Summary
WAVE proteins link upstream signals to actin nucleation by activating the Arp2/3 complex and are at the core of regulatory pathways driving membrane protrusion. They are found in heteropentameric complexes whose role in regulating WAVE function is presently unclear. Here we demonstrate that purified native WAVE complexes are basally inactive; previous reports of constitutive activity are artifacts of in vitro manipulation. Further, the native complexes are not activated by Rac alone. Activation of the WAVE2 complex requires simultaneous interactions with prenylated Rac-GTP and acidic phospholipids, as well as a specific state of phosphorylation. Together these signals promote full activation in a highly cooperative process on the membrane surface, by inducing an allosteric change in the complex rather than by simple recruitment or by dissociation of the subunits. These results explain how the WAVE complex can integrate coincident signals to promote localized actin nucleation during cell motility.
Introduction
During cell motility extracellular signals drive global and dynamic reorganization of the actin cytoskeleton. Immense progress has been made in delineating the initial signaling events at transmembrane receptors, characterizing the downstream signal transduction pathways and establishing the molecular features of actin nucleation. Yet, how multiple signals are integrated to control actin assembly as a coordinated process is still poorly understood.
Of central significance are the WAVE family proteins (and the related WASP proteins), which link upstream signals to actin assembly by stimulating the activity of the Arp2/3 complex, an important actin nucleator (Goley and Welch, 2006; Takenawa and Suetsugu, 2007). WAVE proteins are involved in some of the principal pathways driving membrane protrusion and have a central role in cell motility, cell polarity and morphologic change in perhaps all animals and plants. Often within seconds of a signal, they promote actin polymerization at the plasma membrane. Yet WAVE itself is generally not localized and hence the key to its function is its discrete spatial and temporal regulation. Not surprisingly, the principal upstream signals implicated in WAVE regulation are either components of the plasma membrane or are targeted to it. The small G-protein Rac, an essential player in cell motility and chemotaxis (Heasman and Ridley, 2008), was the first suggested regulator of WAVE (Miki et al., 1998). Rac can recruit WAVE to the plasma membrane and its effects on actin are largely dependent on WAVE (Miki et al., 1998; Steffen et al., 2004). The membrane lipid phosphatidylinositol(3,4,5)trisphosphate (PIP3) is another important regulator of cell polarity and migration (Kolsch et al., 2008) that can recruit WAVE to the plasma membrane and has been implicated in WAVE dependent processes (Oikawa et al., 2004; Sossey-Alaoui et al., 2005). IRSp53, a signaling protein with the ability to deform membranes, has been proposed to modulate WAVE2 activity (Miki et al., 2000; Suetsugu et al., 2006), and phosphorylation of WAVE proteins has also been reported to play a regulatory role (Ardern et al., 2006; Danson et al., 2007; Kim et al., 2006; Leng et al., 2005; Sossey-Alaoui et al., 2007; Stuart et al., 2006). Yet, the precise roles and contributions of these regulators, and most importantly where they act in a complex pathway, are still ill defined.
Like the related WASP proteins, WAVE1, WAVE2 and WAVE3 contain C-terminal VCA domains that can bind both the Arp2/3 complex and actin monomers to promote actin nucleation (Pollard, 2007). But in contrast to WASP and N-WASP, the isolated WAVE polypeptide is not auto-inhibited and hence its VCA domain can constitutively activate the Arp2/3 complex (Machesky et al., 1999). Furthermore, unlike WASP and N-WASP, WAVE proteins lack a G-protein binding domain, so regulation by Rac cannot be direct. However, in cells WAVE proteins are found as a complex (the WAVE complex) with four other proteins: Pir121, Nap 1, Abi-1, HSPC300 or their homologues (Eden et al., 2002; Gautreau et al., 2004). Importantly, Pir121 and its homologue Sra-1 can bind activated Rac (Kobayashi et al., 1998), and Nap 1 can bind the SH3 domain of the adapter protein Nck (Kitamura et al., 1996). Although it is clear that the WAVE complex plays an important regulatory role, its biochemical characterization has been difficult and contentious.
Our group initially found that the WAVE1 complex was inactive in vitro, suggesting that one or more subunits inhibit the VCA domain in trans. Furthermore, the complex could be activated by Rac or Nck, which appeared to promote its dissociation (Eden et al., 2002). Other researchers found that WAVE1 and WAVE2 complexes reconstituted from recombinant proteins were constitutively active, and that Rac binding had no effect on activity (Innocenti et al., 2004). They argued that Rac merely localized the active complexes to the membrane without promoting dissociation (Innocenti et al., 2004; Steffen et al., 2004). In agreement with this view WAVE complexes purified in native form or from cells over-expressing tagged WAVE were also found to be constitutively active (Kim et al., 2006; Suetsugu et al., 2006). However, others still found complexes from cells expressing tagged Abi-1 to be intrinsically inactive (Derivery et al., 2009), and while this manuscript was under review one study of reconstituted complexes suggested that they were inhibited but could be activated with very high concentrations of Rac (Ismail et al., 2009).
Thus the nature of WAVE complex regulation remains a matter of debate, possibly due to differences in experimental conditions for purification or reconstitution. We therefore chose to examine native complexes, being especially careful to preserve their biological properties. We found that purified native WAVE2 and WAVE1 complexes are inactive, in agreement with the original report from our laboratory and other recent work (Derivery et al., 2009; Eden et al., 2002; Ismail et al., 2009), but in contrast to those studies we could not activate the complexes solely with Rac or Nck. Furthermore we could explain previous results by showing that experimental manipulation can result in spontaneous activation. Having established the inactivity of the native complex, we were faced with uncovering the physiological requirements for activation. To do this we developed a gentle and highly efficient immuno-purification method that enabled us to define conditions for activation of the native WAVE2 complex. There are three requirements: 1) binding to prenylated Rac-GTP; 2) binding to lipid vesicles containing acidic phospholipids; 3) a specific state of phosphorylation. What was striking is that all three inputs had to be present simultaneously to generate any activity of the complex; partial activation was not achieved by any subset of these conditions. The WAVE2 complex is activated fully to the level of isolated WAVE2 protein in a highly cooperative process on the lipid membrane surface without dissociation of subunits. These results clarify most of the controversy about WAVE regulation, establish where the key regulators act in the pathway, and show how the WAVE complex integrates coincident signals to promote localized actin nucleation on the plasma membrane.
Results
Native WAVE complexes are basally inactive and are not activated by Rac or Nck
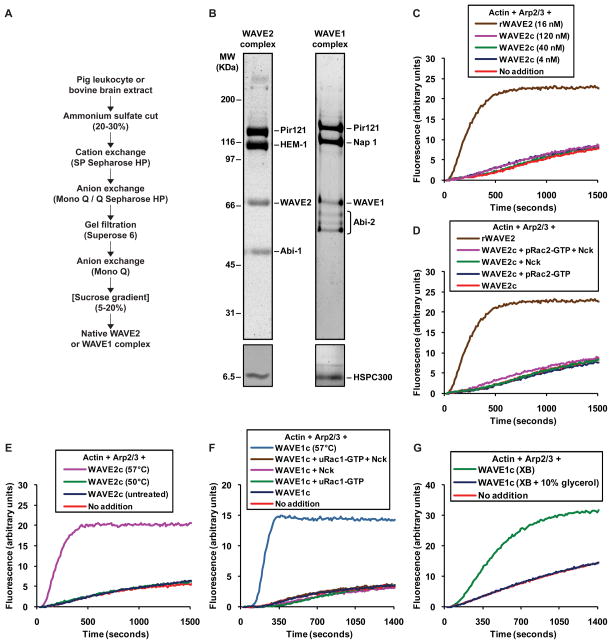
We purified native WAVE2 complex from pig leukocytes (we use the term native to refer to endogenous WAVE complexes that do not contain any recombinantly expressed subunits), whose activity had not been previously studied. A five-step purification (Figure 1A) yielded very pure complex containing the five known subunits (Figure 1B). When we mixed the purified WAVE2 complex with Arp2/3 and actin in the pyrene-actin polymerization assay, we found that it was basally inactive even at the highest concentration tested, 120 nM (Figure 1C). Given that the activity of 0.25 nM isolated recombinant WAVE2 protein can be easily detected (Figure S1A), we conclude that the purified WAVE2 complex has less than 0.2% of the activity of the uncomplexed WAVE polypeptide. We could not activate this complex by addition of constitutively active prenylated Rac2-GTP or Nck (Figure 1D). To rule out the possibility that purification and handling had simply inactivated the WAVE2 subunit, we showed that we could induce activity by partial thermal denaturation of the purified complex. Recombinant WAVE2 protein loses activity when it is heated to 60°C and then cooled prior to being assayed, but is unaffected at lower temperatures (Figure S1B). We found that heating the WAVE2 complex to 57°C generated substantial activity (Figure 1E). Hence the inactive WAVE2 complex must contain WAVE2 protein capable of activity, but this activity is inhibited within the complex.
Figure 1. Native WAVE2 and WAVE1 complexes are basally inactive and are not activated by Rac or Nck.
(A) Scheme used to purify native WAVE2 complex from pig leukocyte extract and native WAVE1 complex from bovine brain extract. The sucrose gradient step indicated in brackets was only included during purification of the WAVE1 complex.
(B) Native WAVE2 complex from pig leukocytes and native WAVE1 complex from bovine brain, resolved on 4–12% Bis-Tris polyacrylamide gels and stained with Coomassie (top panels) or silver (bottom panels). Constituent subunits, identified previously by mass spectrometry, are indicated.
(C–G) The activity of WAVE complexes was tested in polymerization assays containing 1 μM actin/pyrene-actin and 30 nM Arp2/3 complex (C–F) or 2 μM actin/pyrene-actin and 60 nM Arp2/3 complex (G).
(C) Native WAVE2 complex does not stimulate actin polymerization. The indicated concentrations of WAVE2 complex from pig leukocytes (“WAVE2c”) or isolated recombinant WAVE2 protein (“rWAVE2”) were tested. Recombinant WAVE2 protein activates the Arp2/3 complex as expected.
(D) Native WAVE2 complex from pig leukocytes is not activated by Rac2 or Nck. 200 nM constitutively active prenylated Rac2-GTP (“pRac2-GTP”), 200 nM full length Nck or both were added to reactions containing 40 nM WAVE2 complex (“WAVE2c”) as indicated. A reaction containing 16 nM recombinant WAVE2 protein (“rWAVE2”) is shown for reference.
(E) Thermal denaturation activates the WAVE2 complex. WAVE2 complex from pig leukocytes was heated for 10 minutes at the indicated temperature, cooled down to room temperature and 40 nM was immediately added to the assay.
(F) Native WAVE1 complex is inactive and is not activated by Rac1 or Nck. 20 nM WAVE1 complex from bovine brain (“WAVE1c”) was tested alone or with 200 nM constitutively active unprenylated Rac1-GTP (“uRac1-GTP”), 200 nM full length Nck, or both. Thermal denaturation (57°C) revealed the activity of the WAVE1 complex.
(G) Freezing and thawing without cryoprotectants activates the WAVE1 complex. WAVE1 complex from bovine brain was exchanged into buffer XB, freeze thawed directly or after addition of 10% glycerol, and assayed at a final concentration of 20 nM.
WAVE2 and WAVE1 are found in complexes containing different homologues of some of the constituent subunits (Eden et al., 2002; Gautreau et al., 2004). They have different expression patterns and exhibit specialized functions, suggesting possible differences in their regulation (Suetsugu et al., 1999; Suetsugu et al., 2003). Previous studies from our laboratory had shown that the WAVE1 complex is inactive and can be activated by Rac or Nck (Eden et al., 2002). Using a purification strategy similar to that for the WAVE2 complex (Figure 1A), we purified the native WAVE1 complex from bovine brain (Figure 1B), the same source used during the initial characterization of that complex. Like the WAVE2 complex, the purified WAVE1 complex was inactive (Figure 1F) and could not be activated by Rac1-GTP alone or in combination with Nck (Figure 1F). We verified the functionality of Rac and of Nck (Figures S1C and S1D). As with the leukocyte WAVE2 complex, heat denaturation revealed the activity of WAVE1 in the native complex.
Recently Ismail and colleagues reported experiments using WAVE1 complex produced by co-expression of recombinant subunits (Ismail et al., 2009). The complex was basally inactive but could be activated in vitro by addition of unprenylated Rac1 bound to the non-hydrolyzable GTP analog GMPPNP. Of concern was the extraordinarily high concentration of Rac1-GMPPNP required for activation, 5 to 7.5 μM. We tested whether native WAVE1 complex could be activated under similar conditions and found using their protocol that 7.5 μM unprenylated Rac1-GMPPNP did not yield detectable activity (Figure S1E).
Our experiments are also inconsistent with another study showing that native WAVE1 complex purified from rat brains is constitutively active (Kim et al., 2006). Since heat denaturation generates activity, we suspected that adventitious denaturation might have caused a similar effect. During their purification Kim and colleagues froze the complex in buffer without cryoprotectants and thawed aliquots for each assay (Kim et al., 2006). When we freeze-thawed our inactive WAVE1 complex under their conditions we generated substantial activity (Figure 1G). This was prevented by adding 10% glycerol as a cryoprotectant (Figure 1G). Even routine biochemical procedures, such as elution with 0.5 M KCl or prolonged storage on ice, can generate activity (Figure S1F and S1G). The fact that the inactive state of the native complex can be so easily compromised in vitro may explain the observed constitutive activity and activation of the complex reported previously in the literature.
Prenylated Rac-GTP and acidic phospholipids activate the WAVE2 complex in a highly cooperative process
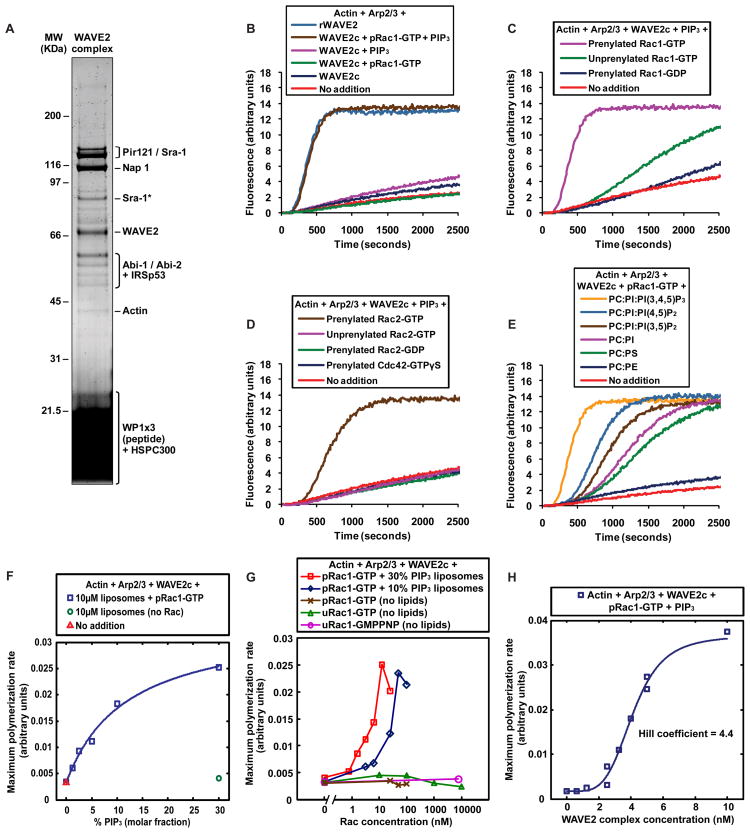
To improve the chances of isolating an inactive but activatable WAVE complex we devised a rapid purification protocol. Using a peptide antibody specific for the WAVE2 protein we isolated the native complex immunoadsorption and eluted with a competing peptide consisting of three tandem repeats of the antigenic sequence. We achieved a similar purity to the conventional purification in less than 24 hours rather than 2 weeks. Immuno-purification allowed us to use smaller amounts of starting material, making it feasible to purify the complex from cultured cells, a more homogeneous source that can be experimentally perturbed. This was useful because it allowed us to test growth factor stimulation, which was known to induce posttranslational modifications in the WAVE2 complex (Danson et al., 2007). We purified the WAVE2 complex from A-431 human epidermoid carcinoma cells stimulated with epidermal growth factor (EGF). The complex contained the five core subunits as confirmed by mass spectrometry (Figure 2A and Table S1). Among other proteins that co-eluted with the complex we identified IRSp53 (Figure 2A, Table S1 and data not shown).
Figure 2. Prenylated Rac-GTP and acidic phospholipids activate the WAVE2 complex cooperatively.
(A) Native WAVE2 complex immuno-purified from the cytosol of EGF stimulated A-431 cells, resolved on a 4–12% Bis-Tris polyacrylamide gel and stained with Coomassie. The main constituent subunits, identified by mass spectrometry (see Table S1), are indicated. The band labeled “Sra-1*” appears to be Sra-1 isoform 2 based on mass spectrometirc analysis and on its apparent molecular weight.
(B–H) The activity of WAVE2 complex immuno-purified from the cytosol of EGF stimulated A-431 cells was tested in polymerization assays containing 1 μM actin/pyrene-actin and 30 nM Arp2/3 complex.
(B) Native WAVE2 complex is fully activated by the combination of prenylated Rac1-GTP and PIP3 liposomes, but not by Rac or liposomes alone. Where indicated, the activity of 5 nM WAVE2 complex (“WAVE2c”) was measured in the presence of either 50 nM constitutively active prenylated Rac1-GTP (“pRac1-GTP”), 10 μM (total lipid) PIP3 liposomes composed of 45% (molar fraction) PC, 45% PI and 10% PIP3 (“PIP3”), or both. Actin polymerization induced by 5 nM isolated recombinant WAVE2 protein (“rWAVE2”) is shown for comparison.
(C) Activation of WAVE2 complex depends on the nucleotide and prenylation state of Rac1. The activity of 5 nM WAVE2 complex was measured in the presence of 10 μM PIP3 liposomes and where indicated 50 nM of either prenylated Rac1-GDP, unprenylated Rac1-GTP or prenylated Rac1-GTP.
(D) Prenylated Rac2-GTP, but not Cdc42, can activate the WAVE2 complex in conjunction with PIP3 liposomes. The activity of 5 nM WAVE2 complex was measured in the presence of 10 μM PIP3 liposomes and where indicated 50 nM of either prenylated Cdc42-GTPγS, prenylated Rac2-GDP, unprenylated Rac2-GTP, or prenylated Rac2-GTP.
(E) Acidic phospholipids other than PIP3 can activate native WAVE2 complex in conjunction with prenylated Rac1-GTP. The activity of 5 nM WAVE2 complex was measured in the presence of 50 nM prenylated Rac1-GTP and where indicated 10 μM (total lipid) liposomes composed of either PC:PE, PC:PS (both 50:50 molar percentage), PC:PI (45:55 molar percentage), PC:PI:PI(3,5)P2, PC:PI:PI(4,5)P2 or PC:PI:PI(3,4,5)P3 (all 45:45:10 molar percentage).
(F) Activation of native WAVE2 complex depends on acidic lipid concentration. Blue squares: maximum actin polymerization rate was measured in reactions containing 5 nM WAVE2 complex, 12.5 nM prenylated Rac1-GTP (“pRac1-GTP”) and 10 μM (total lipid) liposomes containing varying molar fractions of PIP3 (percentage PIP3 is indicated on the x-axis; all liposomes contain 25% PI and the remainder is PC). The data was fitted by least squares to the Langmuir isotherm to obtain the PIP3 concentration required for half maximal activation, 6.1%. Green circle: in the absence of Rac, no activity was observed with 10 μM liposomes containing 30% PIP3. Red triangle: a reaction containing 5 nM WAVE2 complex but no agonists is shown for reference.
(G) Rac alone is not sufficient to activate native WAVE2 complex; activation requires acidic phospholipids and depends on the concentration of prenylated Rac-GTP. Maximum actin polymerization rate was measured in reactions containing 5 nM WAVE2 complex and various concentrations of prenylated Rac1-GTP (“pRac1-GTP”), unprenylated Rac1-GTP (“uRac1-GTP”) or unprenylated Rac1-GMPPNP (“uRac1-GMPPNP”) in the absence of lipids. For prenylated Rac1-GTP, titrations were also done together with 10 μM (total lipid) liposomes containing 30% (molar fraction) PIP3, 45% PC and 25% PI (“30% PIP3 liposomes”), or 10% PIP3, 45% PC and 45% PI (“10% PIP3 liposomes”). To the right of the hatched mark the x-axis is in log 10 scale.
(H) Activation of native WAVE2 complex by prenylated Rac1-GTP and PIP3 liposomes is highly cooperative. Varying concentrations of WAVE2 complex in combination with 50 nM prenylated Rac1-GTP and 10 μM PIP3 liposomes were assayed for stimulation of actin polymerization. The maximum polymerization rate was plotted as a function of the concentration of WAVE2 complex (duplicates are shown for some concentrations) and the data was fitted to the Hill equation as described in supplemental experimental procedures to obtain the Hill coefficient.
This native WAVE2 complex preparation was almost completely inactive (Figure 2B), indicating that immuno-purification did not denature the complex and suggesting that the additional proteins present in this preparation, including IRSp53, were not sufficient for activation. A trace of activity was present in some preparations (Figure 2B), possibly due to a small amount of uncomplexed WAVE2 protein. When we tested several potential activators we found that the combination of prenylated Rac1-GTP and liposomes containing 10% PIP3 (PIP3 liposomes throughout the text) led to robust activation (Figure 2B), comparable to the activity generated by the same molar concentration of isolated recombinant WAVE2 protein. Therefore, we had achieved full activation of an inactive native WAVE2 complex. Individually, prenylated Rac1-GTP or PIP3 liposomes generated no activity (Figure 2B). The observed actin polymerization was due to activation of the complex and not to enhancement of the activity of any free WAVE2 present in the preparation, since addition of PIP3 liposomes and prenylated Rac1-GTP to recombinant WAVE2 protein had essentially no effect on activity (Figure S2A). Similarly, the activity was not due to any contaminating N-WASP, since an inhibitory antibody specific to WAVE2 (Figure S2B) inhibited almost completely the measured activity of the WAVE2 complex (Figure S2C). The activity of the WAVE2 complex was completely dependent on Arp2/3 (Figure S2C).
Activation of the WAVE2 complex depended on the nucleotide state of Rac (Figure 2C). Targeting of Rac to the liposomes was important for activation, since unprenylated Rac1-GTP yielded much less activity than prenylated Rac1-GTP (Figure 2C), even though both unprenylated and prenylated Rac1-GTP were equally active in other assays (Figure S1C). Prenylated Rac was indeed incorporated into the liposomes as determined by co-sedimentation (see below and Figure S3A). Although Rac1 is the most ubiquitous form of Rac, cells of hematopoietic lineage also express high levels of Rac2. Rac2 in combination with PIP3 liposomes activated the WAVE2 complex robustly (Figure 2D), and activation was dependent on the nucleotide and prenylation state of Rac2 (Figures 2D and S1C). Activation was specific for Rac since Cdc42, another GTPase of the Rho family, failed to activate the complex when assayed with PIP3 liposomes (Figure 2D). The functionality of Cdc42 was confirmed by its ability to induce actin assembly in Xenopus egg extracts, an N-WASP dependent process (Lebensohn et al., 2006).
The specificity of PIP3 would be important, since PIP3 is polarized during directed cell migration (Kolsch et al., 2008). Although PIP3 is the most potent activator, we found that other acidic phospholipids can exert similar effects; liposomes containing several negatively charged lipids, including phosphatidylinositol(4,5)bisphosphate (PI(4,5)P2), phosphatydilinositol(3,5)bisphosphate (PI(3,5)P2), phosphoatidylinositol (PI) and even phosphatidylserine (PS) could activate the WAVE2 complex in combination with prenylated Rac1-GTP (Figure 2E). Although the total net charge on the liposomes correlated with activation, these results rule out a specific interaction with the phosphoinositol head group. At sufficiently high concentration even PS, which lacks this head group, can activate the complex efficiently (Figure 2E). On the other hand liposomes composed entirely of lipids with no net charge, such as phosphatidylcholine (PC) and phosphatidylethanolamine (PE), failed to activate the WAVE2 complex in combination with prenylated Rac1-GTP (Figure 2E).
We studied the interplay between Rac and acidic phospholipids by titrating these two effectors. We generated liposomes with a background of PC and PI and varied the molar percentage of PIP3 using a fixed concentration of total lipid and prenylated Rac1-GTP. Activation (measured as maximal actin polymerization rate) as a function of % PIP3 could be fitted to a hyperbolic trace with half-maximal activation at 6.1% PIP3 (Figure 2F). No activation was detected in the absence of Rac, even with 10 micromolar liposomes containing 30% PIP3 (Figure 2F). We also titrated Rac1-GTP with two lipid conditions. With liposomes containing 10% PIP3 half-maximal activation required approximately 25 nM prenylated Rac1-GTP, while at 30% PIP3 half-maximal activation occurred near 6.25 nM prenyalted Rac1-GTP (Figure 2G). In the absence of lipids, up to 100 nM prenylated Rac1-GTP, and up to 10 μM unprenylated Rac1-GTP did not produce detectable activation (Figure 2G). Repeating the exact conditions on the native WAVE2 complex that Ismail and colleagues used to activate the recombinant WAVE1 complex(7.5μM unprenylated Rac1-GMPPNP) (Ismail et al., 2009) we found no measurable activation (Figure 2G). These data highlight the remarkable tightness of the co-requirement of Rac and acidic phospholipids for activation of the native complex.
The activation of the WAVE2 complex as a function of the concentration of the complex was clearly sigmoidal with a Hill coefficient of 4.4, suggesting the highly cooperative assembly of a functional unit (Figure 2H). Activation of the WAVE2 complex happens on the surface of PIP3 liposomes containing prenylated Rac-GTP (see below, Figure 4 and Movie S1). Thus, in addition to providing a chemical or electrostatic signal for activation, the membrane provides a constrained surface for the assembly of supramolecular signaling units in which cooperative interactions between multiple WAVE2 complexes could take place (see Discussion).
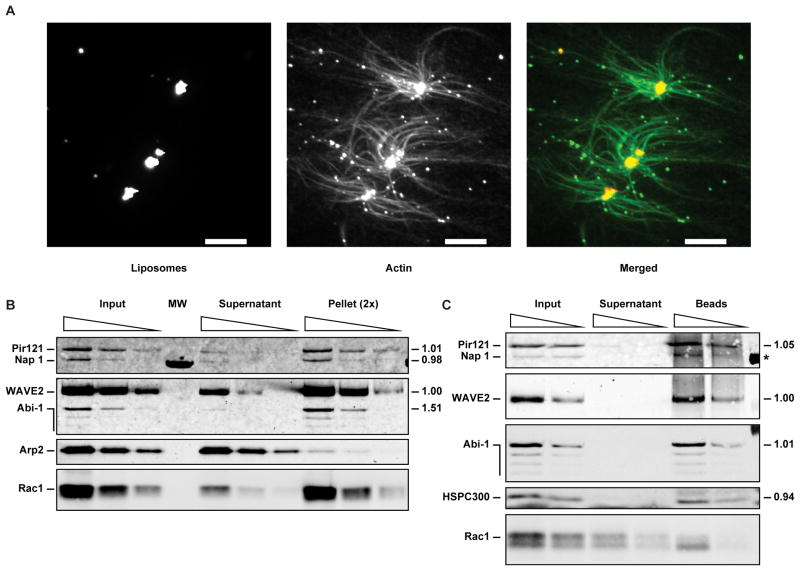
Figure 4. Activation of the WAVE2 complex takes place on the membrane surface without dissociation of constituent subunits.
(A) Actin polymerization induced by WAVE2 complex activation takes place on the membrane surface. Fluorescence images of a reaction where 5 nM WAVE2 complex from serum starved A-431 cells was activated by 50 nM prenylated Rac2-GTP and 2.5 μM liposomes composed of PC, PI, PIP3 and lissamine rhodamine labeled phosphatidylethanolamine (RhPE) (44:45:10:1 molar percentage) in an assay containing 0.7 μM actin, 0.2 μM pyrene-actin, 0.1 μM Alexa 488 actin and 30 nM Arp2/3 complex. The two left panels show maximum intensity projections of confocal z-stacks, and the right panel shows a pseudo-colored, merged image. Image acquisition and processing details are described in supplemental experimental procedures. Scale bar = 5 μm.
(B–C) Liposome co-sedimentation assays contain 30 nM Arp2/3 complex, 5 nM WAVE2 complex from serum starved A-431 cells, 20 μM PIP3 liposomes composed of PC:PI:PIP3:RhPE (44:45:10:1 molar percentage) and 50 nM prenylated Rac1-GTP.
(B) Activation of the WAVE2 complex does not lead to dissociation of constituent subunits. Liposome co-sedimentation assays performed under activating conditions in the presence of PIP3 liposomes and prenylated Rac1-GTP were analyzed by Western blot for Pir121, Nap 1, WAVE2, Abi-1, Arp2 and Rac1. Sequential two-fold dilutions of the input, supernatant and pellet were loaded (the sequential dilutions of the pellet are 2× concentrated relative to the input and supernatant). The stoichiometry (relative to WAVE2) of constituent subunits was calculated from two independent experiments based on the relative amounts of the input signal present in the pellets, and is shown to the right of each blot. MW denotes a lane used for molecular weight standards.
(C) Solubilization of the membrane during activation releases intact WAVE2 complex. The liposome pellet from an assay like the one in part (B) was resuspended and solubilized with 1% Triton X-100, clarified by centrifugation, and WAVE2 was immunoprecipitated. Sequential two-fold dilutions of the IP input, supernatant and beads were analyzed by Western blot for Pir121, Nap 1, WAVE2, Abi-1, HSPC300 and Rac1. The stoichiometry of constituent subunits was determined as in part (B) and is shown to the right of each blot (* the stoichiometry of Nap 1 could not be determined due to the low signal to background ratio).
It had been suggested that WAVE2 complex isolated from the plasma membrane is fully active due to the presence of IRSp53 (Suetsugu et al., 2006). When we rapidly purified native WAVE2 complex from the membranes of EGF stimulated A-431 cells (Figure S2D), we found that like the cytosolic complex it was almost completely inactive despite containing IRSp53, and could also be activated with Rac and PIP3 liposomes (Figure S2E). In contrast, the same conditions have not worked to activate the native WAVE1 complex purified by conventional chromatography (Figure S2F). This suggests that WAVE1 and WAVE2 complexes may be regulated differently, or that the longer conventional purification of the WAVE1 complex may have failed to preserve important posttranslational modifications (see below).
Phosphorylation is required for activation of the WAVE2 complex
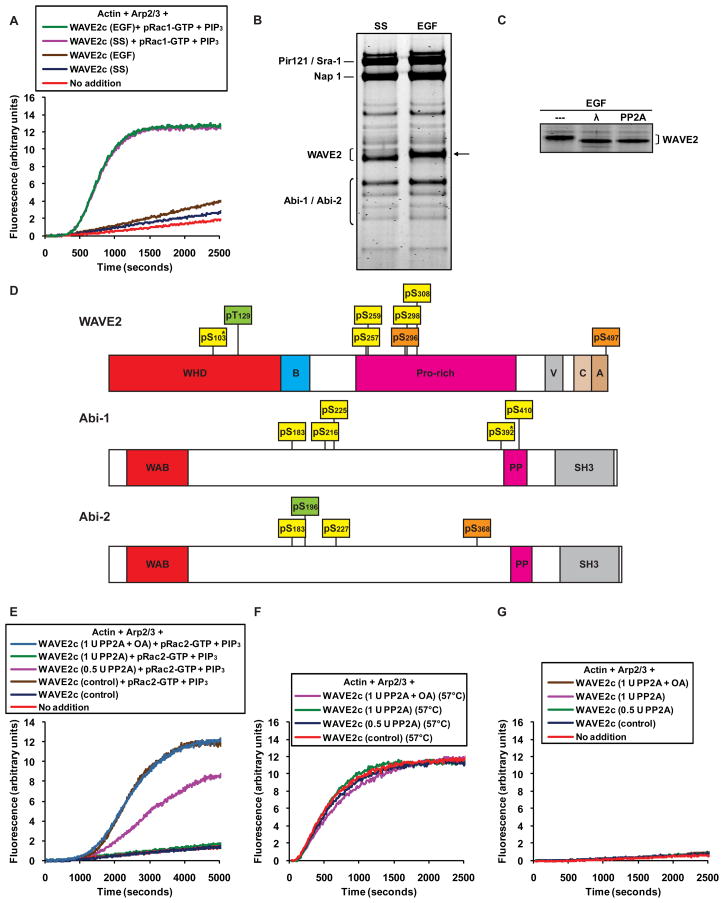
WAVE2 is known to be phosphorylated by MAP kinases in response to growth factor stimulation (Danson et al., 2007) and it was for that reason that we initially purified the complex from EGF stimulated cells (Figure 2). When we subsequently compared complexes from either EGF stimulated or serum starved cells, we found that both complexes were basally inactive and could be equally activated by PIP3 liposomes and Rac (Figure 3A). Yet, the effects of EGF treatment could be detected by a decrease in the electrophoretic mobility of WAVE2 in complexes purified from EGF treated cells (Figure 3B). This shift was due to phosphorylation, since it could be reversed by treatment with lamda protein phosphatase or the serine/threonine phosphatase PP2A (Figure 3C). Therefore EGF-induced phosphorylation is not required for activation of the WAVE2 complex. However, WAVE2 is known to be phosphorylated by other kinases, including Abl (Leng et al., 2005; Stuart et al., 2006), so we asked whether the WAVE2 complex might be phosphorylated independently of EGF. Mass spectrometric analysis of native WAVE2 complexes purified from either serum starved or EGF stimulated cells revealed a total of 12 phosphorylated sites in both conditions (Figure 3D and Table S2): 5 sites in WAVE2 (four serines and one either serine or threonine; see legend to Figure 3 and Table S2), 5 sites in Abi-1 (four serines and one either serine or threonine), and 2 serine sites in Abi-2 (both Abi-1 and Abi-2 were present in our immuno-purified preparations, which presumably consist of a mixture of complexes containing one or the other subunit). In addition there were 2 serine sites in WAVE2 and 1 serine site in Abi-2 detected only in serum starved cells, as well as 1 threonine site in WAVE2 and 1 serine site in Abi-2 detected only in EGF stimulated cells (Figure 3D and Table S2). Only 7 out of these 17 sites seem to have been reported previously and no specific function has been attributed to any of them. Phosphorylation was not detected on Pir121, Nap1 or HSPC300 for either sample.
Figure 3. Phosphorylation is required to activate the WAVE2 complex.
(A, E–G) The activity of native WAVE2 complex from A-431 cells was tested in polymerization assays containing 1 μM actin/pyrene-actin and 30 nM Arp2/3 complex.
(A) Native WAVE2 complexes (5 nM) purified from either serum starved (SS) or EGF stimulated A-431 cells are inactive and can be activated equally by the combination of prenylated Rac1-GTP (50 nM) and PIP3 liposomes (10 μM).
(B) Native WAVE2 complexes immuno-purified from serum starved (SS) or EGF stimulated A431 cells, resolved on a 4–12% Bis-Tris polyacrylamide gel and stained with Coomassie. The reduced electrophoretic mobility of WAVE2 in the EGF lane is indicated with an arrow.
(C) Native WAVE2 complex immuno-purified from EGF stimulated cells was treated with phosphatase buffer (“---“), lamda protein phosphatase (“λ”) or PP2A. The eluted complexes were resolved on a 4–12% Bis-Tris polyacrylamide gel and stained with Coomassie.
(D) Diagram of human WAVE2, Abi-1 and Abi-2 indicating phosphorylated serine (pS) and threonine (pT) residues identified by mass spectrometric analysis of complexes purified from A-431 cells. Residues in yellow boxes were identified in both serum starved and EGF stimulated conditions, residues in orange boxes were identified only in serum starved conditions and residues in green boxes were identified only in EGF stimulated conditions. Residues marked by * could not be assigned unequivocally based on peptide fragmentation data, and equally likely alternative assignments are indicated in Table S2. Abbreviations for protein domains are as follows: WHD, WAVE homology domain; B, basic region; Pro-rich, proline rich region; V, verprolin homology motif; C, central motif; A, acidic motif; WAB, WAVE binding domain; PP, polyproline stretch; SH3, src homology 3 domain.
(E) Phosphatase treatment abolishes the ability of prenylated Rac2-GTP and PIP3 liposomes to activate native WAVE2 complex purified from serum starved A-431 cells. WAVE2 complex was treated with phosphatase buffer (control), 0.5 units of PP2A, 1 unit of PP2A, or 1 unit of PP2A pre-incubated with okadaic acid (OA) (see supplemental experimental procedures for details) as indicated and activation of 5 nM WAVE2 complex was tested in the presence of prenylated Rac2-GTP (50 nM) and PIP3 liposomes (10 μM). Control treated WAVE2 complex in the absence of activators is shown for reference.
(F) Treatment with PP2A does not affect the intrinsic capacity of the WAVE2 protein to stimulate Arp2/3 dependent actin polymerization. Native WAVE2 complexes purified from serum starved A-431 cells were treated as in (D), heated for 10 minutes at 57°C, and 2 nM was tested in the absence of activators.
(G) Treatment with PP2A has no effect on the WAVE2 complex in the absence of activators. Native WAVE2 complexes purified from serum starved A-431 cells were treated as in Figure 3E, and 5 nM was tested in the absence of activators.
To test whether any of these EGF-independent sites is required for activity we exposed the complex isolated from serum starved cells to the phosphatase PP2A. To avoid carry-over of the phosphatase into the polymerization assays, we treated the complex while still bound to the immunoaffinity resin, then washed extensively and eluted. WAVE2 complex treated with control buffer was basally inactive and could be activated by the standard conditions, albeit with slower kinetics than our other immuno-purified preparations, possibly due to the extended incubation at 30°C (Figure 3E). Treatment with PP2A resulted in a marked dose dependent decrease in the ability to activate the complex, entirely abolishing activatability at the highest dose (Figure 3E). This effect could be blocked by the PP2A inhibitor okadaic acid. Phosphatase treatment did not affect the intrinsic activity of the WAVE2 protein, as assayed by partial thermal denaturation (Figure 3F) and had no effect on the WAVE2 complex in the absence of activators (Figure 3G). We conclude that serine and/or threonine phosphorylation of native WAVE2 complex is required for activation by Rac and acidic phospholipids.
Activation of the WAVE2 complex takes place on the membrane surface without dissociation of constituent subunits
Activation of the WAVE2 complex and the resulting actin nucleation might be strictly localized to the membrane surface; alternatively signals at the membrane could play a transient role in stimulating the WAVE2 complex followed by release of the activated complex or dissociation of the WAVE subunit. When we examined activation of the WAVE2 complex microscopically we found that filamentous actin structures radiate from the liposomes themselves (Figure 4A and Movie S1), suggesting that the activated complex promotes localized actin polymerization at the lipid membrane surface. Further, activation of the complex does not appear to be accompanied by dissociation of the constituent subunits, as originally proposed for the WAVE1 complex (Eden et al., 2002). Stoichiometric amounts of Pir121, Nap1, WAVE2 and Abi-1 co-sedimented with PIP3 liposomes and prenylated Rac1-GTP under conditions leading to activation (Figure 4B), suggesting that none of these subunits dissociated from the complex. Under these conditions most of the prenylated Rac1-GTP is incorporated into the liposomes, but the Arp2/3 complex did not co-sediment appreciably (Figure 4B), suggesting that the interaction between the Arp2/3 complex and activated WAVE2 complex is weak.
Because Sra-1/Pir121 can bind Rac-GTP (Kobayashi et al., 1998) and WAVE2 can bind PIP3 (Oikawa et al., 2004), it is conceivable that the complex could have dissociated on the membrane, leaving the sub-complexes independently bound to Rac and PIP3. To test for this possibility we first co-sedimented the WAVE2 complex with PIP3 liposomes and prenylated Rac1-GTP under conditions leading to activation. We then solubilized the liposome pellet with detergent and immunoprecipitated the soluble material with antibody to WAVE2. We found stoichiometric amounts of Pir121, WAVE2, Abi-1 and HSPC300 in the WAVE2 immunoprecipitate (Figure 4C), suggesting that the complex remained intact after activation. Some solubilized prenylated Rac1-GTP co-immunoprecipitated with the complex (Figure 4C), presumably through its interaction with Sra-1/Pir121, further indicating that the complex did not dissociate upon binding to activated Rac. Although we cannot discount the remote possibility that the complex dissociated upon activation and fully re-associated during the experiment, these results disfavor a model of activation through dissociation of the WAVE subunit (Eden et al., 2002). Instead, activation must proceed through an allosteric change in the complex.
The WAVE2 complex binds Rac-GTP or acidic phospholipids independently, but binding to both is required for activation
Although Rac-GTP or PIP3 liposomes were individually unable to activate the WAVE2 complex, previous work suggests that each on its own may bind to the complex (Innocenti et al., 2004; Oikawa et al., 2004); this implies a distinction between recruitment and activation that could be of physiological relevance. To examine these features we co-sedimented the WAVE2 complex with liposomes that contained either uncharged or acidic phospholipids, and either prenylated Rac-GDP or GTP (Table 1). We omitted actin so as not to perturb co-sedimentation. Liposomes containing PC and PE (both uncharged lipids) together with prenylated Rac-GDP brought down a negligible amount of WAVE2 (Table 1). PC:PE liposomes together with prenylated Rac-GTP bound 56% of total WAVE2 while PIP3 liposomes in the absence of Rac bound 69%, but neither activated the complex (Table 1). PIP3 liposomes in combination with prenylated Rac-GTP co-sedimented 91% of the WAVE2 signal and activated the complex robustly (Table 1). Prenylated Rac was incorporated into the liposomes regardless of nucleotide state or liposome composition (Figure S3A). Thus, although there is substantial binding of the WAVE2 complex to either Rac-GTP or acidic phospholipids individually, this binding is insufficient for activation. Simultaneous binding to Rac-GTP and acidic phospholipids results both in more efficient recruitment and activation.
Table 1. The WAVE2 complex binds Rac-GTP or acidic phospholipids independently, but binding to both is required for activation; phosphorylation modulates lipid binding.
Liposome co-sedimentation assays containing 30 nM Arp2/3 complex, 5 nM WAVE2 complex from serum starved A-431 cells, and where indicated 20 μM (total lipid) liposomes composed of PC:PE (50:49 molar percentage) or PIP3 liposomes composed of PC:PI:PIP3 (44:45:10 molar percentage) (all liposomes contained 1% molar fraction Rhodamine PE for visualization purposes), and 50 nM prenylated Rac-GDP or GTP. Percentages of total WAVE2 bound (see experimental procedures for details) are from experiments with Rac2; comparable results were obtained with Rac1. The results of actin polymerization assays of equivalent composition are reported under “Activation”.
| WAVE2 complex | WAVE2 complex, PP2A treated | |||
|---|---|---|---|---|
| % bound | Activation | % bound | Activation | |
| PC:PE liposomes + Rac-GDP | 1 | No | Not tested | |
| PC:PE liposomes + Rac-GTP | 56 | No | 55 | No |
| PIP3 liposomes (no Rac) | 69 | No | 35 | No |
| PIP3 liposomes + Rac-GTP | 91 | Yes | 79 | No |
We asked whether phosphorylation of the WAVE2 complex might affect the interactions with acidic phospholipids and/or Rac. Treatment of the WAVE2 complex with PP2A did not affect the interaction between the WAVE2 complex and PC:PE liposomes containing prenylated Rac-GTP (Table 1). However, PP2A treatment diminished binding between the WAVE2 complex and PIP3 liposomes by half (Table 1). When the phosphatase treated complex was co-sedimented with both PIP3 liposomes and prenylated Rac-GTP binding was also reduced (Table 1). Hence, phosphorylation may regulate WAVE2 complex activation by modulating binding to acidic phospholipids.
Discussion
The physiological state of isolated WAVE complexes
We have demonstrated that both native WAVE1 and WAVE2 complexes, obtained from three different sources (pig leukocytes, bovine brain and A-431 cells), two sub-cellular fractions (cytosol and membranes), two physiological states (serum starved and EGF stimulated), and purified in two different ways (conventional chromatography and immuno-purification) are inactive. Previous reports of constitutive activity are most likely due to experimental artifacts, one of which we could reproduce under controlled conditions (Kim et al., 2006) (Figure 1G). WAVE complexes reconstituted from recombinant subunits or purified from cells over-expressing tagged WAVE may not assemble properly in the first place, which could explain their constitutive activity (Innocenti et al., 2004; Suetsugu et al., 2006). Recent reports suggest that the choice and placement of tags, as well as the procedures used to purify the complex away from unfolded species may help preserve their inactive state (Derivery et al., 2009; Ismail et al., 2009).
When we used native complexes we were unable to reproduce the previously reported activation by addition of Rac alone (Eden et al., 2002; Ismail et al., 2009). Differences in preparation conditions most likely explain the discrepancies. We suspect that in the initial report from our laboratory an unknown feature of the purification or assay conditions may have destabilized the complex, rendering it susceptible to non-physiological activation. In retrospect it is noteworthy that in those experiments activation was achieved with both GDP- and GTP-bound Rac1, suggesting non-specific effects. In the experiments of Ismail and colleagues, imperfect assembly of the WAVE1 complex produced by co-expression of recombinant subunits may have also failed to recapitulate aspects of physiological regulation, resulting in some activation at non-physiologically high Rac concentrations.
Activation of the WAVE2 complex by coincident signals
An extensive body of research indicates that Rac and acidic phospholipids induce actin assembly during lamellipodial extension, cell motility and chemotaxis. It was also evident that WAVE plays an important role in these processes. However until now it was not clear how these signals affected WAVE or how they specifically contributed to the process of actin assembly. Our experiments demonstrating that an inactive WAVE2 complex can be activated fully by the combination of prenylated Rac-GTP and acidic phospholipids establish sufficiency in a reconstituted system of purified components. This conclusion explains many cellular results that suggest that activated Rac on its own is not sufficient to stimulate actin assembly. Expression of a WAVE2 construct harboring mutations in a domain that binds acidic phospholipids diminishes lamellipodia formation induced by constitutively active Rac (Oikawa et al., 2004). FLAIR (fluorescent activation indicator for Rho proteins) experiments show that migrating fibroblasts exhibit a gradient of Rac activity correlated with the direction of movement, but this region of Rac activation is much broader than the region where actin polymerizes (Kraynov et al., 2000). Similarly, in migrating neutrophil-like HL-60 cells Rac activity spatially correlates with but is more diffuse than the domain of the WAVE2 complex (Weiner et al., 2007). Actin polymerization that drives the extension of pseudopods during phagocytosis in leukocytes requires Rac1, but recruitment of constitutively active Rac1 to the plasma membrane that lines an engulfed particle will not maintain actin polymerization after the particle has been internalized (Scott et al., 2005). Instead the disappearance of F-actin from the phagosome parallels the disappearance of PI(4,5)P2. These observations argue for a role of acidic phospholipids such as PI(4,5)P2 as a critical input in vivo. Furthermore, production of acidic phospholipids and activation of Rac are often linked, as in the case of neutrophil chemotaxis. Intracellular signaling networks downstream of chemoattractant receptors engage heterotrimeric G-proteins and Ras, which are thought to activate PI3 kinases and PIP3 production at the leading edge (Charest and Firtel, 2007). This in turn activates Rac through guanine nucleotide exchange factors. Through still poorly defined positive feedback loops, Rac mediated actin assembly further promotes PI3 kinase activity (Charest and Firtel, 2006), producing an area on the membrane enriched in PIP3 and Rac.
An interesting finding is that acidic phospholipids other than PIP3 can support activation of the WAVE2 complex. The relative level of activation of the WAVE2 complex we observed with prenylated Rac-GTP and liposomes of varying phosphoinositide composition (Figure 2E) parallels the binding affinity of recombinant WAVE2 protein for a similar series of phosphoinositides, reported previously by Oikawa and colleagues (Oikawa et al., 2004). Our results suggest that in the absence of PIP3 the WAVE2 complex might still promote actin assembly as long as Rac activation is co-localized with other acidic phospholipids. This is consistent with studies where PIP3 production was abrogated and yet cells were still able to chemotax, although in some cases at slower rates than wild type cells (Ferguson et al., 2007; Hoeller and Kay, 2007).
Regulation of WAVE2 function by IRSp53 has also been suggested. IRSp53 has been shown to enhance the activity of isolated recombinant WAVE2 and of WAVE2 complexes (Miki et al., 2000; Suetsugu et al., 2006). However the preparations of complex used in those experiments had high basal activity, so the effect of IRSp53 on an inactive complex remained to be determined. In our preparations of immuno-purified WAVE2 complex we detected IRSp53 (Figures 2A, S2D and Table S1), and yet these complexes were inactive (Figures 2B and S2E). These results indicate that IRSp53 by itself did not activate at the concentrations present in our preparations, but they do not discount a role for IRSp53 in mediating activation by Rac and acidic phospholipids.
Regulation of WAVE2 complex activity by phosphorylation
Rac and acidic phospholipids are together incapable of activating a WAVE2 complex that is not properly phosphorylated (Figure 3E), rendering phosphorylation a prerequisite for activation. Although phosphorylation of WAVE proteins had been reported, a precise regulatory mechanism had not been defined. With the biochemical reconstitution of WAVE2 complex activation we could show that regulation by phosphorylation is direct and acts at the core of the activation mechanism. Phosphatase treatment also diminished the interaction between the WAVE2 complex and acidic phospholipids (Table 1). This reduction might directly impinge on the efficiency of activation or simply reflect conformational changes important for activation. It will be important in the future to study precise details of this regulation, but it is already clear that unlike the previously described phosphorylation of the VCA domain of WAVE2, which affects its catalytic activity on the Arp2/3 complex (Nakanishi et al., 2007; Pocha and Cory, 2009), the effects we observed are at the level of the activation process itself.
The mechanism of WAVE2 complex activation
Two mechanistic models have been suggested to explain the regulation of WAVE activity: recruitment of an already active complex to sites of actin assembly, and dissociation of the complex to release active WAVE protein (Eden et al., 2002; Innocenti et al., 2004). Our results show that neither of these explanations is correct. Since activation of the WAVE2 complex is not induced by the mere recruitment of the complex to liposomes containing either Rac-GTP or acidic phospholipids alone (Table 1), and it is not mediated by dissociation of the complex subunits (Figures 4B and 4C), activation must involve a conformational change that presumably exposes the VCA domain of WAVE2. With the complex remaining intact, the catalytic activity of WAVE2 would remain strictly localized to sites on the plasma membrane defined by the activators (Figure 4A and Movie S1), allowing precise spatial control of actin assembly and making it possible to attain high local density of activated WAVE2 complexes. This might be crucial for higher order interactions implied by the cooperative nature of the activation process (Figure 2H).
We envision two scenarios that could take place independently or together to produce cooperative activation. Dimerization of WASP and WAVE proteins dramatically increases their affinity for the Arp2/3 complex, substantially enhancing actin assembly (Padrick et al., 2008). Activation of the WAVE2 complex is constrained to the surface of PIP3 liposomes containing Rac, and at sufficiently high WAVE2 complex concentration the proximity of activated WAVE2 molecules could mimic the effect of dimerization, stimulating actin polymerization non-linearly. Alternatively, high concentration of WAVE2 complexes on the liposome surface could induce or stabilize conformations that enhance activity. Regardless of the mechanisms giving rise to it, the observed highly cooperative activation would effectively set a threshold for actin polymerization. Below a certain concentration of complex on the plasma membrane, actin polymerization would be negligible, especially in the presence of abundant actin depolymerizing activities, and this may serve to reduce noise in the system. Once the proper combination of activators leads to the recruitment of sufficient WAVE2 complex to surpass that threshold concentration, higher order interactions might come into play and promote the high levels of actin polymerization necessary for membrane protrusion.
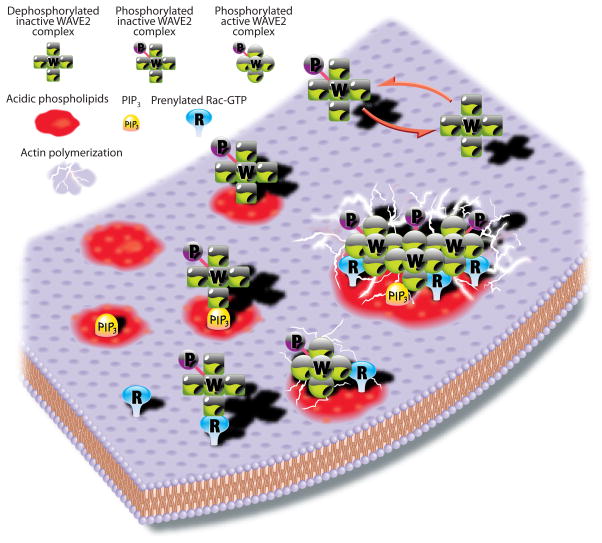
A model for activation of the WAVE complex
In Figure 5 we summarize the main features of WAVE2 complex regulation. We envision several states of the WAVE2 complex (counterclockwise from top right): in the cytoplasm either dephosphorylated or primed for activation by phosphorylation; on the plasma membrane independently bound to acidic phospholipids, including but not limited to PIP3, or bound to prenylated Rac-GTP. None of these conditions is sufficient for activation. Simultaneous binding of the properly phosphorylated complex to both Rac-GTP and acidic phospholipids results in activation, most likely through conformational changes in the complex (depicted by a switch from square to round subunits) leading to Arp2/3 dependent actin polymerization. A high local concentration of activators would lead to the recruitment and activation of multiple WAVE2 complexes in close proximity, resulting in synergistic actin polymerization through cooperative mechanisms.
Figure 5. Model for WAVE2 function.
Counterclockwise from top right, the WAVE2 complex is intrinsically inactive, and must be phosphorylated before it can be activated. However, phosphorylation by itself does not activate the complex. Phosphorylated WAVE2 complex can bind acidic phospholipids, including PIP3, or prenylated Rac-GTP, but binding to either is not sufficient for activation. Binding to both acidic phospholipids and prenylated Rac-GTP activates the WAVE2 complex, most likely though allosteric changes, leading to Arp2/3 dependent actin polymerization. Although PIP3 is most potent, other acidic phospholipids can also activate the complex together with prenylated Rac-GTP. Cooperative association of multiple complexes on the membrane results in greatly enhanced actin nucleation. Key is at the top left corner of the diagram.
This mode of regulation endows the system with special features appropriate for its cellular functions. Unlike other systems where individual signals make incremental contributions to activity, the multiple signals regulating WAVE2 function need to act simultaneously to result in any activation, making this a highly specific coincidence detector. This is important because the signals that control WAVE function, including acidic phospholipids and Rac-GTP, are present throughout the cell and also function in other pathways. However, these signals should only activate the WAVE2 complex where they coincide, generating a very local area of actin polymerization even if their other cellular functions were not so strictly localized. Domains of activity could be sharpened by the highly cooperative nature of activation, which might set a threshold below which activation would be negligible and above which activation would be explosive, especially in the presence of positive feedback loops enhancing the local production of activators. Conversely, the coordinated and cooperative nature of WAVE2 complex regulation would allow for very rapid inactivation by simply lowering the amount of any one of the activators on the membrane, generating true switch-like behavior. Kinases and phosphatases would further tune activity if acting locally, or shut it down entirely throughout the cell if acting globally. This system, with its capacity for explosive activation and rapid inactivation as well as exquisite spatial resolution, is ideally suited for regulating actin dynamics during cell motility and especially chemotaxis, where constant changes in the direction of movement require quick responses, tight localization of protrusions, and the need to effectively discern signal from noise.
In retrospect, the great effort required to reconstitute actin polymerization mediated by the WAVE2 complex was due to the lability of the complex and most especially to the combinatorial nature of inputs that operate in a narrow range of effective concentrations. Yet, achieving full reconstitution was crucial, since it resolved at once the regulatory question of whether a constitutively active WAVE complex is recruited to sites of actin assembly or whether several inputs are necessary to allosterically activate an inactive complex. The quantitative characterization of this system yielded unexpected insights, such as the cooperative nature of activation, the requirement for EGF-independent phosphorylation and the lipid specificity. These results already explain many in vivo findings and should help us construct more realistic and detailed models for the signaling processes that coordinate actin assembly in cell motility and chemotaxis.
Experimental Procedures
Actin polymerization assays
Actin polymerization assays were as described previously (Ho et al., 2006). All reagents were kept on ice except for ddH2O, which was kept at room temperature. 60 μl reactions were set up at room temperature adding reagents in the following order: ddH2O, 10× assay buffer (200 mM Hepes [pH 7.7 at 24°C], 1 M KCl, 10 mM MgCl2, 1 mM EDTA, 10 mM DTT; 10× buffer was diluted to 1× final concentration, subtracting the volume of other reagents already in buffers of comparable composition), ATP (0.2 mM final concentration), Arp2/3 complex (30 nM unless indicated otherwise), WAVE complex or recombinant WAVE protein, liposomes and GTPases. Actin/pyrene-actin (1 μM premixed in an 80% to 20% molar ratio unless indicated otherwise) was added last to start the assay, the reactions were immediately transferred to quartz cuvettes, and pyrene fluorescence was measured at 407 nm with excitation at 365 nm at 24°C in a fluorescence spectrophotometer (Varian Cary Eclipse). Between every assay cuvettes were cleaned with a solution of 2 parts concentrated hydrochloric acid, 1 part concentrated nitric acid, rinsed thoroughly with water followed by ethanol and dried with an air stream. Rinsing the cuvettes with water and ethanol alone built a residue, perhaps of denatured WAVE, that could stimulate the Arp2/3. We also found it necessary to keep the total protein concentration in the reaction above 0.25 mg/ml for optimal activation of WAVE2 complex; this sometimes required adding WP1×3 peptide to 0.25 mg/ml.
Supplemental Experimental Procedures
Antibodies, conventional purification of native WAVE2 complex from pig leukocytes and native WAVE1 complex from bovine brain, immuno-purification of native WAVE2 complex from A-431 cells, phosphatase treatment of WAVE2 complex, non-physiological activation of native WAVE complexes, preparation of recombinant proteins, PAK pull-down assays, preparation of liposomes, preparation of proteins used in actin polymerization assays, data analysis of actin polymerization assays, liposome cosedimentation assays and microscopy and mass spectrometry methods are described in Supplemental Data.
Supplementary Material
Acknowledgments
We thank Henry Ho and Rajat Rohatgi for providing actin, Arp2/3 complex and recombinant N-WASP. We thank Mike Gage for extensive help harvesting cells, Alexandra Lebensohn for help with tissue culture, Jennifer Gallop and Wendy Salmon for help with microscopy, Ran Kafri for help with data analysis, and Ross Tomaino for help with mass spectrometry. We would also like to thank Ian Moores for preparing the illustration in Figure 5. We are very grateful to Henry Ho, Orion Weiner, Timothy Mitchison, Paul Jorgensen, Jennifer Gallop and Alexandra Lebensohn for helpful discussions and for comments on the manuscript, and to Tom Rapoport, Jeffrey Settleman, William Brieher, Richard Deibler, Lea Goentoro, Michael Rape and members of the Kirschner lab for many useful suggestions. This work was supported by a grant from the National Institutes of Health to M.W.K. (GM026875-32).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ardern H, Sandilands E, Machesky LM, Timpson P, Frame MC, Brunton VG. Src-dependent phosphorylation of Scar1 promotes its association with the Arp2/3 complex. Cell Motil Cytoskeleton. 2006;63:6–13. doi: 10.1002/cm.20101. [DOI] [PubMed] [Google Scholar]
- Charest PG, Firtel RA. Feedback signaling controls leading-edge formation during chemotaxis. Curr Opin Genet Dev. 2006;16:339–347. doi: 10.1016/j.gde.2006.06.016. [DOI] [PubMed] [Google Scholar]
- Charest PG, Firtel RA. Big roles for small GTPases in the control of directed cell movement. Biochem J. 2007;401:377–390. doi: 10.1042/BJ20061432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danson CM, Pocha SM, Bloomberg GB, Cory GO. Phosphorylation of WAVE2 by MAP kinases regulates persistent cell migration and polarity. J Cell Sci. 2007;120:4144–4154. doi: 10.1242/jcs.013714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derivery E, Lombard B, Loew D, Gautreau A. The Wave complex is intrinsically inactive. Cell Motil Cytoskeleton. 2009 doi: 10.1002/cm.20342. [DOI] [PubMed] [Google Scholar]
- Eden S, Rohatgi R, Podtelejnikov AV, Mann M, Kirschner MW. Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature. 2002;418:790–793. doi: 10.1038/nature00859. [DOI] [PubMed] [Google Scholar]
- Ferguson GJ, Milne L, Kulkarni S, Sasaki T, Walker S, Andrews S, Crabbe T, Finan P, Jones G, Jackson S, et al. PI(3)Kgamma has an important context-dependent role in neutrophil chemokinesis. Nat Cell Biol. 2007;9:86–91. doi: 10.1038/ncb1517. [DOI] [PubMed] [Google Scholar]
- Gautreau A, Ho HY, Li J, Steen H, Gygi SP, Kirschner MW. Purification and architecture of the ubiquitous Wave complex. Proc Natl Acad Sci U S A. 2004;101:4379–4383. doi: 10.1073/pnas.0400628101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goley ED, Welch MD. The ARP2/3 complex: an actin nucleator comes of age. Nat Rev Mol Cell Biol. 2006;7:713. doi: 10.1038/nrm2026. [DOI] [PubMed] [Google Scholar]
- Heasman SJ, Ridley AJ. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat Rev Mol Cell Biol. 2008;9:690. doi: 10.1038/nrm2476. [DOI] [PubMed] [Google Scholar]
- Ho HY, Rohatgi R, Lebensohn AM, Kirschner MW. In vitro reconstitution of cdc42-mediated actin assembly using purified components. Methods Enzymol. 2006;406:174–190. doi: 10.1016/S0076-6879(06)06014-9. [DOI] [PubMed] [Google Scholar]
- Hoeller O, Kay RR. Chemotaxis in the absence of PIP3 gradients. Curr Biol. 2007;17:813–817. doi: 10.1016/j.cub.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Innocenti M, Zucconi A, Disanza A, Frittoli E, Areces LB, Steffen A, Stradal TE, Di Fiore PP, Carlier MF, Scita G. Abi1 is essential for the formation and activation of a WAVE2 signalling complex. Nat Cell Biol. 2004;6:319–327. doi: 10.1038/ncb1105. [DOI] [PubMed] [Google Scholar]
- Ismail AM, Padrick SB, Chen B, Umetani J, Rosen MK. The WAVE regulatory complex is inhibited. Nat Struct Mol Biol. 2009 doi: 10.1038/nsmb.1587. advanced online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Sung JY, Ceglia I, Lee KW, Ahn JH, Halford JM, Kim AM, Kwak SP, Park JB, Ho Ryu S, et al. Phosphorylation of WAVE1 regulates actin polymerization and dendritic spine morphology. Nature. 2006;442:814–817. doi: 10.1038/nature04976. [DOI] [PubMed] [Google Scholar]
- Kitamura T, Kitamura Y, Yonezawa K, Totty NF, Gout I, Hara K, Waterfield MD, Sakaue M, Ogawa W, Kasuga M. Molecular cloning of p125Nap1, a protein that associates with an SH3 domain of Nck. Biochem Biophys Res Commun. 1996;219:509–514. doi: 10.1006/bbrc.1996.0264. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Kuroda S, Fukata M, Nakamura T, Nagase T, Nomura N, Matsuura Y, Yoshida-Kubomura N, Iwamatsu A, Kaibuchi K. p140Sra-1 (specifically Rac1-associated protein) is a novel specific target for Rac1 small GTPase. J Biol Chem. 1998;273:291–295. doi: 10.1074/jbc.273.1.291. [DOI] [PubMed] [Google Scholar]
- Kolsch V, Charest PG, Firtel RA. The regulation of cell motility and chemotaxis by phospholipid signaling. J Cell Sci. 2008;121:551–559. doi: 10.1242/jcs.023333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraynov VS, Chamberlain C, Bokoch GM, Schwartz MA, Slabaugh S, Hahn KM. Localized Rac Activation Dynamics Visualized in Living Cells. Science. 2000;290:333–337. doi: 10.1126/science.290.5490.333. [DOI] [PubMed] [Google Scholar]
- Lebensohn AM, Ma L, Ho HY, Kirschner MW. Cdc42 and PI(4,5)P2-induced actin assembly in Xenopus egg extracts. Methods Enzymol. 2006;406:156–173. doi: 10.1016/S0076-6879(06)06013-7. [DOI] [PubMed] [Google Scholar]
- Leng Y, Zhang J, Badour K, Arpaia E, Freeman S, Cheung P, Siu M, Siminovitch K. Abelson-interactor-1 promotes WAVE2 membrane translocation and Abelson-mediated tyrosine phosphorylation required for WAVE2 activation. PNAS. 2005;102:1098–1103. doi: 10.1073/pnas.0409120102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky LM, Mullins RD, Higgs HN, Kaiser DA, Blanchoin L, May RC, Hall ME, Pollard TD. Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc Natl Acad Sci U S A. 1999;96:3739–3744. doi: 10.1073/pnas.96.7.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki H, Suetsugu S, Takenawa T. WAVE, a novel WASP-family protein involved in actin reorganization induced by Rac. EMBO J. 1998;17:6932–6941. doi: 10.1093/emboj/17.23.6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki H, Yamaguchi H, Suetsugu S, Takenawa T. IRSp53 is an essential intermediate between Rac and WAVE in the regulation of membrane ruffling. Nature. 2000;408:732–735. doi: 10.1038/35047107. [DOI] [PubMed] [Google Scholar]
- Nakanishi O, Suetsugu S, Yamazaki D, Takenawa T. Effect of WAVE2 Phosphorylation on Activation of the Arp2/3 Complex. J Biochem (Tokyo) 2007;141:319–325. doi: 10.1093/jb/mvm034. [DOI] [PubMed] [Google Scholar]
- Oikawa T, Yamaguchi H, Itoh T, Kato M, Ijuin T, Yamazaki D, Suetsugu S, Takenawa T. PtdIns(3,4,5)P3 binding is necessary for WAVE2-induced formation of lamellipodia. Nat Cell Biol. 2004;6:420–426. doi: 10.1038/ncb1125. [DOI] [PubMed] [Google Scholar]
- Padrick SB, Cheng HC, Ismail AM, Panchal SC, Doolittle LK, Kim S, Skehan BM, Umetani J, Brautigam CA, Leong JM, et al. Hierarchical Regulation of WASP/WAVE Proteins. Mol Cell. 2008;32:426. doi: 10.1016/j.molcel.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pocha SM, Cory GO. WAVE2 is regulated by multiple phosphorylation events within its VCA domain. Cell Motil Cytoskeleton. 2009;66:36–47. doi: 10.1002/cm.20323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD. Regulation of Actin Filament Assembly by Arp2/3 Complex and Formins. Annu Rev Biophys Biomol Struct. 2007;36:451–477. doi: 10.1146/annurev.biophys.35.040405.101936. [DOI] [PubMed] [Google Scholar]
- Scott CC, Dobson W, Botelho RJ, Coady-Osberg N, Chavrier P, Knecht DA, Heath C, Stahl P, Grinstein S. Phosphatidylinositol-4,5-bisphosphate hydrolysis directs actin remodeling during phagocytosis. J Cell Biol. 2005;169:139–149. doi: 10.1083/jcb.200412162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sossey-Alaoui K, Li X, Cowell JK. c-Abl-mediated Phosphorylation of WAVE3 Is Required for Lamellipodia Formation and Cell Migration. J Biol Chem. 2007;282:26257–26265. doi: 10.1074/jbc.M701484200. [DOI] [PubMed] [Google Scholar]
- Sossey-Alaoui K, Li X, Ranalli TA, Cowell JK. WAVE3-mediated Cell Migration and Lamellipodia Formation Are Regulated Downstream of Phosphatidylinositol 3-Kinase. J Biol Chem. 2005;280:21748–21755. doi: 10.1074/jbc.M500503200. [DOI] [PubMed] [Google Scholar]
- Steffen A, Rottner K, Ehinger J, Innocenti M, Scita G, Wehland J, Stradal TEB. Sra-1 and Nap1 link Rac to actin assembly driving lamellipodia formation. EMBO J. 2004;23:749–759. doi: 10.1038/sj.emboj.7600084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart JR, Gonzalez FH, Kawai H, Yuan ZM. c-Abl Interacts with the WAVE2 Signaling Complex to Induce Membrane Ruffling and Cell Spreading. J Biol Chem. 2006;281:31290–31297. doi: 10.1074/jbc.M602389200. [DOI] [PubMed] [Google Scholar]
- Suetsugu S, Kurisu S, Oikawa T, Yamazaki D, Oda A, Takenawa T. Optimization of WAVE2 complex-induced actin polymerization by membrane-bound IRSp53, PIP(3), and Rac. J Cell Biol. 2006;173:571–585. doi: 10.1083/jcb.200509067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu S, Miki H, Takenawa T. Identification of two human WAVE/SCAR homologues as general actin regulatory molecules which associate with the Arp2/3 complex. Biochem Biophys Res Commun. 1999;260:296–302. doi: 10.1006/bbrc.1999.0894. [DOI] [PubMed] [Google Scholar]
- Suetsugu S, Yamazaki D, Kurisu S, Takenawa T. Differential roles of WAVE1 and WAVE2 in dorsal and peripheral ruffle formation for fibroblast cell migration. Dev Cell. 2003;5:595–609. doi: 10.1016/s1534-5807(03)00297-1. [DOI] [PubMed] [Google Scholar]
- Takenawa T, Suetsugu S. The WASP-WAVE protein network: connecting the membrane to the cytoskeleton. Nat Rev Mol Cell Biol. 2007;8:37–48. doi: 10.1038/nrm2069. [DOI] [PubMed] [Google Scholar]
- Weiner OD, Marganski WA, Wu LF, Altschuler SJ, Kirschner MW. An actin-based wave generator organizes cell motility. PLoS Biol. 2007;5:e221. doi: 10.1371/journal.pbio.0050221. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.