Abstract
Purpose
To evaluate the expression of stem cell-related markers at the cellular level in human breast tumors of different subtypes and histologic stage.
Experimental design
We performed immunohistochemical analyses of 12 proteins [CD44, CD24, ALDH1, vimentin, osteonectin, EPCR, caveolin 1, connexin 43, cytokeratin 18 (CK18), MUC1, claudin 7, and GATA3] selected based on their differential expression in breast cancer cells with more differentiated and stem cell-like characteristics in 47 cases of invasive ductal carcinoma (IDC) only, 135 cases of IDC with ductal carcinoma in situ (DCIS), 35 cases of DCIS with microinvasion, and 58 cases of pure DCIS. We also analyzed 73 IDCs with adjacent DCIS to determine the differences in the expression of markers by histology within individual tumors. CD44+/CD24− and CD44−/CD24+ cells were detected using double immunohistochemistry.
Results
CD44 and EPCR expression was different among the four histologic groups and was lower in invasive compared to in situ tumors, especially in luminal A subtype. The expression of vimentin, osteonectin, connexin 43, ALDH1, CK18, GATA3, and MUC1 differed by tumor subtype in some histologic groups. ALDH1 positive cells were more frequent in basal-like and HER2+ than in luminal tumors. CD44+/CD24− cells were detected in 69 % of all tumors with 100% of the basal-like and 52% of HER2+ tumors having some of these cells.
Conclusions
Our findings suggest that in breast cancer the frequency of tumor cells positive for stem cell-like and more differentiated cell markers varies according to tumor subtype and histologic stage.
Keywords: breast cancer, tumor progression, DCIS, heterogeneity
INTRODUCTION
Breast cancer is not a single disease but a heterogeneous group of diseases with different pathologies, biological characteristics, and clinical behavior (1–4). Comprehensive gene expression profiling using DNA microarrays has identified five major molecular subtypes: luminal A, luminal B, HER2+, basal-like, and normal breast-like (2–4). Tumors in different subtypes follow different clinical courses and respond differently to treatment. In general, patients with basal-like tumors have worse prognosis whereas those with luminal A tumors have more favorable outcomes (3, 4). In addition to this inter-tumor heterogeneity, there is also a high degree of intra-tumor diversity that is already present in DCIS (5). Specifically, a single tumor at any given time can contain tumor cell populations with distinct molecular profiles and biological properties.
Currently prevailing models explaining intra-tumor heterogeneity include the clonal evolution and cancer stem cell hypotheses (6, 7). The clonal evolution model emphasizes diversity for heritable traits that in combination with continuous selection allows for the expansion of tumor cells with the most favorable characteristics (8). According to the cancer stem cell hypothesis, cancer stem cells, defined as a subset of tumor cells with stem cell-like features, have the capacity to self-renew and differentiate, giving rise to a heterogeneous tumor cell population (9–12). In breast cancer lin−/CD44+/CD24−/low cells were identified as candidate breast cancer stem cells based on xenotransplant assays in NOD/SCID mice (13). More recently, additional markers including aldehyde dehydrogenase (ALDH) activity (14), CD133 (15, 16), and ITGA6 (17, 18) have also been proposed as putative markers of mammary epithelial and breast cancer stem cells.
In a previous study, we investigated the genome-wide gene expression profiles of lin−/CD44+/CD24−/low (CD44+) and more differentiated luminal epithelial CD44−/CD24+ (CD24+) breast cancer cells and determined that these were consistent with the presumed identity of these cells (19). Specifically, CD44+ cells demonstrated a more mesenchymal stem cell-like profile enriched for genes involved in cell motility, proliferation, and angiogenesis whereas CD24+ cells highly expressed genes implicated in carbohydrate metabolism and RNA splicing. We also found that stem cell-like gene expression patterns in lymph node-negative primary invasive breast tumors correlated with shorter distant metastasis-free survival (19, 20). However, distant metastases were largely composed of more differentiated CD24+ breast cancer cells implying a phenotypic switch during metastatic colonization and tumor growth or clonal selection for CD24+ tumor cells in distant metastases.
In breast cancer cell lines, prior studies revealed an enrichment of CD44+/CD24− and CD44−/CD24+ phenotypes in basal-like and luminal subtypes, respectively (21, 22). Similarly, ALDH1 activity was only detected in basal-like breast cancer cell lines (23). In contrast, most studies analyzing primary human breast cancer samples have not revealed significant association between CD44 and CD24 expression and tumor subtypes (24–26). Only one recent study reported the enrichment of CD44+/CD24− cancer cells in basal-like invasive breast tumors (27). The expression of ALDH1 in invasive breast tumors was also correlated with poor clinical outcome (14), but the association of this with tumor subtypes was not reported.
In this study we investigated the expression of 12 proteins [CD44, CD24, ALDH1, vimentin, osteonectin, EPCR, caveolin 1, connexin 43, cytokeratin 18 (CK18), MUC1, claudin 7, and GATA3], selected based on our gene expression data (19) as differentially expressed in breast cancer cells with more differentiated luminal and stem cell-like characteristics, in 182 IDCs (135 with associated DCIS) and in 93 DCIS (35 with microinvasion) with the goal of correlating the expression of these 12 proteins with clinico-pathologic characteristics of the tumors including molecular subtype and histologic stage. We also analyzed in situ and invasive areas of 73 additional IDCs with adjacent DCIS to define progression-related changes within individual cases. CD44+ stem cell-like and CD24+ more differentiated luminal cancer cells were further defined using double immunohistochemistry.
MATERIALS AND METHODS
Tissue specimens
Three hundred ninety seven (397) cases of surgically resected ductal breast cancers including 289 IDCs with or without adjacent DCIS and 108 DCIS with or without microinvasion were collected in Seoul National University Bundang Hospital during 2003 to 2007. 73 IDCs with adjacent DCIS were evaluated using full tissue sections. Tissue microarrays (TMAs) were constructed from representative tissue column (2.0 mm in diameter for IDC; 4.0 mm in diameter for DCIS), of 216 invasive ductal carcinomas (with or without adjacent DCIS) and 108 DCIS (with or without microinvasion), as previously described (28). The TMAs were prepared with invasive tissue only from the cases of IDC with adjacent DCIS, but the cases of DCIS with microinvasion that were used in the TMAs might have included small numbers of IDC cells in addition to the DCIS cells. All patients except two (99.5%) were female, with a median age of 49 (range, 26–83 years). Clinico-pathologic information was obtained by reviewing pathology reports and hematoxylin and eosin-stained sections. The following histo-pathologic variables were determined in IDCs: tumor subtype, T stage, presence of cancer in ipsilateral axillary lymph nodes, Bloom-Richardson histologic grade, ER, PR and HER2 status, Ki-67, P53, and presence of adjacent DCIS. For DCIS cases with or without microinvasion, we recorded tumor size, nuclear grade, ER, PR and HER2 status, Ki-67 and P53 staining patterns, and presence of microinvasion. All cases were independently reviewed by two breast pathologists (SYP and HEL). This study was approved by the Institutional Review Board of Seoul National University Bundang Hospital (protocol # B-0809-061-301) and Dana-Farber Cancer Institute (protocol #98-229).
Immunohistochemical analyses
Four μ-thick, formalin-fixed, paraffin-embedded tissue sections were cut, dried, deparaffinized, and rehydrated following standard procedures. All sections were subjected to heat-induced antigen retrieval process in citrate (pH 6.0) or Tris-EDTA (pH 9.0) buffer or by protease treatment. Following staining optimization using positive and negative controls, 12 antibodies that provided satisfactory results together with standard prognostic biomarkers were used for the study (Supplementary Table S1). Immunohistochemical staining was performed using a Vectastain Elite avidin-biotin complex detection kit (Vector Laboratories) or DAKO-Envision detection kit (DAKO, Carpinteria, CA) and visualized with 3,3′-diaminobenzidine(Sigma- Aldrich, St. Louis, MO). In double immunohistochemical staining to detected CD44+/CD24− and CD44−/CD24+ cells, CD44 was visualized with Vectastain Elite avidin-biotin complex detection kit and diaminobenzidine as chromogen, whereas CD24 was detected with a polymer-linked alkaline phosphatase-conjugated secondary antibody (DAKO) and visualized with fast red. Sections were counterstained with Mayer’s hematoxylin.
Fluorescence in situ hybridization (FISH) assays for HER2 gene amplification
To determine HER2 gene amplification, the PathVysion (Vysis, Downers Grove, IL) assay was carried out and evaluated as previously described (29).
Definition of breast tumor subtypes
Breast tumor subtypes were defined according to Carey et al. with minor modifications (30). Subtype definitions were as follows: luminal A (ER+ and/or PR+, HER2−), luminal B (ER+ and/or PR+, HER2+), HER2+ (ER−, PR−, HER2+), basal-like (ER−, PR−, HER2−, basal cytokeratin+, and EGFR+/−). Immunostains for ER and PR were performed on full sections and cases with 10% or more positive staining were grouped as positive. EGFR and basal cytokeratins (cytokeratin 5/6, 14, and 17) were evaluated using TMAs or full sections (for 73 IDCs with adjacent DCIS), and cases with any positive membranous staining were grouped as positive using previously established and published criteria (31). HER2 positivity was determined based on FISH results as described (29).
Immunohistochemical scoring
After omitting 34 cases with uninterpretable IHC results, a total of 193 IDCs (139 with DCIS) and 97 DCIS (37 with microinvasion) were informative (interpretable staining) for all antibodies in TMA and used for the analyses. For subtype analyses, a total of 182 IDCs (135 also having DCIS) and 93 cases of DCIS (35 also having microinvasion) were used after omitting 15 cases with unknown tumor subtypes (Table 1). Staining of markers was scored in 10% increments (i.e., 0%, 10%, 20%, etc.) For statistical analysis we coded 12 proteins analyzed into binary (two-valued) covariates (Supplementary Table S2). The definition of “positive” stain for each marker was made before any comparison of groups and depended on the distribution of scores for that marker; in each case our aim was to choose a cutoff for positivity so that at least 10% of cases were categorized as “positive” and at least 10% of cases were categorized as “negative” (because there would be almost no power in this study to detect differences between groups if fewer than 10% of cases were in either category). The cut-off of > 10% satisfied this condition for CD24, CD44, GATA3, connexin 43, claudin7, and ALDH. Osteonectin, vimentin, caveolin1, and EPCR all had fewer than 10% of cases with staining > 10%, so for these markers the cut-off used was >0%. On the other hand, for MUC1 and CK18 the mode of staining was at 90% or 100% (unlike all the other markers that had modes of 0% or 10%), so for these two makers a cut-off of >80% was used and referred to as “highly positive”. Since many investigators use a uniform staining cut-off for all markers, we also redid all analyses using a cut-off of >10% for all markers; the text mentions when this uniform cut-off resulted in a difference in which comparisons were significant. Two additional binary variables created by dividing the cases as having more than the median percentage of stained cells (or not) were defined for two markers (CD44 and GATA3) that appeared to have very spread out distributions (CD44 median was 20% in the two IDC groups combined and 50% in the two DCIS groups combined; GATA3 median was 70% in the two IDC groups combined and 50% in the two DCIS groups combined).
Table 1. Distributions of the tumors in the four histologic groups.
Number of patients (and raw percentage) in each of the histologic group who had each of these tumor subtypes.
| Subtype | Luminal A | Luminal B | HER2+ | Basal-like | Total | ||||
|---|---|---|---|---|---|---|---|---|---|
| IDC alone | 35 | 75% | 1 | 2% | 5 | 11% | 6 | 13% | 47 |
| IDC with DCIS | 85 | 63% | 16 | 12% | 18 | 13% | 16 | 12% | 135 |
| DCIS with microinvasion | 11 | 31% | 6 | 17% | 14 | 40% | 4 | 11% | 35 |
| Pure DCIS | 41 | 71% | 7 | 12% | 9 | 16% | 1 | 2% | 58 |
| Total | 172 | 63% | 30 | 11% | 46 | 17% | 27 | 10% | 275 |
Statistical analysis
Statistical analysis was performed using SAS and StatXact for the analysis. Fisher exact tests were used when comparing the binary categories of expression of proteins between groups. For 73 IDCs with adjacent DCIS, Wilcoxon sign rank tests were used to see if the (paired) difference between the expression levels in the invasive and DCIS components was significantly different from random. Differences of continuous variables between groups were evaluated using Wilcoxon rank sum tests. Significance levels were adjusted for multiple comparisons using Holm’s method. All p-values reported are two-sided.
RESULTS
Histologic groups and distribution of tumor subtypes
We categorized the tumors into four histologic groups: IDC alone, IDC with DCIS, DCIS with microinvasion, and pure DCIS (Table 1). The distribution of the four tumor subtypes differed significantly (p=0.0001) among the four histologic groups, with luminal A tumors being the least common in the DCIS with microinvasion, luminal B tumors being the least common in the IDC alone, HER2+ tumors being the most common in the DCIS with microinvasion, and basal-like tumors being the least common in the pure DCIS groups. The distributions of the four tumor subtypes were not significantly different in the two IDC groups (IDC alone and IDC with DCIS, p=0.81), but were significantly different in the two DCIS groups (pure DCIS and DCIS with microinvasion, p=0.0006). However, the DCIS with microinvasion group could be considered to have both invasion and DCIS, and might be expected to share some characteristics with the two IDC groups. Therefore, we opted to conduct the analysis using four histologic groups.
Expression of CD44, CD24, and ALDH1 in the four histologic groups
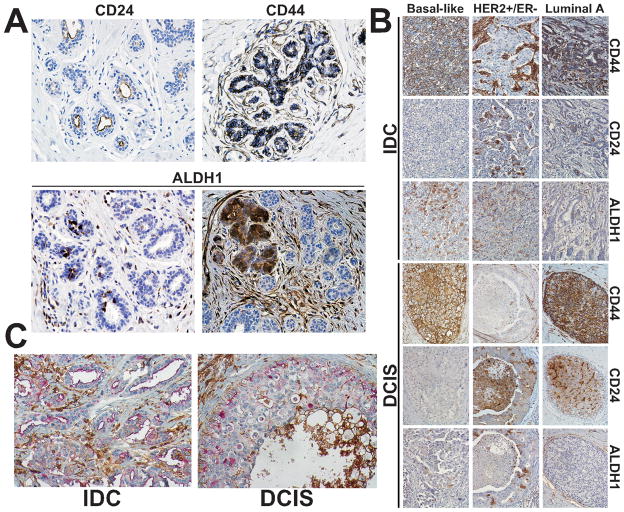
We first analyzed the expression of CD44, CD24, and ALDH1-markers previously shown to be associated with more differentiated luminal epithelial (CD24) and stem cell-like (CD44 and ALDH1) characteristics and used for the purification of these respective cells (14, 19) in 20 normal breast tissue samples to define their normal cellular distribution pattern (Fig. 1A). In normal breast tissue, CD44 was localized to the cell membranes of basal/myoepithelial and a subset of luminal epithelial cells. CD24 expression was occasionally found on the apical membranes of luminal cells. ALDH1 was expressed heterogeneously in both luminal and basal cells and in some lobules all cells were ALDH1+ (Fig. 1A).
Figure 1. Immunohistochemical analyses of CD44, CD24, and ALDH1 in breast cancer.
A, Immunohistochemical staining patterns in normal breast tissue. CD24 and CD44 displayed uniform luminal and basal cell-specific pattern, respectively, whereas ALDH1 expression varied depending on the area of the tissue. B, Representative examples of immunohistochemical analyses of invasive ductal carcinoma (IDC) and ductal carcinoma in situ (DCIS) according to tumor subtype using the indicated antibodies. In luminal A subtype tumor cells positive for CD44 and CD24 (apical membranous pattern) can be seen whereas ALDH1 was not detected. HER2+ tumor is negative for CD44, and positive for CD24 (membrano-cytoplasmic pattern) and ALDH1. In basal-like subtype tumor cells are positive for CD44 and ALDH1, but negative for CD24. C, Representative examples of double immunohistochemistry for CD44 (brown) and CD24 (red) in IDC and in DCIS.
Next, we investigated the expression of these markers in the tumor samples (Fig. 1B; Supplementary Table S3 and S4). Strong membranous CD44 staining was detected in the majority of tumors, although a few cases displayed weak, incomplete membranous staining pattern. CD44 was expressed in 57% of IDC only cases, in 59% of IDCs with DCIS, in 62% of DCIS with microinvasion, and in 85% of pure DCIS (p=0.002). CD24 was expressed in 28% of IDC only tumors, in 38% of IDCs with DCIS, 59% of DCIS with microinvasion, and 62% of pure DCIS (p=0.02). Two different CD24 staining patterns were observed that were almost mutually exclusive: apical membranous (reminiscent of expression pattern observed in normal breast tissue) and circumferential membrano-cytoplasmic. Of the total of 117 cases that were CD24 positive, 35 (30%) showed apical staining and the remaining 82 (70%) showed cytoplasmic staining, but this differed considerably by histologic group. Among the 15, 53, 22, and 27 CD24-positive cases in the four histologic groups, 47%, 26%, 9%, and 44% of the IDC alone, IDC with DCIS, DCIS with microinvasion, and pure DCIS had apical staining (p=0.02). ALDH1 was expressed in few cases (9%, 6%, 3%, and 3% in each of the four histologic groups, p=0.56).
To determine if the expression of these markers may be associated with some characteristics of the tumors, we compared the available clinical/pathologic features of positive and negative cases (Supplementary Table S3). After adjusting for multiple comparisons, the following significant associations were found. In IDC alone cases, a tumor with histologic grade III was more likely to be CD44 positive than a tumor with histologic grade I or II (82% vs. 41%. P=0.005), and an ER negative (ER−) tumor was more likely to be ALDH1 positive than an ER positive (ER+) tumor (24% vs. 0%, p=0.006).
Combined expression of CD44 and CD24 according to tumor subtype and histologic stage
Next, we analyzed the combined expression of CD44 and CD24 in the same tumor according to histologic groups and tumor subtypes (Table 2). Considering only histologic group (the column labeled “Total”), of the four possible combined expression patterns (positivity for both markers, CD44 only, CD24 only, and lack of both CD24 and CD44), the “CD44 only” pattern was the most common one in each histologic stages. However, the four histologic groups did differ significantly (p=0.0001) in pattern, with pure DCIS having the highest percents of CD44 only and of the combination (of CD44 and CD24), with DCIS with microinvasion having the highest percent of CD24 only, and with IDC alone having the highest percent of neither marker. Considering histologic group within each of the four tumor subtypes, the difference in pattern was significant only in the luminal A subtype (which had the largest number of cases).
Table 2. CD24 and CD44 expression pattern defined based on immunohistochemical analyses of adjacent sections for the two markers according to tumor subtype and histologic groups.
Numbers in parentheses indicate column percentage. P-values were calculated using Fisher’s exact tests. NS denotes not significant (crude, unadjusted p>0.05). NSAA denotes not significant after adjustment (using Holm’s method).
| Tumor Subtypes | ||||||
|---|---|---|---|---|---|---|
| Histologic groups | Pattern | Luminal A | Luminal B | HER2+ | Basal-like | Total |
| P = 0.001 | P = NS | P = NS | P = NS | P = 0.0001 | ||
| IDC alone1 P = NS | CD44&CD24 | 7 (20) | 0 (0) | 0 (0) | 0 (0) | 7 (15) |
| CD44 only | 9 (26) | 1 (100) | 3 (60) | 5 (83) | 18 (38) | |
| CD24 only | 4 (11) | 0 (0) | 1 (20) | 0 (0) | 5 (11) | |
| Lack of both | 15 (43) | 0 (0) | 1 (20) | 1 (17) | 17 (36) | |
| Total | 35 (100) | 1 (100) | 5 (100) | 6 (100) | 47 (100) | |
| IDC with DCIS P = NS | CD44&CD24 | 24 (28) | 3 (19) | 4 (22) | 2 (13) | 33 (24) |
| CD44 only | 30 (35) | 6 (37) | 2 (11) | 10 (63) | 48 (36) | |
| CD24 only | 10 (12) | 4 (25) | 5 (28) | 0 (0) | 19 (14) | |
| Lack of both | 21 (25) | 3 (19) | 7 (39) | 4 (25) | 35 (26) | |
| Total | 85 (100) | 16 (100) | 18 (100) | 16 (100) | 135 (100) | |
| CD44&CD24 | 4 (36) | 3 (50) | 3 (21) | 0 (0) | 10 (29) | |
| DCIS with microinvasion P = NSAA | CD44 only | 5 (46) | 2 (33) | 1 (7) | 4 (100) | 12 (34) |
| CD24 only | 2 (18) | 1 (17) | 7 (50) | 0 (0) | 10(29) | |
| Lack of both | 0 (0) | 0 (0) | 3 (21) | 0 (0) | 3 (9) | |
| Total | 11 (100) | 6 (100) | 14 (100) | 4 (100) | 35 (100) | |
| Pure DCIS2 P = NS | CD44&CD24 | 15 (37) | 3 (43) | 3 (33) | 1 (100) | 22 (38) |
| CD44 only | 21 (51) | 3 (43) | 4 (44) | 0 (0) | 28 (48) | |
| CD24 only | 4 (10) | 1 (14) | 0 (0) | 0 (0) | 5 (9) | |
| Lack of both | 1 (2) | 0 (0) | 2 (22) | 0 (0) | 3 (5) | |
| Total | 41 (100) | 7 (100) | 9 (100) | 1 (100) | 58 (100) | |
| Total P = 0.0006 | CD44&CD24 | 50 (29) | 9 (30) | 10 (22) | 3 (11) | 72 (26) |
| CD44 only | 65 (38) | 12 (40) | 10 (22) | 19 (70) | 106 (39) | |
| CD24 only | 20 (12) | 6 (20) | 13 (28) | 0 (0) | 39 (14) | |
| Lack of both | 37 (22) | 3 (10) | 13 (28) | 5 (19) | 58 (21) | |
| Total | 172 (100) | 30 (100) | 46 (100) | 27 (100) | 275 (100) | |
Luminal B subtype was omitted when calculating the p-values, except for Total.
Basal-like subtype was omitted when calculating the p-values, except for Total.
Considering only tumor subtypes, the “CD44 only” pattern was the most common one in each subtype except for HER2+, in which “CD24 only” and lack of both were more common (Table 2). The four tumor subtypes differed significantly (p=0.0006) in pattern, with basal-like tumors having the lowest percent of “CD24 only” and of the combination, with HER2 tumors having the lowest percent of “CD44 only”, and with luminal B tumors having the lowest percent of tumors with neither marker. Considering tumor subtypes within each of the four histologic groups, none were significant (although the subtypes were closest to significantly different in pattern for DCIS with microinvasion).
To investigate the combined expression of CD24 and CD44 within individual tumor cells in further detail, we performed double immunohistochemical analysis (Fig. 1C and Table 3). CD44+/CD24− stem cell-like cells were detected in 64% of IDC only, 64% IDC with DCIS, 74% of DCIS with microinvasion, and 83% of pure DCIS cases. When analyzing by tumor subtype 69% of luminal A, 70% of luminal B, 52% of HER2, and 100% of basal-like tumors contained some CD44+/CD24− cells. CD44−/CD24+ more differentiated luminal epithelial cells were detected in 19% of IDC only, 33% of IDC with DCIS, 63% of DCIS with microinvasion, and 37% of pure DCIS cases. Within subtypes CD44−/CD24+ cells were more common in luminal and HER2+ tumors compared to basal-like cases (Table 3).
Table 3. Presence of CD44+/CD24− and CD44−/CD24+ cells defined by double immunohistochemistry according to tumor subtypes and histologic groups.
Numbers in parentheses indicate column percentage. P-values were calculated using Fisher’s exact tests. NS denotes not significant (crude, unadjusted p>0.05). NSAA denotes not significant after adjustment (using Holm’s method).
| Cell Staining | Tumor Subtypes | Total | |||||
|---|---|---|---|---|---|---|---|
| Luminal A | Luminal B | HER2+ | Basal-like | ||||
| CD44+/CD24− cells | CD44−/CD24+ cells | P = 0.0009 | P = NS | P = NS | P = NS | P < 0.0001 | |
| IDC alone1 P = NS | Positive | Positive | 1 (3) | 0 (0) | 0 (0) | 1 (17) | 2 (4) |
| Positive | Negative | 19 (54) | 1 (100) | 3 (60) | 5 (83) | 28 (60) | |
| Negative | Positive | 6 (17) | 0 (0) | 1 (20) | 0 (0) | 7 (15) | |
| Negative | Negative | 9 (26) | 0 (0) | 1 (20) | 0 (0) | 10 (21) | |
| Total | 35 (100) | 1 (100) | 5 (100) | 6 (100) | 47 (100) | ||
| IDC with DCIS P < 0.0001 | Positive | Positive | 10 (12) | 2 (13) | 2 (11) | 1 (6) | 15 (11) |
| Positive | Negative | 43 (51) | 8 (50) | 5 (28) | 15 (94) | 71 (53) | |
| Negative | Positive | 16 (19) | 6 (38) | 8 (44) | 0 (0) | 30 (22) | |
| Negative | Negative | 16 (19) | 0 (0) | 3 (17) | 0 (0) | 19 (14) | |
| Total | 85 (100) | 16 (100) | 18 (100) | 16 (100) | 135 (100) | ||
| DCIS with microinvasion P = NS | Positive | Positive | 6 (55) | 3 (50) | 3 (21) | 1 (25) | 13 (37) |
| Positive | Negative | 4 (36) | 2 (33) | 4 (29) | 3 (75) | 13 (37) | |
| Negative | Positive | 1 (9) | 1 (17) | 7 (50) | 0 (0) | 9 (26) | |
| Negative | Negative | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| Total | 11 (100) | 6 (100) | 14 (100) | 4 (100) | 35 (100) | ||
| Pure DCIS2 P = NS | Positive | Positive | 12 (29) | 2 (29) | 2 (22) | 0 (0) | 16 (28) |
| Positive | Negative | 23 (56) | 3 (43) | 5 (56) | 1 (100) | 32 (55) | |
| Negative | Positive | 4 (10) | 0 (0) | 1 (11) | 0 (0) | 5 (9) | |
| Negative | Negative | 2 (5) | 2 (29) | 1 (11) | 0 (0) | 5 (9) | |
| Total | 41 (100) | 7 (100) | 9 (100) | 1 (100) | 58 (100) | ||
| Total P = 0.0006 | Positive | Positive | 29 (17) | 7 (23) | 7 (15) | 3 (11) | 46 (17) |
| Positive | Negative | 89 (52) | 14 (47) | 17 (37) | 24 (89) | 144 (52) | |
| Negative | Positive | 27 (16) | 7 (23) | 17 (37) | 0 (0) | 51 (19) | |
| Negative | Negative | 27 (16) | 2 (7) | 5 (11) | 0 (0) | 34 (12) | |
| Total | 172 (100) | 30 (100) | 46 (100) | 27 (100) | 275 (100) | ||
Luminal B subtype was omitted when calculating the p-values, except for Total.
Basal-like subtype was omitted when calculating the p-values, except for Total.
We analyzed associations between tumor characteristics and CD24/CD44 expression patterns in two different ways. Specifically, we determined whether the entire tumor has both CD44 and CD24 staining, but possibly in different cells (maybe different areas of those tumors) based on staining for one marker at a time (Table 2) and we also analyzed CD24 and CD44 expression within individual cells based on double immunohistochemistry (Table 3). In both types of analyses we established four different staining patterns based on positivity/negativity of the tumors for these two markers and the distribution of these four possible patterns were analyzed according to tumor subtype and histologic stage. The distributions of the four types of patterns differed significantly by histologic group (p<0.001, Table 2). Considering histologic group within each of the four tumor subtypes, the pattern of distribution was significantly different by histologic type only within luminal A (the tumor type with the most cases). Overall, the distribution pattern of the four cell types also differed significantly by tumor subtype (p=0.0006, Table 3). Considering tumor subtype within each of the histologic groups, the pattern was significantly different only for IDC with DCIS (the histologic group with the most cases).
Expression of genes differentially expressed between CD44+ and CD24+ breast cancer cells according to histologic stage and tumor subtype
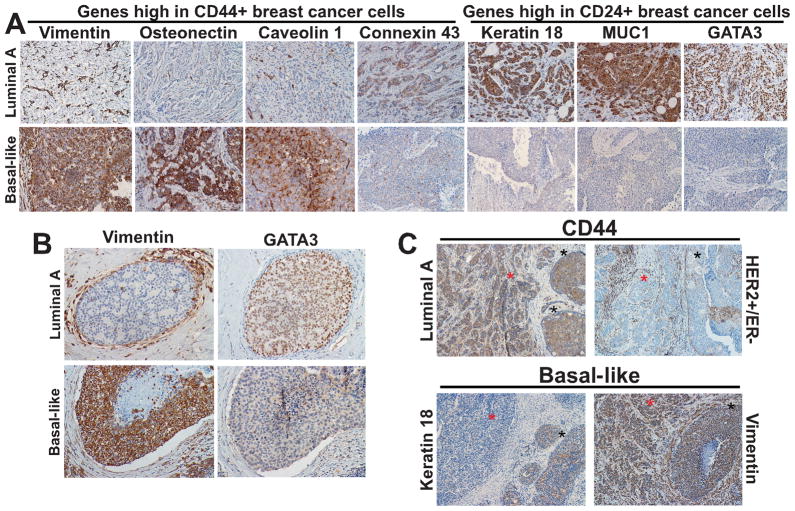
To further investigate inter-tumor cellular heterogeneity for the expression of stem cell-related markers in breast cancer and to correlate this with tumor subtypes, we selected additional genes that we previously identified as differentially expressed between CD44+ and CD24+ breast cancer cells (19) and analyzed their expression by immunohistochemistry in IDCs and in DCIS (Fig. 2A,B, Table 4, and Supplementary Table S4). The proteins analyzed included: vimentin, osteonectin, EPCR, connexin 43, and caveolin 1 (highly expressed in CD44+ breast cancer cells) as well as CD44 itself and ALDH1; and MUC1, CK18, claudin 7, and GATA3 (highly expressed in more differentiated CD24+ luminal cells) as well as CD24 (any type of staining). Expression of each of these markers was represented by binary (two-valued) variables (Supplementary Table S2).
Figure 2. Immunohistochemical analyses of genes differentially expressed between CD44+ and CD24+ breast cancer cells.
A, Representative examples of immunohistochemical staining patterns observed in invasive breast tumors using antibodies against the indicated proteins. A subset of markers that were highly expressed in CD44+ compared to CD24+ breast cancer cells based on our prior study are expressed only in basal-like tumors (vimentin, osteonectin, and caveolin 1), whereas Connexin 43 is expressed both in luminal A and basal-like subtypes. Markers more abundantly expressed in CD24+ compared to CD44+ breast cancer cells (Keratin 18, MUC1, and GATA3) are expressed in luminal A tumors. B, Representative examples of immunohistochemical staining patterns observed in DCIS using antibodies against the indicated proteins. Vimentin is expressed in basal-like but not in luminal A DCIS. Strong vimentin staining is detected in stromal cells in both tumor subtypes. In contrast, GATA3 is expressed in luminal A but not in basal-like DCIS. C, Immunohistochemical analyses of the expression of markers between invasive (red star) and in situ (black star) components of the same tumor. CD44 is expressed in both invasive and in situ components of luminal A subtype. HER2+ tumors are negative for CD44, only stromal staining is detected. In basal-like IDC keratin 18 is expressed only in the in situ component whereas vimentin is expressed in both invasive and in situ areas.
Table 4. Comparison of the expression of 12 proteins between tumor subtypes in each of the four histologic groups.
Numbers in parentheses indicate percentage of patients who were coded as positive for the binary marker variables (column percentage). P-values were calculated using Fisher’s exact tests. Significance levels were adjusted using Holm’s method. NS denotes not significant (crude, unadjusted p>0.05); NSAA denotes not significant after adjustment (using Holm’s method). Luminal B subtype was omitted in IDC alone group because there was only 1 case in this group. Basal-like subtype was omitted in pure DCIS group because there was only 1 case in this group. All DCIS tumors were negative for Caveolin1.
| IDC | IDC alone | IDC with DCIS | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Luminal A (n=35) | Basal-like (n=6) | HER2+ (N=5) | p-value | Luminal A (n=85) | Basal-like (n=16) | HER2+ (n=18) | Luminal B (n=16) | p-value | ||
| CD44+ cell genes | Vimentin | 7 (20) | 6 (100) | 2 (40) | 0.0003 | 10 (12) | 13 (81) | 2 (11) | 1 (6) | <0.0001 |
| Osteonectin | 0 (0) | 3 (50) | 1 (20) | 0.001 | 6 (7) | 4 (25) | 0 (0) | 1 (6) | NS | |
| Connexin 43 | 16 (46) | 2 (33) | 1 (20) | NS | 35 (41) | 7 (44) | 0 (0) | 1 (6) | 0.0001 | |
| ALDH1 | 0 (0) | 2 (33) | 0 (0) | NSAA | 1 (1) | 3 (19) | 4 (22) | 0 (0) | 0.001 | |
| Caveolin1 | 1 (3) | 3 (50) | 0 (0) | NSAA | 0 (0) | 2 (12) | 0 (0) | 0 (0) | NSAA | |
| CD44 | 16 (46) | 5 (83) | 3 (60) | NS | 54 (64) | 12 (75) | 6 (33) | 9 (56) | NS | |
| CD44 median | 14 (40) | 5 (83) | 2 (40) | NS | 44 (52) | 11 (69) | 5 (28) | 6 (38) | NS | |
| EPCR | 1 (3) | 0 (0) | 1 (20) | NS | 2 (2) | 1 (6) | 0 (0) | 1 (6) | NS | |
| CD24+ cell genes | High Pos CK18 | 22 (63) | 0(0) | 0 (0) | 0.0002 | 49 (58) | 1 (6) | 9 (50) | 6 (38) | 0.0008 |
| GATA3 | 29 (83) | 0 (0) | 1 (20) | <0.0001 | 78 (92) | 1 (6) | 6 (33) | 11 (69) | <0.0001 | |
| GATA3 median | 21 (60) | 0(0) | 1 (20) | 0.004 | 56 (66) | 0 (0) | 1 (6) | 6 (38) | <0.0001 | |
| High Pos MUC1 | 23 (66) | 0 (0) | 0 (0) | 0.0002 | 34 (40) | 2 (13) | 6 (33) | 7 (44) | NS | |
| Claudin7 | 9 (26) | 2 (33) | 0 (0) | NS | 28 (33) | 2 (13) | 5 (28) | 6 (38) | NS | |
| CD24 | 11 (31) | 0 (0) | 1 (20) | NS | 34 (40) | 2 (13) | 9 (50) | 7 (44) | NS | |
| DCIS | DCIS with microinvasion | Pure DCIS | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Luminal A (n=11) | Luminal B (n=6) | HER2+ (n=14) | Basal-like (n=4) | p-value | Luminal A (n=41) | Luminal B (n=7) | HER2+ (n=9) | p-value | ||
| CD44+ cell genes | Vimentin | 2 (18) | 2 (33) | 1 (7) | 3 (75) | NSAA | 4 (10) | 1 (14) | 4 (44) | NSAA |
| Osteonectin | 0 (0) | 2 (33) | 1 (7) | 1 (25) | NS | 1 (2) | 1 (14) | 0 (0) | NS | |
| Connexin 43 | 1 (9) | 0 (0) | 0 (0) | 1 (25) | NS | 21 (51) | 1 (14) | 0 (0) | 0.003 | |
| ALDH1 | 0 (0) | 0 (0) | 1 (7) | 0 (0) | NS | 0 (0) | 1 (14) | 1 (11) | NS | |
| CD44 | 9 (82) | 5 (83) | 4 (29) | 4 (100) | NSAA | 36 (88) | 6 (86) | 7 (78) | NS | |
| CD44 median | 4 (36) | 4 (67) | 4 (29) | 3 (75) | NS | 24 (59) | 3 (43) | 4 (44) | NS | |
| EPCR | 4 (36) | 0 (0) | 0 (0) | 0 (0) | NSAA | 4 (10) | 0 (0) | 0 (0) | NS | |
| CD24+ cell genes | High PosCK18 | 4 (36) | 3 (50) | 11 (79) | 0 (0) | NSAA | 32 (78) | 5 (71) | 4 (44) | NS |
| GATA3 | 9 (82) | 3 (50) | 5 (36) | 0 (0) | NSAA | 38 (93) | 5 (71) | 0 (0) | <0.0001 | |
| GATA3 median | 6 (55) | 0 (0) | 0 (0) | 0 (0) | 0.0008 | 29 (71) | 3 (43) | 0 (0) | 0.0001 | |
| High PosMUC1 | 7 (64) | 3 (50) | 5 (36) | 0 (0) | NS | 24 (59) | 3 (43) | 4 (44) | NS | |
| Claudin7 | 5 (45) | 1 (17) | 1 (7) | 1 (25) | NS | 22 (54) | 0 (0) | 2 (22) | NSAA | |
| CD24 | 6 (55) | 4 (67) | 10 (71) | 0 (0) | NS | 19 (46) | 4 (57) | 3 (33) | NS | |
Overall, CD44 (P=0.002) and connexin 43 (P=0.003) staining differed significantly among the four histologic groups; CD44 was higher in pure DCIS tumors whereas connexin 43 was lower in DCIS with microinvasion compared to other histologic groups (Supplementary Table S4). When this analysis was done separately in each of the four tumor subtypes, only the luminal A subtype had any significant associations of positivity of markers and histologic groups. In the luminal A tumor, there was a significant difference in CD44 (p=0.0005) and EPCR (p=0.001) expression; both of them were higher in pure DCIS or DCIS with microinvasion than in IDC tumors or IDC with DCIS tumors (Supplementary Table S4). When the definition of positivity for all markers used a cut-off of > 10% (instead of the cut-offs in Table S2 and the methods section), then overall three other markers would be significantly different by histologic group: vimentin (p=0.003), CK18 (p=0.0001), and MUC1 (p=0.0002). In the luminal A group, EPCR was not significant after correction for multiple comparisons if the > 10% cut-off was used.
We also explored whether the expression of these proteins differed by tumor subtypes within each of the four histologic groups (Table 4). After adjusting for multiple comparisons (separately within each histologic group), vimentin staining differed significantly by tumor subtype in the IDC alone group (p=0.0003) and in the IDC with DCIS group (p<0.0001); for both of these histologic groups basal-like tumors were more likely to be positive than the other subtypes (Table 4). Osteonectin differed significantly by tumor subtype in the IDC alone group (50% of basal-like tumors were positive vs. 0–20% of the other two subtypes). Connexin 43 differed significantly by tumor subtype in the IDC with DCIS (p=0.0001) and pure DCIS (p=0.003) subgroups, with a larger percent of luminal A tumors being positive in the IDC with DCIS group (41%) and pure DCIS group (51%), and a larger percent of the basal-like tumors being positive in the IDC with DCIS group (44%) than the percent that were positive in luminal B and HER2+ tumors. ALDH1 staining differed significantly (p=0.001) according to subtype in the IDC with DCIS group (19% of basal-like, 22% of HER2+, 1% of luminal A, and 6% of luminal B were positive for ALDH1). Highly-positive CK18 differed significantly in the IDC alone and the IDC with DCIS groups (p=0.0002 and p=0.0008, respectively), but there seems to be a difference in the association in these two groups. In both groups, luminal A tumors were very likely to have a highly-positive stain for CK18 (63% and 58% of the cases) and basal-like tumors were very unlikely to have a highly-positive stain (0% and 6%), but in IDC alone cases the HER2+ tumors were also unlikely to be highly-positive (0%) whereas in the IDC with DCIS cases the HER2+ tumors were likely to be highly-positive (50%). GATA3 differed significantly by tumor subtype in three of the histologic groups (IDC alone, IDC with DCIS, and pure DCIS, each p<0.0001). In each case, luminal A tumors were very likely to be positive for GATA3; luminal B tumors were very likely to be positive, HER2+ tumors had a lower percent of positive cases; and basal-like tumors were very unlikely to be positive (Table 4). All four histologic groups had significant tumor type differences in GATA3 when the IHC scoring was split at the median staining level (all p values ≤ 0.004). For this GATA3 variable, luminal A cases were very likely to have staining above the median (55% to 71%); in two of the groups, luminal B cases were moderately likely to have staining above the median (38% and 43% in IDC with DCIS and in pure DCIS, but 0% in DCIS with microinvasion); HER2+ cases were less likely to have staining above the median (20%, 38%, 0%, and 0%); and basal-like cases never had staining above the median (Table 4). The three tumor subtypes available in the IDC alone group differed significantly in percentage with highly-positive MUC1 staining (p=0.0002), with 66% of the luminal A cases being highly positive staining and none of the basal-like tumors or HER2+ tumors being highly positive. If in this group MUC1 was categorized as positive if the staining was > 10%, then the three subtypes would have positive staining for 100% of luminal A and HER2+ cases and 83% for basal-like cases, and this was not significant.
Gene expression patterns of invasive and in situ components of the same tumor
To determine if the differences seen in the expression of some proteins between IDCs and DCIS may reflect a potential divergence in expression associated with in situ to invasive breast carcinoma progression, we examined the expression of selected markers in tumors with invasive and in situ areas present on the same slides. Markers to be included in this analysis were selected based on the trend of their expression pattern when all IDC and all DCIS were combined in each of the tumor subtypes. Thus, CD44 was analyzed in 23 luminal A tumors, CD44 and CK18 in 25 HER2+ cases, and ALDH1, caveolin 1, vimentin, and CK18 in 25 basal-like tumors (Supplementary Table S5). In luminal A tumors CD44 had significantly higher expression levels (p=0.01) in the in situ than in the invasive components (48% of patients had more expression in the in situ component with a median difference in expression scores of 0.40; 17% had more expression in the invasive component with a median difference in expression scores of 0.10; and 35% had the same expression code for both components). This pattern was not seen in HER2+ tumors, in which CD44 expression was higher in the invasive component for 32% of patients and in the in situ component for 28% of patients. CK18 in basal-like tumors had significantly higher expression levels (p=0.005) in the in situ compared to the invasive components (40% of patients had more expression in the in situ component with a median difference of 0.275; 8% had more expression in the invasive component with a median difference of 0.075; and 52% had the same expression code for both components). This pattern was not seen in HER2+ tumors, in which CK18 expression was higher in the invasive component in 12% and higher in the in situ component in 24% of cases (with median differences in both cases of 0.20). In contrast, in basal-like tumors the expression of ALDH 1 and caveolin 1 were highly concordant in the invasive and in situ areas (80% of patients had the same expression score) and no tests were done (because the power is zero for a Wilcoxon sign rank test based on 5 non-zero differences).
DISCUSSION
Breast tumors display heterogeneity both among tumors in different individuals (e.g., inter-tumor heterogeneity) (3, 4) and in tumor cell subpopulations within tumors (e.g., intra-tumor heterogeneity) (5). The molecular mechanisms underlying inter-tumor heterogeneity are poorly defined; distinct cell-of-origin and tumor subtype-specific transforming events are potential explanations (1, 32). Intra-tumor heterogeneity on the other hand can be explained by three potential mechanisms which are not independent of each other: (1) genetic diversity and genomic instability combined with clonal selection, (2) differences in cellular phenotypes including stem cell-like and more differentiated characteristics, and (3) epigenetic diversity and plasticity (1, 12, 32, 33). We and others have been investigating the relative contribution of all three mechanisms to intra-tumor heterogeneity in breast cancer and the relevance of these to tumor progression and therapeutic response. We previously described the comprehensive gene expression, genetic, and DNA methylation profiles of CD44+ stem cell-like and CD24+ more differentiated luminal epithelial breast cancer cells and found cell type-specific differences for all molecular characteristics analyzed (19, 20). The relative frequency of CD44+ and CD24+ cells in breast tumors (defined based on the presence of cell type-specific gene expression signatures within tumors) was correlated with distant metastasis-free survival (19, 20). Interestingly, CD44+ cell-signature in primary invasive tumors was associated with higher risk of distant metastasis but distant metastases were enriched for more luminal epithelial CD24+ cells implying a phenotypic switch during tumor progression or clonal selection for cells with CD24+ phenotype (19, 20). Correlating with this, recent studies described that epithelial-to-mesenchymal transition (EMT) generates cells with CD44+ stem cell-like characteristics (34), whereas in distant metastases a reversion of this process mesenchymal-to-epithelial transition (MET) have also been reported in bladder (35) and colon (36) carcinomas. However, because distant metastases are frequently detected as recurrences following chemotherapeutic treatment, these observations may not necessarily reflect the natural history of the disease and further studies are needed to define the potential roles of EMT and MET in tumor progression. To investigate if phenotypic divergence for stem cell-like and more differentiated luminal markers might also be observed between in situ and invasive breast carcinomas in patients prior to systemic treatment, we analyzed the expression of a panel of markers previously associated with CD44+ stem cell-like and more differentiated CD24+ luminal cell characteristics in breast cancer.
We analyzed the expression of each of the markers in a series of IDCs and DCIS in relation to clinicopathologic features focusing especially on breast tumor subtype. Confirming the results of recent reports (21, 22, 27) the frequency of CD44+/CD24− cells defined based on double immunohistochemisty was the highest in basal-like tumors compared to the other three subtypes. High-grade IDC only tumors were also more likely to be CD44 positive than lower grade one. However, the expression of CD44 was lower in IDCs and microinvasive DCIS compared to pure DCIS suggesting a decrease in the number of CD44+ cells as tumors progress.
The expression of ALDH1, another marker associated with stem cell-like features, was significantly more common in ER− tumors in the cases of IDC alone or IDC with DCIS, confirming previous findings (14). We also found that in IDC with DCIS tumors, ALDH1+ cells were the least frequent in luminal A and B tumors compared to basal-like and HER2+ subtypes, correlating with a recent study in breast cancer cell lines (23). CD44+/CD24− cells and ALDH1+ cells were more frequently found in the basal-like than in luminal subtypes and CD44+/CD24− cells but not ALDH1+ cells were less common in HER2+ than in basal-like cases. In normal breast tissue, CD44+/CD24− cells were limited to the basal layer whereas ALDH1+ cells were found in both basal and luminal compartments suggesting that ALDH might be a marker of both bipotential mammary epithelial stem cells and luminal lineage committed progenitors. However, other interpretations of these results, including the likelihood that ALDH+ cells defined by enzymatic activity assay and immunohistochemistry may not completely overlap, are possible. The difference in the relative frequency of CD44+/CD24− and ALDH1+ cells between basal-like and HER2+ tumor subtypes may reflect their distinct cell-of-origin or the alteration of stem cell-like gene expression programs due to tumor subtype-specific transforming events. Correlating with this latter hypothesis a recent study described that the exogenous expression of HER2 increases the frequency of ALDH+ cells in breast cancer cell lines (37).
We found two different patterns of CD24 expression in breast cancers; apical membranous and circumferential membrano-cytoplamic, as reported in previous studies (24–26). Whereas apical membranous CD24 localization was found in normal luminal cells and well-differentiated luminal A tumors, circumferential membranous and cytoplasmic CD24 expression was more common in luminal B and HER2+ subtype IDCs, suggesting an association of membrano-cytoplasmic CD24 staining with more aggressive tumor characteristics. We also detected a shift of CD24 staining pattern from apical membranous to membrano-cytoplasmic during the progression of DCIS to IDC. Membrano-cytoplasmic (or cytoplasmic) CD24 expression has been shown to be associated with tumor progression and poor prognosis (24–26). It has also been reported that ectopic introduction of CD24 into breast carcinoma cells increases tumor cell proliferation, and stimulates cell motility and invasion (38). Although the biological significance of the membrano-cytoplasmic localization of CD24 remains unclear, it seems to be associated with invasive and aggressive tumor cell behavior. Thus, it would be important to establish whether cells with membrano-cytoplasmic expression of CD24 retain a differentiated, luminal epithelial cell phenotype.
Among the 12 genes differentially expressed between CD44+ and CD24+ cells analyzed by immunohistochemistry the expression of vimentin, GATA3, and EPCR is noteworthy. Vimentin expression was found predominantly in basal-like IDCs. Vimentin expression is one of the characteristics of epithelial-mesenchymal transition (EMT) that may play a role in metastatic progression (39). Basal-like breast tumors were reported to show high expression of multiple EMT markers including vimentin and to have intrinsic phenotypic plasticity for mesenchymal transition (40). Thus, there appears to be a link among stem cell-like phenotype, EMT, and basal-like subtype of breast cancer. However, not all basal-like tumors were uniformly vimentin positive suggesting further heterogeneity even within this tumor subtype. Correlating with this, a recent study described the subclassification of basal-like tumors based on claudin expression pattern (41, 42).
The GATA3 transcription factor that was shown to be essential for luminal epithelial cell differentiation (43, 44) in mice, was frequently expressed in luminal and rarely detected in basal-like breast tumors; a pattern consistent with previous studies (2–4). Based on these findings, lack of GATA3 expression may be responsible for the decrease of CD24+ more differentiated luminal epithelial cells and abundance of CD44+ stem cell-like populations in basal-like breast cancers.
EPCR (also called PROCR) is a receptor for protein C and has been shown to increase the invasiveness of breast cancer cells (45) and identified as a marker of hematopoietic, embryonic, and neural stem cells in mice (46–48). Similar to CD44, we observed an enrichment for EPCR+ cells in basal-like tumors and the number of these cells was lower in invasive compared to in situ carcinomas within the luminal A subtype. Thus, EPCR may identify cells with progenitor characteristics in human breast tissue as well.
Although the expression of some markers, such as CD44 and EPCR in luminal subtype, was different between DCIS and IDCs, the expression of the other markers was similar in the two histologic stages. The expression of connexin 43 was lower in microinvasive DCIS compared to IDCs and pure DCIS potentially implicating connexin 43 in the progression to invasion. The expression CD44 was also lower in the invasive compared to in situ areas of the same tumor in the luminal A but not HER2+ cases. The decreased frequency of CD44+ cells during in situ to invasive breast carcinoma transition relates to the decreased number of these cells in distant metastases compared to primary invasive tumors (19, 20). Thus, the frequency of CD44+ cells may gradually decline during breast tumor progression. Similarly, CK18 expression was higher in in situ compared to invasive areas in basal-like but not HER2+ subtype tumors. However, there were very few cells positive for CK18 in basal-like tumors, thus, the possibility of sampling error cannot be excluded. In contrast, the frequency of cells positive for ALDH1 and caveolin 1 appeared to be the same in the in situ and invasive areas of the same tumor. The observed differences in the frequency of cells with stem cell-like and more differentiated cell phenotypes may suggest subtype-specific tumor progression pathways.
In summary, our study revealed high degree of diversity for the expression of several selected markers in different tumor subtypes and histologic stages. The expression of CD44 appeared to decrease with tumor progression. The expression of vimentin and osteonectin was more frequent in basal-like subtype, while CK18, GATA3 and MUC1 expression was more common in luminal subtypes. CD44+/CD24− cells were most common in basal-like tumors, whereas the frequency of ALDH+ cells was the highest in HER2+ and basal-like tumors. Alterations in the frequency and distribution of cells with stem cell-like and more differentiated luminal cell features among tumors according to subtype and tumor progression stage have implications for therapeutic strategies aimed at the selective targeting of these distinct tumor cell populations.
Supplementary Material
Acknowledgments
Grant support: Funded by the National Cancer Institute SPORE in Breast Cancer (CA89393) at Dana-Farber Harvard Cancer Center (CA89393) and the Dana-Farber/Harvard Cancer Center (CA006516) grants to RG and KP, Breast Cancer Research Foundation and Department of Defense (W81XWH-07-1-0294) grants to KP, and Susan G. Komen Foundation fellowship to MS.
We thank members of our laboratories for their critical reading of this manuscript and useful discussions. We are grateful to Drs. Donald Kufe (Dana-Farber Cancer Institute) and Charles Esmon (University of Oklahoma Health Sciences Center) for their generous gift of MUC1 and EPCR antibodies, respectively.
Footnotes
STATEMENT OF TRANSLATIONAL RELEVANCE
The presence of cancer cells with stem cell-like characteristics within primary invasive breast tumors have been shown to correlate with higher risk of distant metastasis and poor clinical outcome. However, distant metastases frequently are composed of tumor cells with more differentiated luminal epithelial features. The goal of this study was to analyze if similar divergence in cellular phenotypes also occurs between in situ and invasive breast carcinomas and if this is associated with a particular tumor subtype. Tumor cell phenotypic plasticity and intra-tumor diversity have significant impact on the efficacy of cancer preventative and therapeutic strategies.
References
- 1.Polyak K. Breast cancer: origins and evolution. J Clin Invest. 2007;117:3155–63. doi: 10.1172/JCI33295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 3.Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sorlie T, Wang Y, Xiao C, et al. Distinct molecular mechanisms underlying clinically relevant subtypes of breast cancer: gene expression analyses across three different platforms. BMC Genomics. 2006;7:127. doi: 10.1186/1471-2164-7-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allred DC, Wu Y, Mao S, et al. Ductal carcinoma in situ and the emergence of diversity during breast cancer evolution. Clin Cancer Res. 2008;14:370–8. doi: 10.1158/1078-0432.CCR-07-1127. [DOI] [PubMed] [Google Scholar]
- 6.Campbell LL, Polyak K. Breast tumor heterogeneity: cancer stem cells or clonal evolution? Cell Cycle. 2007;6:2332–8. doi: 10.4161/cc.6.19.4914. [DOI] [PubMed] [Google Scholar]
- 7.Shackleton M, Quintana E, Fearon ER, Morrison SJ. Heterogeneity in cancer: cancer stem cells versus clonal evolution. Cell. 2009;138:822–9. doi: 10.1016/j.cell.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 8.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–8. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 9.Clarke MF, Fuller M. Stem cells and cancer: two faces of eve. Cell. 2006;124:1111–5. doi: 10.1016/j.cell.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 10.Polyak K, Hahn WC. Roots and stems: stem cells in cancer. Nat Med. 2006;12:296–300. doi: 10.1038/nm1379. [DOI] [PubMed] [Google Scholar]
- 11.Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755–68. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 12.Rosen JM, Jordan CT. The increasing complexity of the cancer stem cell paradigm. Science. 2009;324:1670–3. doi: 10.1126/science.1171837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–8. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ginestier C, Hur MH, Charafe-Jauffret E, et al. ALDH1 Is a Marker of Normal and Malignant Human Mammary Stem Cells and a Predictor of Poor Clinical Outcome. Cell stem cell. 2007;1:555–67. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wright MH, Calcagno AM, Salcido CD, Carlson MD, Ambudkar SV, Varticovski L. Brca1 breast tumors contain distinct CD44+/CD24− and CD133+ cells with cancer stem cell characteristics. Breast Cancer Res. 2008;10:R10. doi: 10.1186/bcr1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiao Y, Ye Y, Yearsley K, Jones S, Barsky SH. The lymphovascular embolus of inflammatory breast cancer expresses a stem cell-like phenotype. Am J Pathol. 2008;173:561–74. doi: 10.2353/ajpath.2008.071214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raouf A, Zhao Y, To K, et al. Transcriptome analysis of the normal human mammary cell commitment and differentiation process. Cell stem cell. 2008;3:109–18. doi: 10.1016/j.stem.2008.05.018. [DOI] [PubMed] [Google Scholar]
- 18.Lim E, Vaillant F, Wu D, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15:907–13. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- 19.Shipitsin M, Campbell LL, Argani P, et al. Molecular definition of breast tumor heterogeneity. Cancer Cell. 2007;11:259–73. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 20.Bloushtain-Qimron N, Yao J, Snyder EL, et al. Cell type-specific DNA methylation patterns in the human breast. Proc Natl Acad Sci U S A. 2008 doi: 10.1073/pnas.0805206105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fillmore CM, Kuperwasser C. Human breast cancer cell lines contain stem-like cells that self-renew, give rise to phenotypically diverse progeny and survive chemotherapy. Breast Cancer Res. 2008;10:R25. doi: 10.1186/bcr1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sheridan C, Kishimoto H, Fuchs RK, et al. CD44+/CD24− breast cancer cells exhibit enhanced invasive properties: an early step necessary for metastasis. Breast Cancer Res. 2006;8:R59. doi: 10.1186/bcr1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Charafe-Jauffret E, Ginestier C, Iovino F, et al. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Res. 2009;69:1302–13. doi: 10.1158/0008-5472.CAN-08-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kristiansen G, Winzer KJ, Mayordomo E, et al. CD24 expression is a new prognostic marker in breast cancer. Clin Cancer Res. 2003;9:4906–13. [PubMed] [Google Scholar]
- 25.Abraham BK, Fritz P, McClellan M, Hauptvogel P, Athelogou M, Brauch H. Prevalence of CD44+/CD24−/low cells in breast cancer may not be associated with clinical outcome but may favor distant metastasis. Clin Cancer Res. 2005;11:1154–9. [PubMed] [Google Scholar]
- 26.Mylona E, Giannopoulou I, Fasomytakis E, et al. The clinicopathologic and prognostic significance of CD44+/CD24(−/low) and CD44−/CD24+ tumor cells in invasive breast carcinomas. Hum Pathol. 2008;39:1096–102. doi: 10.1016/j.humpath.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 27.Honeth G, Bendahl PO, Ringner M, et al. The CD44+/CD24− phenotype is enriched in basal-like breast tumors. Breast Cancer Res. 2008;10:R53. doi: 10.1186/bcr2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park SY, Kim HS, Hong EK, Kim WH. Expression of cytokeratins 7 and 20 in primary carcinomas of the stomach and colorectum and their value in the differential diagnosis of metastatic carcinomas to the ovary. Hum Pathol. 2002;33:1078–85. doi: 10.1053/hupa.2002.129422. [DOI] [PubMed] [Google Scholar]
- 29.Hyun CL, Lee HE, Kim KS, et al. The effect of chromosome 17 polysomy on HER-2/neu status in breast cancer. J Clin Pathol. 2008;61:317–21. doi: 10.1136/jcp.2007.050336. [DOI] [PubMed] [Google Scholar]
- 30.Carey LA, Perou CM, Livasy CA, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. Jama. 2006;295:2492–502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 31.Nielsen TO, Hsu FD, Jensen K, et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res. 2004;10:5367–74. doi: 10.1158/1078-0432.CCR-04-0220. [DOI] [PubMed] [Google Scholar]
- 32.Heppner GH, Miller BE. Tumor heterogeneity: biological implications and therapeutic consequences. Cancer Metastasis Rev. 1983;2:5–23. doi: 10.1007/BF00046903. [DOI] [PubMed] [Google Scholar]
- 33.Charafe-Jauffret E, Monville F, Ginestier C, Dontu G, Birnbaum D, Wicha MS. Cancer stem cells in breast: current opinion and future challenges. Pathobiology. 2008;75:75–84. doi: 10.1159/000123845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mani SA, Guo W, Liao MJ, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–15. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chaffer CL, Thompson EW, Williams ED. Mesenchymal to epithelial transition in development and disease. Cells Tissues Organs. 2007;185:7–19. doi: 10.1159/000101298. [DOI] [PubMed] [Google Scholar]
- 36.Graff JR, Gabrielson E, Fujii H, Baylin SB, Herman JG. Methylation patterns of the E-cadherin 5′ CpG island are unstable and reflect the dynamic, heterogeneous loss of E-cadherin expression during metastatic progression. J Biol Chem. 2000;275:2727–32. doi: 10.1074/jbc.275.4.2727. [DOI] [PubMed] [Google Scholar]
- 37.Korkaya H, Paulson A, Iovino F, Wicha MS. HER2 regulates the mammary stem/progenitor cell population driving tumorigenesis and invasion. Oncogene. 2008;27:6120–30. doi: 10.1038/onc.2008.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baumann P, Cremers N, Kroese F, et al. CD24 expression causes the acquisition of multiple cellular properties associated with tumor growth and metastasis. Cancer Res. 2005;65:10783–93. doi: 10.1158/0008-5472.CAN-05-0619. [DOI] [PubMed] [Google Scholar]
- 39.Thiery JP. Epithelial-mesenchymal transitions in development and pathologies. Curr Opin Cell Biol. 2003;15:740–6. doi: 10.1016/j.ceb.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 40.Sarrio D, Rodriguez-Pinilla SM, Hardisson D, Cano A, Moreno-Bueno G, Palacios J. Epithelial-mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer Res. 2008;68:989–97. doi: 10.1158/0008-5472.CAN-07-2017. [DOI] [PubMed] [Google Scholar]
- 41.Creighton CJ, Li X, Landis M, et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci U S A. 2009;106:13820–5. doi: 10.1073/pnas.0905718106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hennessy BT, Gonzalez-Angulo AM, Stemke-Hale K, et al. Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer Res. 2009;69:4116–24. doi: 10.1158/0008-5472.CAN-08-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kouros-Mehr H, Slorach EM, Sternlicht MD, Werb Z. GATA-3 maintains the differentiation of the luminal cell fate in the mammary gland. Cell. 2006;127:1041–55. doi: 10.1016/j.cell.2006.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Asselin-Labat ML, Sutherland KD, Barker H, et al. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat Cell Biol. 2007;9:201–9. doi: 10.1038/ncb1530. [DOI] [PubMed] [Google Scholar]
- 45.Beaulieu LM, Church FC. Activated protein C promotes breast cancer cell migration through interactions with EPCR and PAR-1. Exp Cell Res. 2007;313:677–87. doi: 10.1016/j.yexcr.2006.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ivanova NB, Dimos JT, Schaniel C, Hackney JA, Moore KA, Lemischka IR. A stem cell molecular signature. Science. 2002;298:601–4. doi: 10.1126/science.1073823. [DOI] [PubMed] [Google Scholar]
- 47.Blanpain C, Lowry WE, Geoghegan A, Polak L, Fuchs E. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell. 2004;118:635–48. doi: 10.1016/j.cell.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 48.Ramalho-Santos M, Yoon S, Matsuzaki Y, Mulligan RC, Melton DA. “Stemness”: transcriptional profiling of embryonic and adult stem cells. Science. 2002;298:597–600. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.