 30mg/mg/day of HU for four years, 8 months and demonstrated a 10-fold increase in p55 levels as compared to the −(HU) sample from the untreated group (lane 1).
Fig. 3.
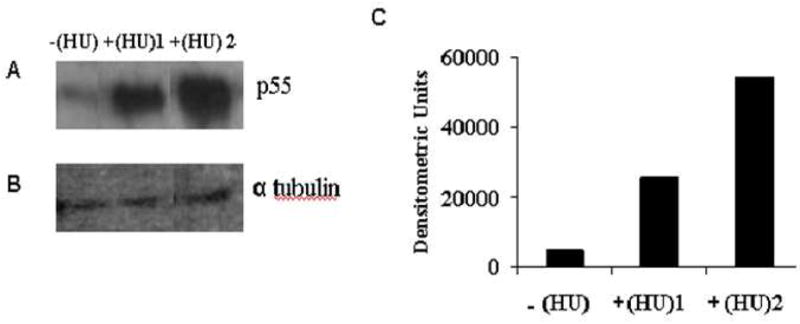
Western blot to confirm increased levels of palmitoylated membrane protein (p55) in SS RBC membranes from representative SS patients on HU therapy. Panel A Lane1 - SS RBC membrane sample from the untreated group of SS patients represented as −(HU). Lane 2 and 3 - SS RBC membrane samples from two SS different patients, +(HU)1 and +(HU)2 in the treatment group. Panel B. α tubulin loaded as internal control. Panel C. Densitometric analysis of band intensities. +(HU)1 and +(HU)2 samples showed a 5 fold and 10 fold increase in p55 levels respectively as compared to −(HU) sample.
4. Discussion
Though the in vivo effects of HU on the status of RBC dehydration [4], adhesion to endothelium [5], deformability [6] and NO production [15] have been studied, very few protein targets involved in these pathways have been identified. The goal of the present study was to identify protein markers of HU therapy in vivo. Since, in the in vivo system, the main target of HU is the bone marrow, protein changes at the level of protein expression as well as post-translational modification can be analyzed. Hence these changes are a reflection of changes throughout erythropoiesis as well as after RBC maturation (enucleation). In our previous in vitro protein profiling studies, we analyzed HU-mediated quantitative protein changes in membranes of mature, enucleated sickle RBCs exposed to increasing HU concentrations. Due to lack of new protein synthesis in these cells, we mainly anticipated identifying post-translational protein modifications caused by HU. We were successful in demonstrating increased tyrosine phosphorylation of catalase after in vitro exposure to low concentrations of HU (50 and 100μM). The current study focused on in vivo proteomic analysis of sickle RBC membranes obtained from patients receiving an average dose of 400μM HU. This higher HU concentration achieved in vivo, caused a significant increase in RBC membrane skeletal components as well as glycolytic enzymes and a significant decrease in a few protein repair and degradation components. Ankyrin, protein 4.1, β actin, anion exchanger Band 3, palmitoylated membrane protein (p55) and stomatin were the major RBC membrane skeletal components that were identified as targets of HU therapy. Of these proteins, 4.1 and actin are members of the RBC membrane skeleton that covers the cytoplasmic surface of the RBC plasma membrane. The ‘spectrin-actin-protein 4.1 ternary complex’ is responsible for regulating the elasticity and flexibility of the RBC. The membrane skeleton is attached to the plasma membrane through the binding of protein 4.1 to the transmembrane protein glycophorin C and also through the binding of ankyrin to β spectrin and transmembrane protein band 3 [16]. Previous studies reported improved rheological properties of the sickle RBC membrane in some patients receiving HU therapy [6]. A study undertaken to measure rigidity index (measure of cell rigidity) and elastic sheer modulus (measure of membrane deformability) demonstrated better values after HU therapy [17]. Oxidation of actin [18] and protein 4.1 [19] has been directly observed in the erythrocyte membrane skeleton of patients with SCD. Our laboratory demonstrated that a locked RBC membrane skeleton, as a result of oxidatively damaged actin and spectrin, forms the molecular basis of the abnormal sickle shape [20, 21]. Thus, the increased content of crucial ternary complex proteins like protein 4.1 and actin could signify beneficial effects of HU therapy and is likely to have important implications in regulating RBC shape and flexibility in the course of the disease. Also, HU mediated increase in glycolytic enzymes G3PDH and fructose bisphosphate aldolase could satisfy the increased demand for ATP synthesis under conditions of oxidative stress.
Of all the HU-mediated protein changes, the marked increase in erythrocyte membrane protein p55 may signify an important consequence of HU therapy as it was the only common HU target identified in our in vitro and in vivo analyses of HU induced changes. The increased levels of p55 were further confirmed through immunoblot analysis. P55 is a peripheral membrane protein, most abundantly palmitoylated within human RBCs and constitutively expressed through erythropoiesis [22]. Interestingly, the primary structure of p55 consists of the PDZ domain, the SH3 domain, the tyrosine phosphorylation domain and carboxy-terminal guanylate kinase-like domain. Presence of the N-terminal SH3 domain makes it an important candidate protein in signal transduction [23]. It is suggested that p55 binds in precise proportions to the protein 4.1-glycophorin C complex, linking the plasma membrane to the RBC membrane skeleton and maintaining the stability and mechanical properties of the RBC plasma membrane [24]. Considering the possible roles of protein p55 in the RBC membrane skeletal network, and in the formation of a ternary complex with protein 4.1 and glycophorin C, the significant increase in p55 levels at low and high HU doses may represent a key response to HU therapy.
A significant decrease in chaperonin TCP1 subunit 1 and 26S proteasomal subunit α type 4 suggested increased oxidative stress in SS patients on high HU doses. Anti-oxidant enzymes catalase and thioredoxin peroxidase demonstrated a decrease by more than 2.5 fold in three SS patients out of the five under study; however, these changes were not statistically significant.
It would be interesting to note that patients 1180 and P1185 did not show substantial increases in HbF levels (9.5% and 6% respectively) characteristic of SS patients on HU therapy. However, these patients showed excellent clinical improvement after HU therapy and hence were included in the study. Also, we understand that there is variation in the treatment regimen and HbF responses within the treatment group. However, it has been demonstrated in previous literature that HU induced changes like decreased adhesion of sickle erythrocytes to laminin and thrombospondin are observed 1–2 months after initiating therapy and these changes precede substantial increases in HbF [5]. Hence it is possible that HU-mediated proteomic changes observed in our study are independent of HbF levels and reflect immediate (after few months of treatment) as well as long-term responses to HU (after few years of treatment).
A limitation of our study is that the number of patients used is small (5 patients in each of treatment and control groups). Certainly our findings would need to be confirmed in a large study consisting of groups of patients that are comparable except for the HU treatment.
To date 751 proteins have been identified and their interactome network was published by Goodman et al [25]. It will be interesting to determine which HU dependent protein changes are at nodes that represent articulation points within the RBC interactome network. A more recent review with up-to-date coverage of the RBC proteome, as well as the proteome of other blood cells, has recently appeared (26).
Acknowledgments
We thank Dr. David Kakhniashvili and Dr. Anita Hryniewicz-Jankowska at the Department of Molecular and Cell Biology, The University of Texas at Dallas for their valuable discussion and assistance in the proteomic techniques. We thank Michael Henson and Leah Adix at Children’s Medical Center Dallas for procurement of specimens from patients.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ingram VM. A specific chemical difference between the globins of normal human and sickle-cell anemia haemoglobin. Nature. 1956;178:792–794. doi: 10.1038/178792a0. [DOI] [PubMed] [Google Scholar]
- 2.Rodgers GP. Overview of pathophysiology and rationale for treatment of sickle cell anemia. Semin Hematol. 1997;34:2–7. [PubMed] [Google Scholar]
- 3.Charache S, Terrin ML, Moore RD, Dover GJ, Barton FB, Eckert SV, MacMahon RP, Bonds DR. Effect of hydroxyurea on the painful crises in sickle cell anemia. Investigators of the Multicenter Study of Hydroxyurea in Sickle Cell Anemia. N Engl J Med. 1995;332:1317–1322. doi: 10.1056/NEJM199505183322001. [DOI] [PubMed] [Google Scholar]
- 4.Orringer EP, Blythe DS, Johnson A, Philips G, Jr, Dover GJ, Parket JC. Effects of hydroxyurea on hemoglobin F and water content in the red blood cells of dogs and of patients with sickle cell anemia. Blood. 1991;78:212–216. [PubMed] [Google Scholar]
- 5.Hillery CA, Du MC, Wang WC, Scott JP. Hydroxyurea therapy decreases the in vitro adhesion of sickle erythrocytes to thrombospondin and laminin. British J Haematol. 2000;109:322–327. doi: 10.1046/j.1365-2141.2000.02040.x. [DOI] [PubMed] [Google Scholar]
- 6.Ballas SK, Dover GJ, Charache S. Effect of hydroxyurea on rheological properties of sickle cell erythrocytes in vivo. Am J Hematol. 2001;67:151–156. doi: 10.1002/ajh.2830320206. [DOI] [PubMed] [Google Scholar]
- 7.Ghatpande SS, Choudhary PK, Quinn CT, Goodman SR. Pharmaco-Proteomic study of hydroxyurea-induced modifications in the sickle red blood cell membrane proteome. Exp Biol Med. 2008;233:1510–1517. doi: 10.3181/0805-S-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fibach E, Burke LP, Schechter AN, Noguchi CT, Rodgers GP. Hydroxyurea increases fetal hemoglobin in cultured erythroid cells derived from normal individuals and patients with sickle cell anemia or beta-thalassemia. Blood. 1993;81:1630–1635. [PubMed] [Google Scholar]
- 9.Hughes KMH, Kakhniashvili DG, Neelam S, Goodman SR. The isolation of reticulocyte-free human red blood cells. Exp Biol Med. 2007;232:1470–1476. doi: 10.3181/0706-RM-163. [DOI] [PubMed] [Google Scholar]
- 10.Kakhniashvili DG, Bulla LA, Jr, Goodman SR. The human erythrocyte proteome: Analysis by ion trap tandem mass spectrometry. Mol Cell Proteomics. 2004;3:501–509. doi: 10.1074/mcp.M300132-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Dudoit S, Shaffer JP, Boldrick JC. Multiple hypotheses testing in microarray experiments. Statistical Science. 2003;18:71–103. [Google Scholar]
- 12.Development Core Team. R Foundation for Statistical Computing. Vienna, Austria: 2007. R: A language and environment for statistical computing. URL http://www.R-project.org. [Google Scholar]
- 13.Charache S, Dover GJ, Moore RD, Eckert S, Ballas SK, Koshy M, Milner PF, Orringer EP, Phillips G, Jr, Platt OS, Thomas GH. Hydroxyurea: Effects on hemoglobin F production in patients with sickle cell anemia. Blood. 1992;79:2555–2565. [PubMed] [Google Scholar]
- 14.Willison KR, Horwich AI. Structure and function of chaperonins in Archaebacteria and eukaryotic cytosol. In: Ellis RJ, editor. The Chaperonins. San Diego: Academic Press; 1996. p. p107. [Google Scholar]
- 15.King SB. The nitric oxide producing reactions of hydroxyurea. Curr Med Chem. 2003;6:437–452. doi: 10.2174/0929867033368213. [DOI] [PubMed] [Google Scholar]
- 16.Sangerman Jose, Kakhniashvili D, Brown A, Shartava A, Goodman SR. Spectrin ubiquitination and oxidative stress: Potential roles in blood and neurological disorders. Cell Molec Biol. 2001;6:607–636. [PubMed] [Google Scholar]
- 17.Athanassiou G, Moutzouri A, Kourakli A, Zoumbos N. Effect of hydroxyurea on the deformability of the red blood cell membrane in patients with sickle cell disease. Clin Hemorheol Microcirc. 2006;35:291–295. [PubMed] [Google Scholar]
- 18.Bencsath FA, Shartava A, Monteiro CA, Goodman SR. Identification of the disulfide-linked peptide in irreversibly sickled beta-actin. Biochemistry. 1996;35:4403–4408. doi: 10.1021/bi960063n. [DOI] [PubMed] [Google Scholar]
- 19.Schwartz RS, Rybicki AC, Heath RH, Lubin BH. Protein 4.1 in sickle erythrocytes. Evidence for oxidative damage. J Biol Chem. 1987;262:15666–15672. [PubMed] [Google Scholar]
- 20.Shartava A, Monteiro CA, Bencsath FA, Schneidner K, Chait BT, Gussio R, Casoria-Scott LA, Shah AK, Heuerman CA, Goodman SR. A posttranslational modification of beta-actin contributes to the slow dissociation of the spectrin-protein 4.1-actin complex of irreversibly sickled cells. J Cell Biol. 1995;128:805–818. doi: 10.1083/jcb.128.5.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shartava A, Korn W, Shah AK, Goodman SR. Irreversibly sickled cell beta-actin: defective filament formation. Am J Hematol. 1997;55:97–103. doi: 10.1002/(sici)1096-8652(199706)55:2<97::aid-ajh8>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 22.Ruff P, Speicher DW, Husain-Chishti A. Molecular identification of a major palmitoylated erythrocyte membrane protein containing the src homology 3 motif. Proc Natl Acad Sci USA. 1991;88:6595–6599. doi: 10.1073/pnas.88.15.6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alloisio N, Dalla Venezia N, Rana A, Andrabi K, Texier P, Gilsanz F, Cartron JP, Delaunay J, Chishti AH. Evidence that red blood cell protein p55 may participate in the skeleton-membrane linkage that involves protein 4.1 and glycophorin C. Blood. 1993;82:1323–1327. [PubMed] [Google Scholar]
- 24.Marfatia SM, Lue RA, Branton D, Chishti AH. In vitro binding studies suggest a membrane-associated complex between erythroid p55, protein 4.1, and glycophorin C. J Biol Chem. 1994;269:8631–8634. [PubMed] [Google Scholar]
- 25.Goodman SR, Kurdia A, Ammann L, Kakhniashvili D, Daescu O. The human red blood cell proteome and interactome. Exp Biol Med. 2007;232:1391–1408. doi: 10.3181/0706-MR-156. [DOI] [PubMed] [Google Scholar]
- 26.Liumbruno G, D’Alessandro A, Grazzini G, Zolla L. Blood-related proteomics. J Proteomics . 2009 Jun 27; doi: 10.1016/j.jprot.2009.06.010. in press. [DOI] [PubMed] [Google Scholar]