Translocation is one of the most frequently occurring human chromosomal aberrations. Balanced carriers usually manifest no phenotype but experience problems with reproduction. These include infertility, recurrent abortion, and offspring with chromosomal imbalance. The constitutional t(11;22)(q23;q11) is a balanced translocation between chromosomes 11 and 22, with breakpoints at bands 11q23 and 22q11. It is the only known recurrent non-Robertsonian translocation and represents a good model for studying translocations in humans (1). The recurrent nature of this translocation prompted us to examine t(11;22) breakpoints for a specific genomic structure. The analysis of many unrelated t(11;22) cases revealed that the breakpoints occur within palindromic AT-rich repeats (PATRRs) on 11q23 and 22q11 (PATRR11 and PATRR22) (2–4). The majority of the breakpoints are localized at the center of the PATRRs, suggesting that the center of the palindrome is susceptible to double-strand breaks (DSBs), thereby inducing illegitimate chromosomal rearrangement (3, 5). Recent findings of PATRR-like sequences at the breakpoints of other translocations support the possibility that palindrome-mediated chromosomal translocation is a general pathway for human genomic rearrangements (6–8).
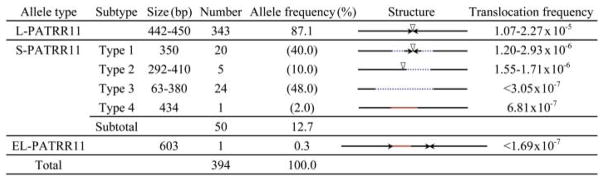
The PATRR11 is variable in size in normal healthy individuals (Fig. 1 and table S1). The most common allele is a ~450–base pair (bp) PATRR11 (L-PATRR11) that forms a nearly perfect palindrome (5). Several types of short variants were identified (S-PATRR11) that appear to be derived from the longer version primarily by deletion near the symmetric center of the palindromic structure. We can classify the S-PATRR11s into four groups. The most frequent 350-bp variant, S1-PATRR11, has a 50-bp deletion at both of the palindromic arms but still remains completely symmetrical. S2-PATRR11 has an asymmetric deletion at its center, but the new center manifests a symmetric palindrome. S3-PATRR11 does not possess palindromic features by virtue of a deletion at the center of the palindrome. We identified a rare 434-bp S-PATRR11, which sustained an asymmetric central deletion followed by the insertion of an AT-rich sequence of unknown origin (S4-PATRR11). We also identified another rare allele with a duplication of the proximal arm, which constitutes a 603-bp asymmetric palindrome (EL-PATRR11). On the basis of the palindrome-mediated mechanism of the translocation, it is reasonable to hypothesize that the polymorphism of the PATRR11 could affect the frequency of de novo t(11;22) translocations.
Fig. 1.
Polymorphic PATRR11 alleles and their translocation frequencies. Arrows indicate each unit of inverted repeats. Vertical arrowheads indicate the center of the palindrome. Dotted blue lines show deleted regions, whereas red lines indicate insertions. Calculated frequencies of de novo translocation produced from each allele are shown at the right.
Translocation-specific polymerase chain reaction (PCR) detects de novo t(11;22) translocations in normal sperm samples from healthy individuals with a normal karyotype (9). We analyzed sperm DNA from individuals with various genotypes for the PATRR11. Five men homozygous for the L-PATRR11 genotype produced de novo translocations at a frequency ranging between 1.52 × 10−5 and 1.57 × 10−4 (table S2). A heterozygote for the L-PATRR11 and S1- or S2-PATRR11 alleles produced de novo translocations at a similar overall frequency, as did homozygotes for the L-PATRR11. Sequence analysis of the PCR products revealed that products derived from the symmetrical S1- or S2-PATRR were less frequently observed than products originating from the L-PATRR11, with a low estimated frequency of de novo translocations (~10−6), suggesting that the frequency of de novo translocation depends on the size of the PATRR. On the other hand, although two individuals heterozygous for L-PATRR11 and S3-PATRR11 produced de novo translocations at a similar overall frequency, sequence analysis revealed that virtually all of the de novo translocations appeared to originate from the L-PATRR11. This finding suggests that the asymmetric S-PATRR does not produce de novo translocations (table S2).
Sperm from one individual, who was a rare compound heterozygote for the S1- and S3-PATRR11, contained de novo translocations at a very low frequency. Sequence analysis of the junction fragments showed that the translocations were exclusively derived from the S1-PATRR11. Because both the S2- and S4-PATRR11s alleles produced translocations at a low frequency (~10−6 and ~10−7, respectively), a compound heterozygote for this genotype had a very low translocation frequency overall. We identified another individual heterozygous for the S3-PATRR and EL-PATRR. This individual is the only person who carried asymmetric PATRR11s on both alleles. He did not produce any de novo translocations (table S2). The frequencies for each allele are summarized in Fig. 1.
This work demonstrates genetic variation over more than three orders of magnitude in the susceptibility for generating the recurrent translocation in humans. Our results point to the importance of genomic sequence variation on the frequency of chromosomal rearrangements, a class of human mutation that is generally thought to be random. Our data imply that genetic variation plays an important role in this process.
Supplementary Material
Footnotes
Supporting Online Material
www.sciencemag.org/cgi/content/full/311/5763/971/DC1
Materials and Methods
References and Notes
- 1.Zackai EH, Emanuel BS. Am J Med Genet. 1980;7:507. doi: 10.1002/ajmg.1320070412. [DOI] [PubMed] [Google Scholar]
- 2.Kurahashi H, et al. Hum Mol Genet. 2000;9:1665. doi: 10.1093/hmg/9.11.1665. [DOI] [PubMed] [Google Scholar]
- 3.Edelmann L, et al. Am J Hum Genet. 2001;68:1. doi: 10.1086/316952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tapia-Paez I, et al. Hum Genet. 2001;109:167. [Google Scholar]
- 5.Kurahashi H, Emanuel BS. Hum Mol Genet. 2001;10:2605. doi: 10.1093/hmg/10.23.2605. [DOI] [PubMed] [Google Scholar]
- 6.Kurahashi H, et al. Am J Hum Genet. 2003;72:733. doi: 10.1086/368062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nimmakayalu MA, Gotter AL, Shaikh TH, Emanuel BS. Hum Mol Genet. 2003;12:2817. doi: 10.1093/hmg/ddg301. [DOI] [PubMed] [Google Scholar]
- 8.Gotter AL, et al. Hum Mol Genet. 2004;13:103. doi: 10.1093/hmg/ddh004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kurahashi H, Emanuel BS. Nat Genet. 2001;29:139. doi: 10.1038/ng1001-139. [DOI] [PubMed] [Google Scholar]
- 10.The authors wish to thank T. Mori and E. Hosoba for technical assistance. These studies were supported by a grant-in-aid for Scientific Research, Genome, and 21st Century Center of Excellence program from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (H.K.) and by a grant (CA39926) from the NIH (B.S.E.).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.